Abstract
It has been reported that miR-124a is abundant in the central nervous system including the eye, and is related to neurogenesis in several species. However, the role of miR-124a in the eye remains unclear. In this study, we show that the expression of miR-124a in Xenopus laevis begins along the neural fold, including the protruding eye anlagen, at a low level at around stage 18; its expression level gradually increases in the neural tube and the eye as embryos develop into later stages and then maintains at a high level in eye to adult stages. Microinjection of a miR-124a precursor at the 8-cell stage leads to malformation of the optic nerve and optic cup, indicating the importance of maintaining low levels of miR-124a during early embryonic development. In addition, miR-124a overexpression markedly down regulates the expression of its predicted targets Lhx2, Hairy2, Gli3, NeuroD1 and Otx2 in/around the eye anlagen, and the interaction of miR-124a with the 3′ UTR of Lhx2 represses gene expression as shown by luciferase assays. Moreover, excess miR-124a inhibits cell proliferation in the eye of Xenopus embryos during retinogenesis. These results indicate that miR-124a acts as a post-transcriptional regulator in the genetic network controlling eye morphogenesis and neurogenesis. The mechanism of miR-124a’s early interaction with the genetic network may also persist in its later role in the maturing and adult eye and brain.
Keywords: miR-124a, Eye, Lhx2, Morphogenesis, Cell proliferation, Xenopus laevis
1. Introduction
MicroRNAs (miRNAs) are recently identified small non-coding RNA molecules which regulate gene expression at the post-transcriptional level by either repressing translation or promoting mRNA degradation (Lewis et al., 2003; Eulalio et al., 2007; Hofacker, 2007). Thousands of miRNAs have been identified in vertebrate genomes and they have been estimated to regulate up to 30% of the genes in the genome (Wienholds and Plasterk, 2005; Lewis et al., 2005). Many mammalian miRNAs are highly or specifically expressed in neural tissues and approximately 70% of experimentally detectable miRNAs are expressed in the brain (Cao et al., 2006), suggesting that miRNAs play important roles in neural development and regulation of the adult nervous system. However, the function of most miRNAs remains unclear, and an extensive analysis is necessary to reveal their precise roles in vivo.
To reveal the role of miRNAs in the central nervous system (CNS), especially in the eye, we carried out a microarray screening for miRNAs expressed in the eye using retinal small RNAs isolated from adult mice, rats and zebrafish. miR-124a is one of the miRNAs identified in retinas from all three species (unpublished data). In a further screening using a functional assay in Xenopus laevis, we found that microinjection of miR-124a precursors in the anterior part of the Xenopus embryo led to eye anomalies (unpublished data), suggesting that miR-124a plays an important role in eye development.
miR-124a is a group of highly conserved microRNAs abundant in the CNS including the eye (Deo et al., 2006; Darnell et al., 2006; Wienholds et al., 2005; Kloosterman et al., 2006; Sweetman et al., 2006). In situ hybridization with a miR-124a probe on coronal sections of the adult mouse brain shows that miR-124a is expressed throughout most parts of the brain, including the cerebral cortex and hippocampus. However, its signal is absent from the white matter and appears to localize primarily to the cytoplasm (Deo et al., 2006). In the chick, miR-124a is also expressed strongly in the brain, especially in the hindbrain, midbrain, lateral regions of the spinal cord and the pituitary rudiment (Darnell et al., 2006; Sweetman et al., 2006). miR-124a has also been shown to be expressed in the eye. It is detected strongly in most cells in the neural retina but not in the pigmented epithelium (RPE) (Deo et al., 2006). Northern blotting has demonstrated that miR-124a is also expressed in the mouse lens (Frederikse et al., 2006). Another study of embryonic development using Northern blotting showed that miR-124a expression emerges at the end of the neurula (after stage 18) and remains continuously detectable till the tadpole stage (stage 42) in X. laevis (Watanabe et al., 2005). Furthermore, its expression level increases as embryos develop into later stages (Krichevsky et al., 2003; Miska et al., 2004). However, a detailed report of the location of miR-124a in the CNS, especially in the eye, during development is still unavailable.
The prevalence of miR-124a expression in the developing and adult CNS suggests that miR-124a plays a pivotal role in the CNS and neurogenesis. Introducing miR-124 into a human cell line causes the expression profile to shift towards that of the brain (Lim et al., 2005), and its overexpression together with that of miR-9 in neural progenitors prevents gliogenesis (Krichevsky et al., 2006). RE1 silencing transcription factor (REST), a transcriptional repressor, inhibits the expression of neuronal genes and miR-124a in non-neuronal cells, allowing the persistence of non-neuronal transcripts (Conaco et al., 2006). On the other hand, REST and its cofactor complex are also targets of miR-124a, suggesting a double-negative feedback loop between REST and miR-124a in stabilizing and maintaining neuronal gene expression (Wu and Xie, 2006). In addition to REST, small C-terminal domain phosphatase 1 (SCP1) is another anti-neural factor expressed in non-neuronal tissues, which is reported to be an miR-124 target during neurogenesis of the developing chick neural tube (Visvanathan et al., 2007). Another recent finding also reports that miR-124 promotes the differentiation of progenitor cells to mature neurons by directly targeting PTBP1 (PTB/hnRNP I) mRNA, which encodes a global repressor of alternative premRNA splicing in non-neuronal cells (Makeyev et al., 2007). However, it has also been reported that neither inhibition nor overexpression of miR-124 alone significantly altered neuronal fate (Cao et al., 2007; Conaco et al., 2006). Therefore, the exact role of miR-124a in neurogenesis remains to be elucidated. Moreover, although a high level of miR-124a expression has been detected in the retina in several species, the role of miR-124a in retina has not yet been revealed.
In this paper, using X. laevis as an animal model, we have studied the role of miR-124a in the developing eye and brain. Our results indicate that miR-124a is able to act on the genetic network involved in the early morphogenesis of the eye, and that maintaining a low level of miR-124a at early stages is necessary for proper cell proliferation and eye morphogenesis. We also show that Lhx2, a gene reported to be involved in eye development (Porter et al., 1997; Zuber et al., 2003; Ando et al., 2005; Seth et al., 2006), is a potential endogenous target for miR-124a.
2. Results
2.1. miR-124a shows a dynamic expression pattern in the developing and adult eye of X. laevis
Sequence analysis of validated and predicted miR-124a precursor and mature molecules of human, mouse, zebrafish, Xenopus tropicalis and X. laevis showed that the mature sequence of miR-124 is highly conserved between species. Except for miR-124 from X. laevis, which has a single nucleotide difference, all the other species share identical mature miR-124 sequences (Fig. S1). To check if its expression is different from that reported for other species (Deo et al., 2006; Frederikse et al., 2006) and to reveal its potential function in the Xenopus retina, we examined miR-124a expression in X. laevis. Whole mount in situ hybridization with a LNA-probe showed that the earliest miR-124a expression was detectable along the entire neural fold including the anterior eye anlagen at around stage 18 (Fig. 1A). During the optic vesicle stage (stage 23) (Fig. 1B), the expression of miR-124a was stronger in the brain than in the optic vesicle and posterior neural tube. Starting from stage 33 (optic cup stage) (Fig. 1C), a high level of miR-124a expression was observed in the anterior CNS which includes the brain, eye and anterior spinal cord, and miR-124a remained highly expressed in the entire CNS from stage 37 (Fig. 1D) to late tadpole stages (Fig. 1E and data not shown). This expression pattern suggests that miR-124a plays a pivotal role in the development of the central nervous system.
Fig. 1.
Expression pattern of miR-124a during the development of Xenopus laevis. (A–G) Expression of miR-124a (dark blue) as detected by whole mount in situ hybridization in Xenopus embryos at stage 18 (A), stage 23 (B), stage 33 (C), stage 37 (D), stage 46 (E), and dissected adult retinas (F) and brain (G), shown from the anterior (A), lateral (B–D), or dorsal views (E and G). (H–L) Restricted location of miR-124a in the eye (H–K) and brain (L) is shown on transversal (H–K) or coronal sections (L) at stage 33 (H), stage 37 (I), stage 40 (J), and stage 46 (K and L). The pink arrow indicates the most peripheral part of the dorsal ciliary marginal zone. Scale bars: 300 μm (A–E); 1mm (F and G); 100 μm (H–L). Ce, cerebellum; Die, diencephalon; Mes, mesencephalon; OB, olfactory bulb; Rho, rhobemcephalon; Tel, telencephalon.
Sections of the embryos at eye-level showed that miR-124a was exclusively expressed in the neural retina with almost no signal detected in the most peripheral ciliary marginal zone (CMZ) at all the optic cup stages checked from stages 33 to 46 (Fig. 1H–K, data not shown). From stage 40 (Fig. 1J and K), the region where miR-124a was highly expressed was restricted further to the peripheral retina around the CMZ. In the brain, miR-124a was mainly expressed at the periphery of the olfactory bulb, telencephalon, mesencephalon and rhombencephalon (Fig. 1G and L).
In the adult, miR-124a expression was detectable in the brain and the neural retina. As shown in Fig. 1, miR-124a was highly expressed in the dissected neural retina (Fig. 1F) and in brain regions including the olfactory bulb, dorsal and rostroventral telencephalon, thalamus, hypothalamus and cerebellum (Fig. 1G and data not shown). No signal was detected in the adult retina hybridized with the probe for miR-133a, a negative control which is not expressed in retina (data not shown).
2.2. Down-regulation of miR-124a in the Xenopus eye
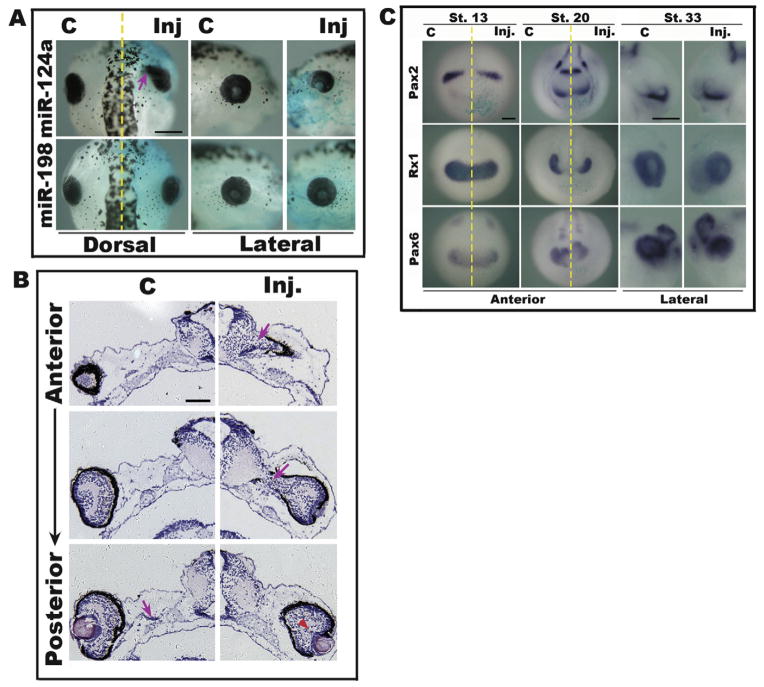
To investigate the function of miR-124a in the eye, we first used 2′-O-methyl antisense oligonucleotides (anti-miR-124a) to block miRNA function. Different doses (0.1–0.5 pmol) of 2′-O-methyl anti-miR-124a were injected into one dorsal-animal blastomere at the 8-cell stage to restrict the change in miR-124a level to one side of the dorsal head that included the eye region. Beta-galactosidase (beta-gal) was co-injected as a tracer. An inhibitor of miR-198, which is not expressed in the eye, or a control molecule (Ambion), was injected into the embryo at the same doses as negative controls. The injected embryos were grown to stages 33 or 46 to check the efficiency of inhibition and the phenotype (Fig. 2). We used in situ hybridization to assess changes in miR-124a levels in the eye. Results showed that 0.2 pmol 2′-O-methyl anti-miR-124a markedly reduced the miR-124a signal in the eye and brain at stage 33 (Fig. 2E), while at stage 46 only a very few embryos showed slight decreases in miR-124a expression in the brain (Fig. 2D). The same dose of miR-198 or a control inhibitor did not affect the expression of miR-124a (Fig. 2I and data not shown), indicating that 2′-O-methyl anti-miR-124a could specifically and efficiently reduce or mask endogenous miR-124a during early developmental stages. However, this inhibition of miR-124a did not lead to any obvious morphological changes in the development of Xenopus embryos, including the eye (Fig. 2A–C). Further histological analysis using transversal sections of the injected embryos showed no observable differences between the eye structures on the injected and control sides (Fig. 2F–H), as was the case for embryos injected with miR-198 or control inhibitor (data not shown). Embryos injected with either the miR-124a inhibitor or the two controls at a higher dosage showed similar developmental defects, suggesting non-specific phenotypes induced by overdose toxicity.
Fig. 2.
Down-regulation of miR-124a shows no obvious effect on the morphogenesis of the Xenopus eye. Embryos were unilaterally injected with 0.2 pmol 2′-O-methyl antisense oligonucleotides against miR-124a (A–H) or a control miRNA, miR-198 (I). (A–C) The eyes/head of an injected Xenopus embryo at stage 46 shown fromthe dorsal (A) or lateral (B and C) view. (D and E) The expression of miR-124a (dark blue) detected by whole mount in situ hybridization at stage 46 (D) and stage 33 (E). The eyes/heads of embryos are shownfrom the dorsal view. (F–H) The structure of the eyes of an injected embryo at stage 46 shown in a transversal section (5 μm) stained with hematoxylin (blue)–eosin solution (pink). (G) and (H) show the eyes in (F) under a higher magnification. (I) Expression of miR-124a (dark blue) was not affected by the microinjection of miR-198 inhibitor at stage 33. Scale bars: 300 μm (A–E, I); 100 μm (F); 50 μm (G and H). The yellow dashed line indicates the midline of the embryos separating the uninjected control side (C) and the injected side (Inj). The injected side is traced by the light blue staining of beta-galactosidase co-injected with miR-124a or miR-198. L, lens; NR, neural retina; RPE, retina pigmented epithelium.
We then attempted to inhibit miR-124a using LNA-modified antisense oligonucleotides (Cao et al., 2007). Injection of 0.0675 pmol LNA-anti-miR-124a was sufficient to reduce the endogenous miR-124a signal and induce eye defects (size reduction) (Fig. S2). However, these effects were also observed in embryos injected with the control LNA inhibitor of miR-198. Lower dosage of either of the above LNA molecules gave no morphological effects on injected embryos (Fig. S2, and data not shown). These results indicate that the LNA-anti-miR-124a was not a specific inhibitor for miR-124a under the experimental conditions used here. Further experiments are necessary to develop a more efficient strategy for knocking down miR-124a expression without non-specific toxicity.
2.3. Overexpression of miR-124a leads to dose-dependent eye anomalies
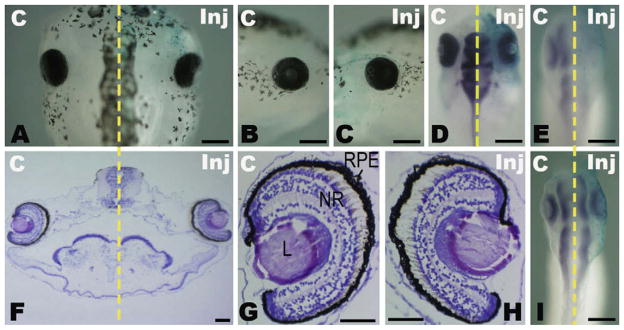
We examined the effects of miR-124a overexpression in Xenopus embryos by injecting at the early embryonic stage. As miR-124a expression is low at early stages and becomes high later, miR-124a microinjection would lead to a relative stronger change of miR-124a level at early stages than at later stages therefore should give a stronger overexpression effect at early stages. Precursor molecules of miR-124a (pre-miR-124a) were injected into one dorsal-animal blastomere at the 8-cell stage and the injected embryos were grown up to stage 46 for morphological analysis. Microinjection of 0.025 pmol pre-miR-124a induced eye anomalies, especially defects of the optic nerve (Fig. 3A). As shown in Table 1, approximately half (54%) of the injected embryos showed enlarged and pigmented optic nerves on the injected side, and these eyes were located closer to the midline of the body than those on the control side. With a lower dose of pre-miR-124a (0.0125 pmol), markedly fewer (14%) injected embryos showed abnormal optic nerves, while with a higher dosage (0.05 pmol), the proportion of Xenopus embryos with this phenotype increased to 73%, indicating that the eye defect induced by miR-124a overexpression was dose-dependent. In addition, when 0.025 pmol pre-miR-124awas co-injected with the same dose of 2′-O-methyl anti-124a, the percentage of embryos with optic stalk anomalies was reduced from 54% to 0%, while the inhibitor alone did not affect normal eye morphogenesis (Table 1). Our results indicate that eye anomalies induced by miR-124a precursor molecules could be rescued by its inhibitor. Embryos microinjected with precursor molecules of the control miRNA, miR-198 (pre-miR-198), or another negative control (pre-control) from Ambion did not cause any morphological defects at the above doses (Fig. 3A and data not shown). These results demonstrate that miR-124a overexpression specifically induced malformation of the eye.
Fig. 3.
Effects of miR-124a overexpression on eye development in Xenopus. miR-124a or control (miR-198) miRNA precursors (0.025 pmol) were microinjected into one Xenopus dorsal-animal blastomere at the 8-cell stage. (A) The head region of the injected Xenopus embryo grown to stage 46 is shown from the dorsal or lateral view. The pink arrow marks an enlarged and pigmented optic stalk-like structure induced by miR-124a injection. (B) The structure of wild-type and abnormal eyes of a stage 46 embryo injected by miR-124a is shown on a series of transverse sections (5 μm) from anterior to posterior. The sectionswere stained with hematoxylin (blue)–eosin solution (pink). Pink arrows indicate the optic nerve. The red arrowhead shows the reduced retina ganglion layers. (C) The expression (dark blue) of Pax2, Rx1, and Pax6 after miR-124a overexpression was detected by whole mount in situ hybridization at stages 13, 20, and 33. Stage 13 and 20 embryos are shown from the anterior view, and the eyes of stage 33 embryos are shown from the lateral view. Scale bars: 300 μm (A and C); 100 μm (B).
Table 1.
miR-124a overexpression induced specific optic nerve anomalies with a dose-dependent effect.
| RNA injected | Total number of injected embryos | Percentage of embryos with an abnormal optic nerve (%) |
|---|---|---|
| 0.05 pmol pre-miR-124a | 66 | 73 |
| 0.025 pmol pre-miR-124a | 63 | 54 |
| 0.0125 pmol pre-miR-124a | 59 | 14 |
| 0.025 pmol pre-miR-124a + 0.025 pmol anti-miR-124a | 92 | 0 |
| 0.025 pmol anti-miR-124a | 75 | 0 |
| 0.025/0.05 pmol control precursor (negative control 1) | 50 | 0 |
| 0.025/0.05 pmol pre-miR-198 (negative control 2) | 50 | 0 |
Abbreviations: pre-miR-124a, precursor molecules of miR-124a; pre-miR-198, precursor molecules of miR-198; anti-miR-124a, 2′-O-methyl antisense oligonucleotides of miR-124a.
Note: Data are representative of three independent experiments.
To further characterize the eye defects induced by miR-124a overexpression, the injected embryos were sectioned at eye-level. As shown in Fig. 3B, primary eye morphogenesis with a distinct lens, retina and optic nerve was already complete on the control side. Retina cells were well organized from proximal to distal as the retinal pigment epithelium (RPE), the outer nuclear layer (ONL), the inner nuclear layer (INL) and the retinal ganglion cells (RGC). On the side injected with pre-miR-124a, the eye still kept the primary structures of the RPE, neural retina and lens, but the RPE and neural retina expanded into the prospective optic nerve region and formed a significantly larger and shorter optic stalk-like structure. Moreover, sequential transversal sections showed that this enlarged ‘optic stalk’ was located more anterior to the optic cup, but not at the middle-level of the eye as on the control side (Fig. 3B, indicated by pink arrows). The distal retinas in the reduced optic cup, which showed disorganization of cell layers in the neural retina and a significant reduction of distal cell layers (including INL and RGC), were also less invaginated than controls (Fig. 3B, red arrowhead). In more severely affected embryos, distal retinas were less or even not invaginated, and lens formation was also inhibited in uninvaginated retinas (data not shown). These defects in optic nerve and retina formation suggest that excess miR-124a expression during the early embryonic stage disturbs eye morphogenesis, including the formation of the optic vesicle and cup, which in turn affects subsequent lens formation and layer organization in the neural retina.
As defects in the division of the optic stalk and retina and the subsequent formation of the optic cup and lens suggest the disturbance of proximodistal patterning of the eye, we examined expression of genetic markers for the proximal and distal eye to further characterize the timing and properties of the effects induced by miR-124a overexpression. Pax2 is a commonly used marker of the optic stalk and fissure, while Rx1 and Pax6 mark the developing retina which is distally located to the optic stalk (Liu et al., 2001; Lupo et al., 2005). When retina layers are formed, the expression domains of Rx1 and Pax6 become restricted to the proximal (ONL, outer part of INL) and distal layers (inner part of INL and RGC), respectively (Zuber et al., 2003; Wang and Harris, 2005). The expression of these genes was detected in 0.05 pmol pre-miR-124a-injected embryos at stages 13, 20 and 33, corresponding to early eye-field specification, optic vesicle and optic cup stages, respectively (Fig. 3C, Table S1). At stage 13, when miR-124a expression was not yet detectable, Pax2 was not clearly expressed in the eye field. The expression of Rx1 and Pax6, which were localized in the eye field at this stage, were not markedly changed in most of the injected embryos, indicating that excess miR-124a does not disturb eye-field specification. At stage 20 when miR-124a starts to be expressed in the entire neural tube (including the eye anlagen), the size of the Pax2 expression domain in the eye, including the ventral protruding optic vesicle, was not obviously affected, while distinct reduction was observed in the Rx1-and Pax6-marked retina domains in around 90% of the embryos. Moreover, the expression of Pax2 (in 60% embryos) and Pax6 in the presumptive eye and brain was more posterior on the injected side than on the control side. Therefore, miR-124a overexpression represses the extension of the neural tube, especially the protrusion of the optic vesicle. The repression of extension might become more evident in structures anterior than posterior to the eye during later development, which in the end led to a relative anterior ‘shift’ of the optic stalk observed in the miR-124 injected side at stage 46. At stage 33 when miR-124a was strongly expressed in both the eye and the brain, the expression of Pax2 in the optic stalk and fissure was still not markedly changed on the injected side, while the Rx1 and Pax6 domains that mark the retina region were still reduced in around 80% of embryos. Moreover, the expression domain of Pax6, which is normally restricted to the most distal retina, was more strongly reduced than that of Rx1. These effects are consistent with results from the histological analyses on retinal sections discussed above, and indicate that the inner cell layers of the distal retina are more severely affected than the outer layers. The lack of effect on Pax2 expression suggests that the optic fissure may have expanded with the abnormal retina into the optic stalk region. As Pax2 is also expressed in the optical fissure, the lack of effect of miR-124a on Pax2 expression observed here may be due to a masking effect of expression in the optical fissure.
2.4. miR-124a overexpression changes expression patterns of candidate target genes in the eye
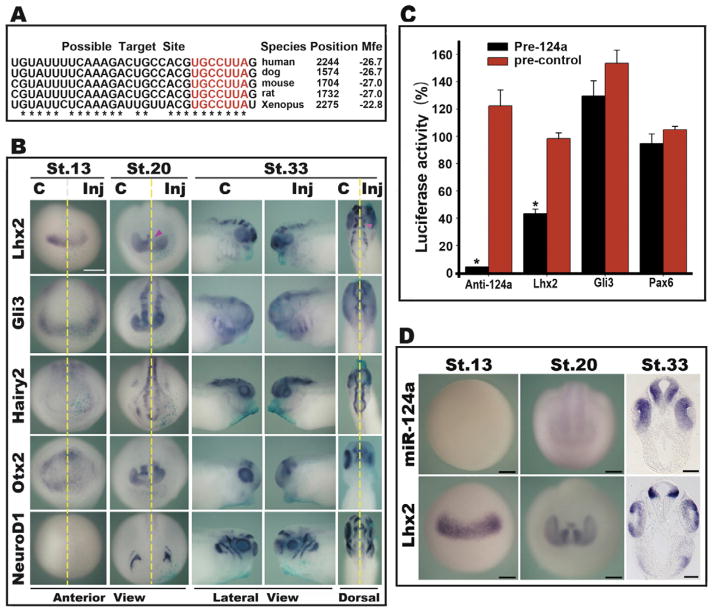
In order to identify the target(s) of miR-124a in the eye, we screened the candidate target genes predicted by MiRanda (John et al., 2004), miRBase (Griffiths-Jones et al., 2006), Target-Scan (Lewis et al., 2005) and PicTar (Krek et al., 2005) in vertebrates. Genes involved in eye development and neurogenesis were selected and used to search for potential target sites in the 3′ UTR of X. laevis transcripts by RNAhybrid with a requirement of 7-nt match to the seed of miRNA (position 2–8) (Lewis et al., 2003; Xie et al., 2005). To increase the reliability of prediction results, at least one perfect (no U:G pair) 6-nt match to position 2–7 of miRNA seed was also required for transcripts considered to be potential targets here (Rehmsmeier et al., 2004; Lewis et al., 2005). Our results of online searching showed that Lhx2, Gli3, Hairy2, Otx2 and NeuroD1 are predicted targets of miR-124a in mammal and X. tropicalis. Moreover, mRNA transcripts of X. laevis Lhx2, Gli3 and Hairy2 also contained potential target sites for miR-124a as predicted by RNAhybrid (Table S2). In particular, one conserved candidate binding site of miR-124a was found in the 3′ UTR of human, mouse, rat, dog and X. laevis Lhx2 (Fig. 4A).
Fig. 4.
miR-124a and its candidate targets. (A) An evolutionarily conserved miR-124 target site is found at the 3′ UTR of Lhx2 mRNAs in different species. Seed sequences are marked in red. Homologous sites are marked by asterisks. (B) Expression of the predicted targets of miR-124a: Lhx2, Hairy2, Gli3, NeuroD1 and Otx2 were detected by whole mount in situ hybridization. Embryos were injected with 0.05 pmol miR-124a precursor molecules into one dorsal-animal blastomere at the 8-cell stage and collected at stage 13 (anterior view), stage 20 (anterior view) or stage 33 (lateral view and dorsal view). The pink arrowhead indicates the reduced expression of Lhx2 on the injected side. Scale bars: 500 μm. (C) Luciferase assays were carried out in HEK293 cells using the pCS2-Luc-3′ UTR reporters of Lhx2, Gli3, and the positive (anti-miR-124a) and negative (Pax6) controls. Luciferase activity after transfection of the miR-124a precursor (pre-124a) or control precursor (pre-control) was normalized by that of the control cells without precursor transfection. Data are expressed as the means of three independent transfections ±SD, each carried out in triplicate. Asterisks indicate where the miR-124a transfected group was significantly different from the control precursor group (P < 0.05). (D) In situ hybridization detection of miR-124a and Lhx2 expression in Xenopus embryos at stage13 (anterior view), stage 20 (anterior view) and stage 33 (transection at the eye-level). Scale bars: 300 μm (stage 13, stage 20); 100 μm (stage 33).
To test whether expression of these five candidate genes could be affected by miR-124a overexpression at the mRNA level, we performed in situ hybridization at the stages indicated above following injection of miR-124a precursor molecules (0.05 pmol) (Fig. 4B and Table S1). At stage 13, expression of all the candidate genes on the control side became clear in or around the eye field located in the anterior neural plate, except that NeuroD1 was not yet expressed. On the injected side, Lhx2 and Otx2 were down-regulated in 82% and 57% of embryos, but miR-124a overexpression did not affect the expression of Hairy2 and Gli3 at stage 13. At the neural tube stage (stage 20) when endogenous miR-124a and the transcripts of all five genes were detectable in or around the normal primary optic vesicle, the expression domains of these candidate miR-124a targets were all reduced in more than half of the injected embryos. In addition, the expression of Lhx2 and Hairy2 was also reduced in the arch-encephalon. However, changes in expression of all the candidates at this stage tended to be restricted to specific regions instead of an overall reduction of the expression level throughout the miR-124a-overexpressed region traced by beta-gal staining. At the optic cup stage (stage 33), the reduction in expression of Lhx2, Hairy2, NeuroD1 and Otx2 in the eye was still observed in more than 70% of the embryos, whereas it was difficult to distinguish the change in Gli3 expression between the injected and control sides due to its low expression in the normal eye. In addition, the expression domain of Otx2 in the eye, which will represent the distal central retina later (Wang and Harris, 2005), was dramatically reduced on the injected side, indicating an inhibition of distal retina formation coincident with the results of the above analysis at both the histological and molecular levels. The expression of Lhx2 and Hairy2 in the brain was also significantly reduced or distorted, which is consistent with the decrease of their expression in the arch-encephalon induced by miR-124a overexpression at st.20.
The above results indicate that miR-124a overexpression leads to dynamic changes in down-regulation of the expression of these five candidate genes. The expression of these genes is significantly inhibited at stage 20, when endogenous miR-124a expression starts. These observations suggest that miR-124a is able to regulate the genetic network involved in eye development. Moreover, Lhx2 and Otx2 mRNAs are negatively regulated by miR-124a even at stage 13, and this early effect indicates inhibition by the degradation of these two mRNAs even though the target site of Otx2 was not predicted in the reported 3′ UTR of Xenopus transcripts. The temporal and spatial restriction of the effects on other candidate target genes also implies that miR-124a may act alongside other mechanisms to regulate the expression of these genes.
2.5. miR-124a targets the 3′ UTR of Lhx2 mRNA
To determine if miR-124a can act directly on the predicted targets discussed above, we performed a luciferase reporter assay using the Xenopus genes Lhx2 and Gli3, the two with the best seed match (Table S2, perfect 2–8 seed match). The 3′ UTRs of these two genes containing the predicted miR-124a binding sites were separately fused to the luciferase coding region. The 3′ UTR of Pax6, which does not contain perfect seed match and is not predicted to be targeted by miR-124a (Table S2), was fused to the luciferase reporter gene as a negative control and the antisense sequence of miR-124a (anti-124a) was used as a positive control. As shown in Fig. 4C, transfection of control miRNA precursor did not reduce the luciferase activity of any of the four constructs. When the miR-124a precursor was introduced, the luciferase activity of the positive control (anti-124a) and Lhx2 reporters was down-regulated to 4% and 43%, respectively. Expression of the Gli3 reporter and the negative control (Pax6), on the other hand, was not significantly changed. These results demonstrate that miR-124a can interact with the 3′ UTR of Lhx2 and downregulate its expression.
To explore the functional relationship between miR-124a and Lhx2, we compared their expression patterns in Xenopus embryos at different stages by in situ hybridization. As shown in Fig. 4D, while miR-124a showed low expression at early stages and high expression at late stages in the eye, Lhx2 showed almost the reverse expression pattern during eye development. At stage 13 before miR-124a was expressed, Lhx2 mRNA was clearly detected in the eye field. At stage 20 when the miR-124a signal became detectable in the eye anlagen, Lhx2 expression appeared to be lower in the overlapping ventrodistal region. At stage 33 when the miR-124a signal became strong in the central retina, Lhx2 expression was more restricted and exhibited a low level of expression in this location. On the other hand, in the peripheral CMZ, where miR-124awas absent, Lhx2 was expressed at a higher level. These temporal and spatial complementary expression patterns of Lhx2 and miR-124a suggest that Lhx2 could be an endogenous target of miR-124a during eye development, and that the expression of miR-124a leads to degradation of Lhx2 transcripts.
2.6. miR-124a overexpression inhibits cell proliferation during retinogenesis
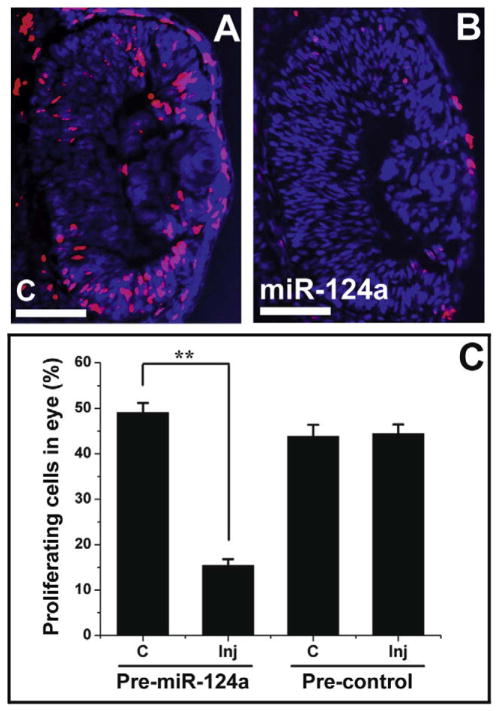
It has been shown that overexpression of Lhx2 in the head results in increased cell proliferation, and that knockdown of Lhx2 leads to deficient cellular proliferation resulting in retarded development of the eye and brain (Ando et al., 2005; Monuki et al., 2001). It is interesting therefore to see whether miR-124a overexpression affects cell proliferation in the eye. We carried out BrdU incorporation analysis at stage 33 of retinogenesis in Xenopus embryos injected with 0.025 pmol pre-miR-124a or pre-control, and found a significant decrease in the number of dividing (BrdU-positive) cells on the injected side of the embryo compared with the control side (Fig. 5). As shown in Fig. 5B, the average ratio of proliferating cells in all retina cells (Hoechst stained) of embryos injected with pre-miR-124a, was 49.1 ± 2.1% (mean ± SE) in the uninjected control side at stage 33, while that in the injected side decreased significantly to 15.5 ± 1.3%. In embryos injected with the control precursor, however, the average ratio of BrdU-positive cells in the injected retina was 44.4 ± 2.0%, not significantly different from that (43.8 ± 2.6%) in the control side. These results indicate that excessive miR-124a inhibits retina cell proliferation in Xenopus embryos, and suggests a role for miR-124a in controlling the proliferation of retinal progenitors during eye development.
Fig. 5.
miR-124a overexpression inhibits cell proliferation during retinogenesis. miR-124a or control miRNA precursor (0.025 pmol) was co-injected with GFP mRNA into one dorsal-animal blastomere of a Xenopus embryo at the 8-cell stage, and cell proliferation was detected by BrdU incorporation. (A and B) Transversal section of a stage 33 embryo at the level of the eye showing that the number of proliferating cells (BrdU-positive, red) on the miR-124a overexpressed side (miR-124a) was much lower than that on the control side (C). All cell nuclei were counterstained with Hoechst 33258 (blue). Scale bar: 100 μm. (C) The ratio of proliferating retinal cells (BrdU-positive/Hoechst-labeled cells) in either the injected side (inj) or the control side (C) of embryos injected with miR-124a (pre-miR-124a) or control (pre-control) precursors. Data are expressed as means ± SE from twelve sections of four embryos. A significant difference (**P < 0.01 by one-way ANOVA followed by Duncan’s test) between the mean ratios between the injected and control sides was only detected for pre-miR-124a injected embryos.
3. Discussion
3.1. The role of miR-124a in Xenopus eye development
In this paper, we have used the Xenopus eye as a model system to study the role of miR-124a. Although there have been reports that miR-124a is expressed in the adult brain and eye in several species including X. tropicalis (Deo et al., 2006; Darnell et al., 2006; Wienholds et al., 2005; Kloosterman et al., 2006; Sweetman et al., 2006), our functional analysis data provides the first evidence that confirms the involvement of miR-124a in eye development and identifies its potential targets in X. laevis.
The role of miR-124a in the eye was first revealed by an expression analysis of miR-124a. In situ hybridization experiments showed that the expression pattern of miR-124a is dynamic throughout the developing eye and brain, beginning at a low level along the entire neural fold at around mid-neurula and increasing to a high level in the mature brain and eye. The onset of miR-124a expression observed here and its location in the mature CNS (including the eye) is similar to previous results obtained by other methods or in other species (Aboobaker et al., 2005; Darnell et al., 2006; Kloosterman et al., 2006; Watanabe et al., 2005; Wienholds et al., 2005). The presence of miR-124a in the developing neural tube and eye suggests it has an early role in the CNS development of X. laevis. This hypothesis is also supported by our observation that early introduction of excess miR-124a leads to malformation of the optic nerve and retina. Moreover, these eye defects are related to an early reduction of the eye anlagen and a later repression of the distal structures of the eye, as shown by the effects of a set of eye markers on expression and reduction of retinal cell proliferation during retinogenesis. These results indicate that miR-124a is able to act on the genetic network involved in the early morphogenesis of the eye, and that maintaining a low level of miR-124a at early stages is necessary for proper cell proliferation and eye formation.
In this work, the loss-of-function analysis showed no significant disturbance of eye morphogenesis, though endogenous miR-124a had clearly been inhibited at least until the early optic cup stage. This result is consistent with two previous reports: miR-124a knockdown does not have any significant effect on differentiating neural precursors (Krichevsky et al., 2006) and does not affect neuronal differentiation in the developing chick neural tube (Cao et al., 2007). It has been reported that less than 10% of individual miRNA knockouts result in clear developmental or morphological defects in Caenorhabditis elegans (Miska et al., 2007), and most miRNAs might act redundantly with other miRNAs as fine-tuning regulators (Cao et al., 2007; Baroukh et al., 2007). Our results support the hypothesis that miR-124a might not act as a crucial regulator of early developmental events, and it will be interesting to determine if some fine structures or characteristics of the eye are altered by miR-124a down-regulation.
Although our results tend to indicate that miR-124a may not be necessary for early eye development in Xenopus, we cannot rule out the possibility that the early expression of miR-124a is not inhibited completely by the miR-124a inhibitor and that only a small amount of miR-124a is enough to maintain normal function in vivo. Furthermore, our loss-of-function approach using antisense oligonucleotides did not generate significant knockdown effects at the late embryonic stage (Fig. 2), so we cannot exclude the possibility that miR-124a might be necessary during late eye development in Xenopus. Our results of both gain- and loss-of-function analyses also reveal that early microinjection of miRNA precursors or inhibitors is efficient for early but not for late disturbance of miR-124a levels, which are present at low levels during early stages and at higher levels during later stages of development.
The LNA-modified miRNA inhibitor showed a strong non-specific effect in this study, in contrast to previously reported experiments (Cao et al., 2007; Visvanathan et al., 2007; Corsten et al., 2007; Elmén et al., 2008). This might be due to the lower incubating temperature (22 °C) used for growth of Xenopus embryos than that (no lower than 37 °C) for chick and mouse embryos, or for cultured cells. In our experience with in situ hybridization, an appropriate hybridization temperature is critical for the specific binding of the antisense LNA probe to its target. In a similar way, the interaction of antisense LNA inhibitors and endogenous miRNAs may be sensitive to temperature fluctuations. A more effective inhibitor and a better approach for disturbing the strong expression of miR-124a at late developmental stages are needed to better characterize miR-124a function in X. laevis.
3.2. The mechanism of the role of miR-124a in eye development
The effects of miR-124a overexpression suggest a critical role for its target(s) in retinogenesis and eye morphogenesis, especially in the events involved in protrusion of the optic vesicle and in the formation of the optic stalk and optic cup. Results of computer predictions imply that the mechanism underlying the role of miR-124a in the eye involves multiple targets. Our results for miR-124a target identification show that the 3′ UTR of Lhx2 could be directly targeted by miR-124a in vitro, and that its expression in vivo was downregulated by miR-124a overexpression. Moreover, the location of Lhx2 transcripts and miR-124a in the eye are somewhat complementary throughout early eye development, and early effects of miR-124a overexpression down regulates Lhx2 RNA level at stage 13. These results suggest that Lhx2 could be an endogenous target for miR-124a in the eye, and down-regulation of the expression of Lhx2 by miR-124a might be due to, or at least include, mRNA degradation. It has been reported that in the Lhx2 knockout mouse, eye development arrests prior to formation of an optic cup although specification of the optic vesicle occurs (Porter et al., 1997). In Zebrafish, bel (Lhx2) mutant eyes are shorter in the dorsoventral axis and wider in the mediolateral axis than wild-type eyes (Seth et al., 2006). In addition, Lhx2 can regulate eye size by regulating cell proliferation in zebrafish embryos (Ando et al., 2005). These results are consistent with the effects of miR-124a overexpression in Xenopus, implying that down-regulation of Lhx2 by miR-124a is involved in malformation of the eye.
For the other predicted targets, the 3′ UTR of Gli3 did not show significant interaction with miR-124a to downregulate gene expression in vitro. Hairy2, Otx2 and NeuroD in X. laevis either contain weaker or no potential target sites within their reported 3′ UTR sequences (Table S1), making it less likely for them to be inhibited by miR-124a in this region. However, the effect of miR-124a overexpression on Otx2 expression was similar to that on Lhx2 expression and even stronger at stage 33 (Fig. 4). It is possible that some target sites have not been identified by the target prediction algorithms (including the transcript identification) used here, or that the analyzed transcript do not include all the 3′ regulatory regions of these genes, or that miR-124a might interact with other regions of these genes to exert its regulatory role (Lytle et al., 2007; Duursma et al., 2008). In addition, as miRNAs can mediate down-regulation of target gene activity by either translational inhibition or target mRNA cleavage, we cannot exclude the possibility that miR-124a repressed their translation. Other factors may also interact with miR-124a to regulate the expression of these genes in restricted regions during development. Furthermore, as Otx2, Pax6 and Lhx2 are present in a genetic network controlling eye-field specification (Zuber et al., 2003), cross regulation might also be involved in the change of their expression profiles. Extensive rescue experiments might be needed to specify the key factors responsible for the phenotype induced by miR-124a overexpression.
Some other genes have been reported to be targeted by miR-124, such as the anti-neural factors REST (Conaco et al., 2006) and SCP1 (Wu and Xie, 2006), neural progenitor enriched laminin_1 and integrin_1 (Cao et al., 2007) in the spinal cord, and a pancreatic-related Foxa2 (Baroukh et al., 2007). Since the roles of these targets of miR-124 in the eye are still not clear, it will be interesting to determine whether these genes are involved in the malformation of the eye by miR-124a overexpression in Xenopus. These reports also suggest that miR-124a represses both non-neuronal and neuronal progenitor fates, and hence helps to induce neurogenesis/differentiation and maintain neuronal fate in the CNS. In addition to Lhx2, NeuroD1 and Hairy2a, which are involved in neurogenesis, were predicted to be targets of miR-124a and the expression of these two genes was down-regulated by miR-124a overexpression. NeuroD1 is expressed in the primary neuron and promotes its differentiation (Cho and Tsai, 2004), and Hairy2a has been shown to repress the expression of pro-neuronal genes in non-neuronal cells surrounding developing neurons and play a role similar to REST and SCP1 (Klose and Bird, 2003). Our BrdU analysis results show that miR-124a overexpression inhibited cell proliferation in the eye during retinogenesis, which is consistent with the loss-of-function effects of Lhx2 and Hairy2a (Ando et al., 2005; Monuki et al., 2001; Nichane et al., 2008). In addition, as Lhx2, Gli3, Otx2 and NeuroD1 are also involved in regulation of retinal cell fate (Viczian et al., 2006; Perron et al., 2003; Wang and Harris, 2005), miR-124a may be involved in retinal cell determination by its interaction with these genes. The above evidence suggests that miR-124a might also play an important role in neurogenesis during eye development. Moreover, miR-124a might act in a similar manner in other parts of the central nervous system. Further investigation is needed to confirm these hypotheses and a more extensive screening is also necessary to identify other possible targets of miR-124a in the eye.
In summary, we have provided evidence that miR-124a antagonizes Lhx2 and is involved in the early development of the vertebrate eye. Early overexpression of miR-124a causes decreased cell proliferation during retinogenesis and malformation of Xenopus eyes. The mechanism by which miR-124a interacts with the genetic network at early stages of development may persist through later development into adult stages, and may be involved in other parts of the central nervous system.
4. Experimental procedures
4.1. LNA-modified probe preparation
The sequence of the LNA probe for mature miR-124a was 5′-TGGCATTCACCGCGTGCCTTAA-3′. Probes were synthesized using EXIQON (miRCURY™detection probe) and labeled with a DIG oligonucleotide 3′-end labeling kit (Roche), then purified with Sephadex G25 MicroSpin columns (Amersham Biosciences). Approximately 100 ng/ml of labeled probe was used in the in situ hybridization experiments (Wienholds et al., 2005; Kloosterman et al., 2006).
4.2. In situ hybridization
Embryos were obtained and staged as previously described (Nieuwkoop and Faber, 1967). In situ hybridization of the transcription factors Otx2, Rx1, Pax6, Pax2, Gli3, NeuroD1, Hairy2 and Lhx2 was performed on whole embryos of Xenopus as previously described (Harland, 1991), except for the following modifications: embryos which were pre-stage 33, those from stage 33 to stage 46, and dissected adult brain and eye, were treated with proteinase K for 10, 45, 60 min, respectively. The temperature of hybridization and subsequent washing steps was adjusted to approximately 22 °C below the predicted melting temperatures of the LNA-modified probes. In addition, a digoxigenin (DIG)-labeled oligonucleotide corresponding to the lacZ′ region in the pUC and M13 plasmids (30-mer, 5′-p TTGGGTAACGCCAGGGTTTTCCCAGTCACG OH-3′) was used as a negative control.
4.3. Microinjection
Both LNA antisense DNA and 2′-O-methyl antisense RNA oligonucleotides were used for loss-of-function analysis. The sequences were designed to target the conserved mature sequence of miR-124a (5′-UAAGGCACGCGGUGAAUGCC-3′) or that of a control miRNA, miR-198 (5′-GGUCCAGAGGGGA GAUAGG-3′). The LNA antisense DNA was obtained from the same sources as the probes mentioned above. The 2′-O-methyl antisense RNA (Anti-miR™miRNA Inhibitor) and the other negative control for the 2′-O-methyl oligonucleotide were obtained from Ambion (Cat. Nos. 17000 and 17010, respectively).
Pre-mmu-miR-124a-2 shares higher sequence identity than other miR-124a homologues to the reported Xenopus miR-124a (Xtr-miR-124) (Fig. S1), so the synthetic Pre-miR™miRNA precursor molecule for pre-mmu-miR-124a-2 (Ambion Cat. No. 17110) was used for overexpression analysis. The Pre-miR™miRNA precursor molecule for hsa-miR-198 (Ambion Cat. No. 17100) and a control sequence (Ambion Cat. No. 17110) were used as negative controls.
In both analyses, embryos were co-injected with 100–500 pg beta-gal or 200–400 pg GFP mRNA as a lineage tracer; and embryos injected with beta-gal were stained as previously described (Andreazzoli et al., 1999). Capped mRNAs were synthesized from linearized plasmid templates using mMESSAGE mMACHINE kits (Ambion). Oligonucleotides or mRNAs were injected into one Xenopus dorsal-animal blastomere at the 8-cell stage using an Eppendorf FemtoJet (Hamburg, Germany), and embryos were then cultured as previously described (Lupo et al., 2005).
4.4. Bromodeoxyuridine (BrdU) administration
BrdU (Sigma B9285) was administered to X. laevis embryos injected with 0.025 pmol pre-miR-124a or pre-control molecules, by immersion in a solution of 10 mMBrdU in phosphate buffered saline (PBS) for 30–60 min at room temperature (19–22 °C) as described by Quick and Serrano (2007). Three hours after exposure to BrdU, embryos were grown to the optic cup stage (stage 33) and stored in 20% sucrose after being treated with 4% para formaldehyde (PFA) (Quick and Serrano, 2007; Zuber et al., 1999). For sectioning analysis, samples were embedded in OTC, serially sectioned at a thickness of 12 μm and stained immunohistochemically using mouse anti-BrdU monoclonal antibodies. Hoechst33258 (Sigma) was used for counter-staining of all cell nuclei.
Counts of BrdU-positive and Hoechst-labeled cells in the optic cup were obtained from twelve sections of four injected embryos by tracing of digitized images projected on a computer monitor. The ratio of proliferating cells in the eye was calculated by dividing the number of BrdU-positive cells by all the cells counted (Hoechst-labeled cells) in the eye on either the injected side or the control side. Statistical analysis was performed using one-way ANOVA followed by the Duncan test. Comparisons of means of proliferating cell ratios were made between the uninjected and injected eye of either pre-miR-124a or pre-control injected embryos. Differences between groups were considered significant when P < 0.05. At least three independent experiments were performed.
4.5. Histology and imaging
The injected and control embryos for phenotype analysis were grown to late tadpole stage (about stage 46) and stored in 100% ethanol after being fixed in 4% formaldehyde in MEM (MOPS/EGTA/magnesium sulfate phosphate buffer) (Harland, 1991). For sectioning, samples were embedded in paraffin, serially sectioned at a thickness of 5 μm for histological analysis of the overexpression phenotype, or 15 μm for expression analysis, and then stained with hematoxylin–eosin solution.
Images were taken using an OLYMPUS SZX12 (Japan) stereomicroscope with a digital acquisition system (Olympus C4040). Sections were photographed on an inverted microscope (OLYMPUS IX71, Japan) or a compound microscope (Nikon FXA, Japan) using DIC optics or fluorescent filters. The fluorescent images taken with different filters were merged by Adobe Photoshop CS.
4.6. Luciferase assays
The firefly luciferase reporter genes were constructed using the pCS2-Luc vector and the 3′ UTR sequences of Xenopus Lhx2, Gli3 and Pax6, following PCR amplification using the primers listed in Table S3. The reporter construct of Pax6 and that with the antisense sequence of mature miR-124a were constructed as negative and positive controls, respectively. HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum. 5 · 104 cells/well were seeded in 24-well plates. After 24 h in culture, the cells were transfected using Lipofectamine 2000 (Invitrogen) with a mixture containing 1 μg/ml of firefly luciferase reporter plasmid, 20 nM miR-124a or control precursor, and 20 ng/ml of Renilla reniformis luciferase encoding plasmid (pRL-TK, Promega). Cells transfected without precursor served as a control for normalization. Luciferase activity was measured 24–48 h post-transfection using a Dual-luciferase assay system (Promega). All transfections were repeated independently at least three times.
4.7. Informatics
Mouse, human, zebrafish and X. tropicalis miRNA precursor and mature sequences were downloaded from the miRNA Registry database (http://www.sanger.ac.uk/Software/Rfam/mirna/). The X. laevis sequence was obtained from Watanabe et al. (2005). The mRNA sequences of Lhx2 (AY141037), Gli3 (NM_001087971), Hairy2 (AF139914), Otx2 (NM_001090691, U19813), NeuroD (U28067) and Pax6 (U77532) were obtained from the NCBI nucleotide database.
The predicted binding sites of miR-124a in transcripts of X. laevis were analyzed by RNAhybrid (Rehmsmeier et al., 2004) using the mature sequence of X. laevis miR-124a. RNAhybrid was run with either perfect (no U:G in seed) or imperfect (U:G allowed in seed) seed match; helix constraint in seed was set either from position 2 to 7 or 2 to 8 of the miRNA sequence. miR-124a target candidates in other species were searched from the online sources of MiRanda (www.microrna.org), mirBase (microrna.sanger.ac.uk/targets/v5/), Target-Scan (www.targetscan.org/vert_50/) and PicTar (www.pictar.org).
Acknowledgments
We are grateful to Prof. Giuseppeina Barsacchi’s lab for generously providing plasmids Rx1, Otx2, Hairy2a; William A. Harris/Christine Holt’s labs for Lhx2, and NeuroD and Muriel Perron for Gli3. We thank Xiumei Wang for technical assistance, Xiangqian Guo for support on computer prediction analysis, Giuseppe Lupo, William A. Harris, Marie-Laure Baudet for helpful discussions, and Ruimin Zheng, Xudong Zhao of IBP core facilities centre for technical support. This work was supported by NSFC (Nos. 30771129, 30530280) and National Basic Research Program of China (973 Program 2005CB522804).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mod.2009.08.002.
References
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H, Kobayashi M, Tsubokawa T, Uyemura K, Furuta T, Okamoto H. Lhx2 mediates the activity of Six3 in zebrafish forebrain growth. Dev Biol. 2005;287:456–468. doi: 10.1016/j.ydbio.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Angeloni D, Menna E, Barsacchi G. Role of XRx1 in Xenopus eye and anterior brain development. Development. 1999;126:2451–2460. doi: 10.1242/dev.126.11.2451. [DOI] [PubMed] [Google Scholar]
- Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, Van Obberghen E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Tsai MJ. The role of BETA2/NeuroD1 in the development of the nervous system. Mol Neurobiol. 2004;30:35–47. doi: 10.1385/MN:30:1:035. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. MiR-148 targets human DNMT3b protein coding region. RNA. 2008;14 (5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederikse PH, Donnelly R, Partyka LM. MiRNA and Dicer in the mammalian lens: expression of brain-specific miRNAs in the lens. Histochem Cell Biol. 2006;126:1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved wholemount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. How microRNAs choose their targets. Nat Genet. 2007;39:1191–1192. doi: 10.1038/ng1007-1191. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Klose R, Bird A. Molecular biology. MeCP2 repression goes nonglobal. Science. 2003;302:793–795. doi: 10.1126/science.1091762. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lupo G, Marchitiello A, Gestri G, He RQ, Banfi S, Barsacchi G. Expression of the Xvax2 gene demarcates presumptive ventral telencephalon and specific visual structures in Xenopus laevis. Mech Dev. 2001;100:115–118. doi: 10.1016/s0925-4773(00)00505-0. [DOI] [PubMed] [Google Scholar]
- Lupo G, Liu Y, Qiu R, Chandraratna RA, Barsacchi G, He RQ, Harris WA. Dorsoventral patterning of the Xenopus eye: a collaboration of Retinoid, Hedgehog and FGF receptor signaling. Development. 2005;132:1737–1748. doi: 10.1242/dev.01726. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104 (23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, et al. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32 (4):591–604. doi: 10.1016/s0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Nichane M, Ren X, Souopgui J, Bellefroid EJ. Hairy2 functions through both DNA-binding and non DNA-binding mechanisms at the neural plate border in Xenopus. Dev Biol. 2008;322(2):368–380. doi: 10.1016/j.ydbio.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North Holland; Amsterdam, The Netherlands: 1967. [Google Scholar]
- Perron M, Boy S, Amato MA, Viczian A, Koebernick K, Pieler T, Harris WA. A novel function for Hedgehog signalling in retinal pigment epithelium differentiation. Development. 2003;130 (8):1565–1577. doi: 10.1242/dev.00391. [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Quick QA, Serrano EE. Cell proliferation during the early compartmentalization of the Xenopus laevis inner ear. Int J Dev Biol. 2007;51:201–209. doi: 10.1387/ijdb.062176qq. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Culverwell J, Walkowicz M, Toro S, Rick JM, Neuhauss SC, Varga ZM, Karlstrom RO. Belladonna/(Ihx2) is required for neuralpatterning and midline axon guidance in the zebrafish forebrain. Development. 2006;133:725–735. doi: 10.1242/dev.02244. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Rathjen T, Jefferson M, Wheeler G, Smith TG, Wheeler GN, Münsterberg A, Dalmay T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev Dyn. 2006;235:2185–2191. doi: 10.1002/dvdy.20881. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Bang AG, Harris WA, Zuber ME. Expression of Xenopus laevis Lhx2 during eye development and evidence for divergent expression among vertebrates. Dev Dyn. 2006;235(4):1133–1141. doi: 10.1002/dvdy.20708. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434 (7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Perron M, Philpott A, Bang A, Harris WA. Giant eyes in Xenopus laevis by overexpression of Xoptx2. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]