Abstract
Research in nucleic acids has made major advances in the past decade in multiple fields of science and technology. Here we discuss some of the most important findings in DNA and RNA research in the fields of biology, chemistry, biotechnology, synthetic biology, nanostructures and optical materials, with emphasis on how chemistry has impacted, and is impacted by, these developments. Major challenges ahead include the development of new chemical strategies that allow synthetically modified nucleic acids to enter into, and function in, living systems.
The fields of research involving nucleic acids have been marked by amazing progress in the past decade. This recent work has been notable for both its diversity—covering many varied fields of research—and its breathtaking pace, with huge numbers of scientists and engineers diving into the pool.
Research and technology development involving DNA and RNA has been broadly attractive for many reasons. First is the relevance to biology and medicine; thus many of the developments have involved new basic findings in biology. Moreover, many of the technological developments are aimed at eventual application in medicine (either diagnostic or therapeutic). Second is the variety of predictable structures formed by DNA. This has encouraged not only chemists and biologists (who have a longer history with DNA) to make new tools, but also others in the fields of physics and engineering to enter the field as well. Third is the widespread availability of DNA: a large number of commercial sources now offer oligonucleotides of widely varied sequence and length and with a wide variety of modifications to be ordered at reasonable cost and shipped quickly. Moreover, high-throughput synthesizers have made large numbers of DNAs available at an attainable cost as well.
Natural discoveries inspire chemistry
Although nucleic acids have been studied for decades, there is room for surprising new discoveries in this field. The last ten years have uncovered several new and highly important fundamental aspects of the biology of nucleic acids, and many of these are inspiring chemically oriented research. One of the biggest findings in all of biology in the past decade has been the realization that there are many thousands of RNAs in the cell that are not classical messenger RNAs (mRNAs) or transfer RNAs (tRNAs) but which have biological functions nevertheless.1 These RNAs range from very long (stretching thousands of nucleotides over multiple genes) to very short (20 nucleotides (nt) or less), and are encoded by either strand (i.e. sense or antisense) of the genomic DNA. The presence of the additional complexity afforded by cellular networks of interacting RNAs and proteins has helped explain how humans can be so complex with a relatively small genome (much smaller than some plants, for example).
Gaining a good deal of attention among chemists have been the various small noncoding RNAs (ncRNA) that are under study.2 These RNAs are often as short as 20 nt and do not encode any polypeptide sequence (hence the name). The most important examples include miRNAs (micro RNAs) and siRNAs (short interfering RNAs). These latter RNAs have become the standard tool for downregulating genes in biological studies. They are most active as short RNA duplexes, and given the relative accessibility of synthetic RNA, they have been widely studied by chemists and biologists. Many examples of chemically modified siRNAs have been described over the past decade.3 Modifications have been aimed at improved cellular uptake, enhanced stability against degradation, and altered activity in gene suppression. Chemically synthesized siRNAs are under intense study as drug candidates, and thus have entered the medicinal chemistry realm.4
Another fascinating class of small non-coding RNA is the riboswitches, which started as designed oligoribonucleotides that could switch activity (such as ribozyme activity) in response to this binding.5 Subsequently, searches were carried out for small-molecule binding sequences within the genomes of organisms, and they were found in bacteria (and later in a wide variety of organisms). Their function as genetic switches have been explored in recent years.
Developments in synthesis and modification
The chemistry of DNA modification, which blossomed in the 1980s, has undergone some important new advances in the past ten years or so. A number of new conjugation methodologies have been developed recently that have supplanted some of the earlier approaches by offering greater simplicity, milder conditions, and higher yields. These allow the researcher to add fluorescent tags, affnity tags, electrochemical reporters, and groups to aid cellular uptake. For example, metal-mediated carbon-carbon bond forming chemistries have shown real utility in recent years.6
One of the most important new chemistries for synthesis of DNA conjugates has been the [3+2] alkyne-azide cyclization (Huisgen-Sharpless ‘‘click’’ chemistry).7 The reaction is mild and proceeds readily in water. Importantly, the reaction is strongly ‘‘bioorthogonal’’—that is, the reacting groups do not react with typical functional groups found in biological systems. The development of this chemistry for DNA and RNA conjugation has led to the availability of ‘‘clickable’’ reagents (for example, fluorophores with azides or alkynes attached) that can be readily attached to an oligonucleotide with the proper functional group.8 The reaction is typically carried out with Cu[I] catalysis, although recent application of strained alkynes has opened the possibility of carrying out the reaction without this metal as well.9
Useful chemically modified nucleotide monomers were explored widely this past decade. One example is designed monomers that allow one to generate unstable reactive intermediates in DNA, for the purpose of studying their biochemistry and biology. Examples include precursors for photochemical generation of free radicals on deoxyribose10 and monomer nucleotides of putative damaged nucleotides.11 Marx has shown that C-4′ alkylated nucleotides can be highly effcient polymerase substrates that can enhance fidelity of replication.12 Hocek has demonstrated methods for rapidly introducing aldehydes on DNA bases, which yields a ready site for addition of labels.6c
Another important chemical advance in nucleic acids research has been the development of improved chemistry for the synthesis of RNA. The chemistry of Scaringe, who developed a new set of protecting groups for RNA in the late ‘90s (Fig. 1),13 was especially important in the past ten years because of the strong demand for siRNAs among biologists.
Fig. 1.
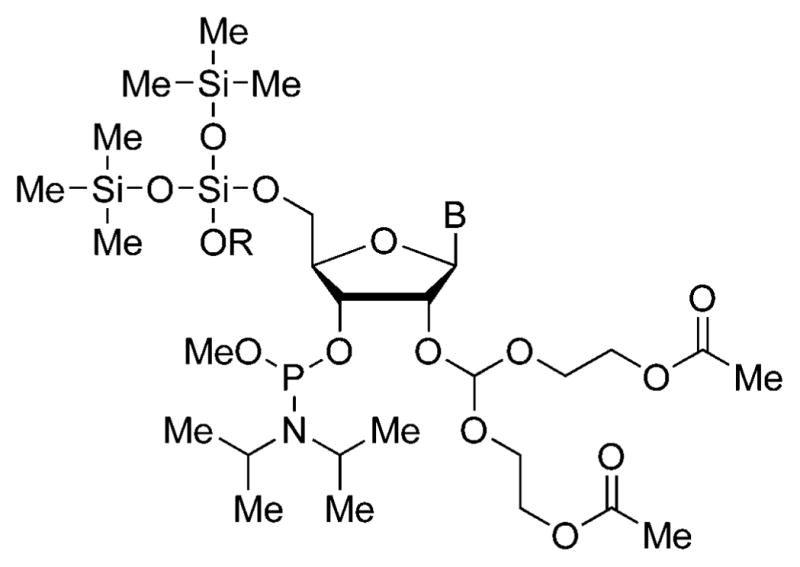
RNA synthesis building block developed by Scaringe.
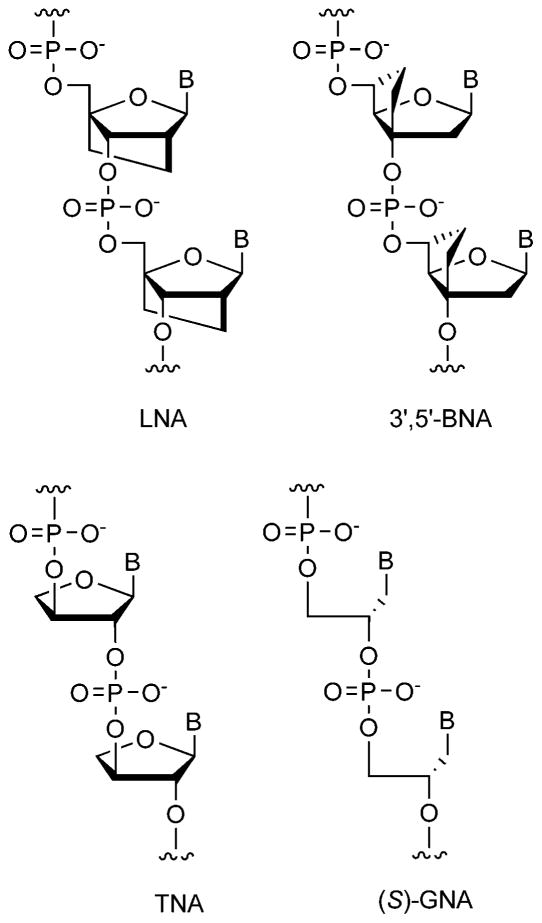
The last decade has also seen vigorous development of DNA analogs having useful or unusual properties. Notable examples studied most in the past ten years include conformationally locked analogs LNA (locked nucleic acids),14 BNA (bicyclo-DNA),15 TNA (threose nucleic acids),16 GNA (glycerol nucleic acids),17 and xDNA.18 The first two, LNAs and BNAs (Fig. 2), bind very strongly to DNA/RNA complements, due largely to rigidification of the otherwise relatively flexible ribose ring. Although developed prior to this decade, LNAs in particular have seen much study recently, with many biological applications reported, and derivatization with fluorophores and other moieties.19 TNA, having a smaller sugar than natural nucleic acids, is of special interest from the molecular evolution standpoint; despite its simplicity, it hybridizes well to complements and can even be a substrate for polymerase enzymes.20 And speaking of simplicity, one of these modifications, GNA, is remarkable because it hybridizes exceptionally strongly given its lack of a ribose (or indeed, any) ring. It is also quite simple to synthesize from simple chiral starting materials, allowing modified bases to be installed easily.21 Finally, a different kind of DNA analog that has a natural phosphodiester backbone but with an entirely non-natural base pairing scaffold was developed during this decade, namely expanded DNA (xDNA); it is described in more detail below (see Synthetic Biology).
Fig. 2.
Structures of backbone analogs of DNA studied recently.
Nucleic acid biotechnologies
Of all the technologies built around DNA in this past decade, perhaps the highest impact came (and is continuing to come) from the new generation of high-throughput sequencing methodologies, most of which required substantial chemical innovation. Venter made a groundbreaking demonstration during the human genome project that ‘‘shotgun’’ sequencing approaches (combining many short reads and sorting them out computationally) could work to yield complex genomes quickly.22
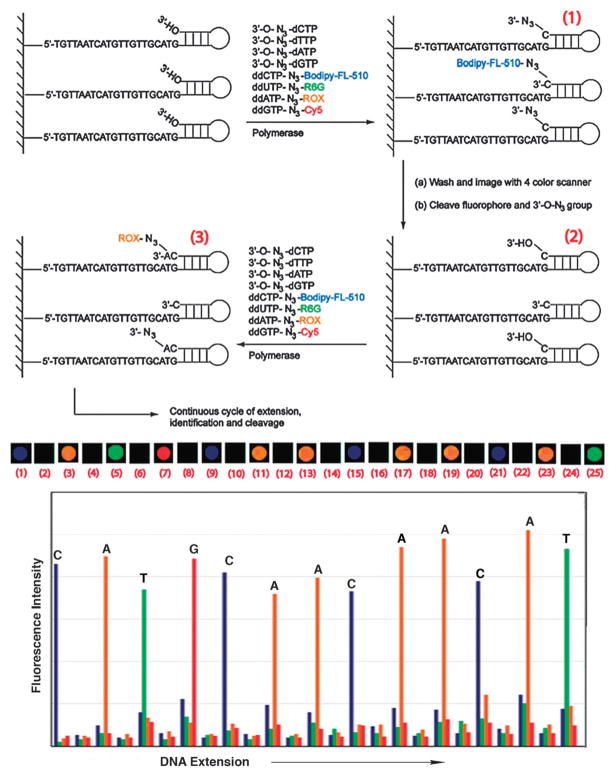
This realization rapidly led to the development during this past decade of several high-throughput strategies for sequencing genomes more rapidly and cheaply. Examples of some of these approaches include the pyrosequencing approach; sequencing by synthesis (SBS) and by hybridization (SBH) approaches in which chemically blocked nucleotides are added (with their signal) by polymerase, then chemically deblocked to allow the next step (Fig. 3); and the SOLiD approach, which involves ligation of nucleotide dimers.23 Even more effcient methodologies are under continuing development by chemists, biologists, physicists, materials scientists and engineers.
Fig. 3.
Example of sequencing by synthesis (SBS). Adapted with permission from PNAS, ref. 23a; Copyright 2006 National Academy of Sciences, USA
In recent years there has been an increase in interest in not only the genome, but also epigenetics, and in particular, the state of CpG methylation, as specific methylation sites in the genome have become linked with cancer.24 Chemists have reported a number of methods for detection of methylation of cytosine, including a bisulfite assay25 and osmium oxidation.26 Approaches to identifying methylation state at particular sites have also become commercialized recently. In addition, although it has been known for decades that 5′-hydroxymethylC exists as a common base in some bacteriophages, recent work has shown that it occurs to some extent even in mammalian tissues,27 and the oxidative mechanism of its synthesis is being uncovered.28
In addition to full sequencing methodologies, this was the decade of massive development of methods for detecting and identifying short nucleic acid sequences in solution. Hundreds of new types of probes were developed; indeed, it seems now that every chemical journal issue offers one or two more papers describing new DNA detection methods. The majority of these have made use of fluorescence, either in labels directly attached to probes or associated with the DNAs otherwise.29 It is clear that most of the hundreds of probes are unlikely to be developed for widespread use, and there are far too many to describe here. However, we will mention four approaches, either because they employed a dominating concept or offered real novelty in approach. These are molecular beacons (MBs), electronic DNA detection, nanoparticles, and templated chemistry. Although MBs were initially developed nearly two decades ago,30 their simplicity of design (based on a stem-loop structure) has remained appealing. This past decade saw many dozens of reports of variations of the beacon design, and uses in detecting not just nucleic acids but also proteins.31 Electronic DNA detection involves generating a change in an electron current signal upon hybridization; usually this involved attachment of a redox-active group (such as ferrocene) to a structured DNA.32 The approach is appealing not only because it can offer high sensitivity, but also because it marries well with the ongoing rapid development in solid-state electronics.
Nanoparticle-based DNA detection methods also came into their own in the last few years;33 the particles offered unusual fluorescence or optical reporting activities and high sensitivities in some applications (Fig. 4). This of course required substantial development of chemical methods for conjugating DNAs to such inorganic particles.
Fig. 4.
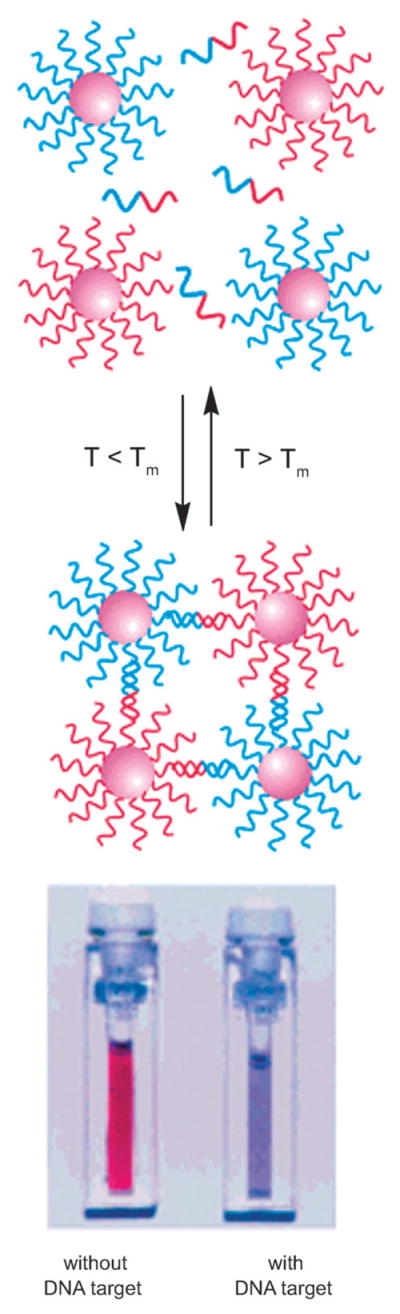
Detection of DNA with inorganic nanoparticles. Adapted with permission from ref. 33a. Copyright 2005 American Chemical Society.
Finally, nucleic acid templated chemistry underwent major developments this decade. The approach commonly involves the use of two modified DNA (or PNA) probes that carry reactants; hybridization of these adjacent to one another on a complementary DNA (the analyte) results in a reaction that can yield a fluorescent signal. Several labs (including ours) demonstrated the use of templated fluorogenic reactions for highly sensitive and selective identification of DNAs in solution (Fig. 5),29b,34 and recently, researchers have shown the approach to function in intact bacterial and human cells (where RNA is the template) as well.35 DNA-templated chemistry has also moved beyond uses in DNA detection, by offering a way to encode chemical libraries and allow them to undergo selection and amplification.36 Moreover, Liu demonstrated the use of DNA templated chemistry in the discovery of new aqueous bond-forming reactions.37
Fig. 5.
Examples of DNA detection with templated chemical reactions.
Synthetic biology
The field of Synthetic Biology has exploded as the need for modified cells and organisms in medicine, environmental sciences and energy fields has become widely recognized. As a whole, the field is defined by biologically functioning systems that have synthetic (designed) components.38
For biologists, who take a top-down approach starting with already-functioning cells, this field entails adding, subtracting and mixing existing proteins and pathways to alter phenotypic function. For chemists, who like to work from the bottom up, the field (‘‘chemical synthetic biology’’) means designing the basic chemical components from scratch—new amino acids, new nucleotides, altered carbohydrates, modified metabolites, substrates and cofactors. These new components can then be inserted into live pathways for either basic science study or for technological utility. Necessarily this field has focused to a major degree on nucleic acids.
One of the biggest breakthroughs in this area was the demonstration of assembly of a functioning genome by Venter.39 Venter extended the known methodologies of synthetic gene assembly (which prepares DNAs of ca. 1000 bp) on a massive scale, making a 1 megabase bacterial genome that was shown to function correctly. This required important developments in chemical DNA synthesis, gene synthesis, enzymatic repair methods, artificial chromosomes, and others, and required a large team of biologists to complete.
On the more chemical side, there has been a good deal of development of new functional base pairs and augmented genetic alphabets for DNA. Benner has continued the development of base pairs that utilize rearranged hydrogen bonding patterns.40 Several groups described base pairs consisting of two ligands, with a metal acting as a bridge between the two.41 Kool, Romesberg and Hirao, among others, demonstrated dozens of examples of replicable base pairs that lack hydrogen bonding and even many that lack purine and pyrimidine architectures.42 Some examples have shown impressive abilities to be functional in PCR reactions alongside natural DNA pairs.43 The work has culminated recently in examples of unnatural bases that function to encode information in cells, by being read correctly by the cellular replication machinery.44 One of the future uses of new genetic alphabets will be the biological encoding of many new designer amino acids into protein.
Some chemists have been inspired to go beyond the purine-pyrimidine architecture to avoid natural DNA base pair structure altogether. This has produced base pairing designs—new genetic sets—that yield full helices that are larger than the natural one. The first of these was xDNA, which is analogous to DNA but with bases enlarged by benzo-homologation.18,45 It is composed of eight letters rather than Nature’s four, and is not only more stable than natural DNA, but is inherently fluorescent. Other examples of new DNA-like genetic architectures have been described, including base pairs with four hydrogen bonds46 and bases with ethynyl homologation.47 Although such fully unnatural designs might generally be expected to interact with natural enzymes poorly, experiments show that this might not always be the case. For example, our laboratory demonstrated that xDNA can be replicated in vitro by some DNA polymerases.48 Even more surprising was the recent observation that xDNA bases can correctly encode genetic information in a living cell.49
Nanostructures and materials
A growing trend recently, followed by chemists and materials scientists, has been the building of functioning nano-structures from DNA. The field was seeded almost entirely from the early work of Seeman,50 and has branched and blossomed in many directions recently. The early parts of the decade saw rapid development of strategies for formation of tiled 2-D DNA structures,51 which yielded important insights into how to build complex structures from simple repeatable helical units and crossover junctions. Seeman also showed how to build 3D closed structures (such as polyhedrons) from DNA by developing strategies for corners and edges.52 More chemists have recently entered the design arena and have made chimeric structures containing DNA and small molecules,53 DNA and metal complexes54 and DNA and inorganic nanoparticles.55 Over time, designs for forming an impressive array of structures, such as tubes56 and buckyballs,57 from DNA have been demonstrated.
One of the most influential recent developments has been the ‘‘DNA origami’’ designs of Rothemund, who used single-stranded phage DNA combined with hundreds of different oligonucleotides to make an impressive array of closed structures (Fig. 6).58 One highly appealing aspect of this approach is that half of the DNA is easy and cheap to obtain (circular phage DNA), and the closed nature of that DNA makes the ultimate structures more stable. Many new examples and applications following this approach have emerged and no doubt will continue to appear.59 Synthetic DNA can be used not only to build static structures, but also mobile ones. This has led to the development of a number of robots that move or perform work in response to a stimulus or molecular fuel.60
Fig. 6.
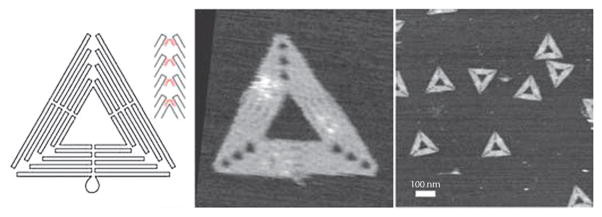
DNA origami shapes: folding path and AFM images. Adapted with permission from Macmillan Publishers Ltd: Nature, ref. 58, Copyright 2006.
Finally in the field of functional nanomaterials and devices has come the recent development of DNA as a scaffold for structures with useful optical properties. Several laboratories have demonstrated methods for incorporating several fluorophores into DNA oligonucleotides, and have explored the unusual fluorescence emission properties that result from photophysical interactions between them.61 Researchers have reported ‘‘superquenching’’ emissive molecules,62 white light-emitting molecules,63 photonic wires,64 polychromatic enzyme sensors,65 multicolor biological dyes (Fig. 7),66 and sensors of organic vapors built from these modified DNAs.67
Fig. 7.
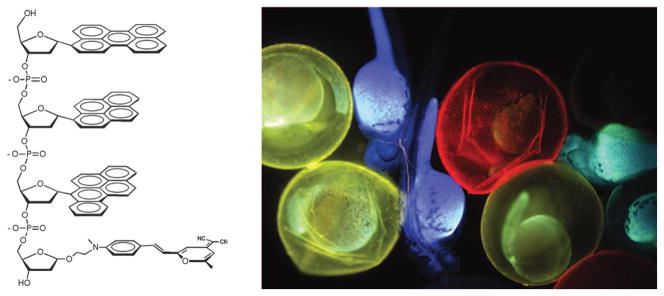
Example of DNA oligomer bearing multiple fluorophores and application in imaging biological systems. Adapted with permission from ref. 66. Copyright 2009 American Chemical Society.
The next decade
Extrapolating recent trends allows us to make some predictions about the coming decade of chemical research in the nucleic acids. One of the biggest trends among chemists has been the increasing interest in biology and medicine. In the next ten years we will see a move toward biotechnological applications in vivo—that is, in cells and whole organisms. Intracellular applications allow researchers and clinicians to avoid removing the contents from the cell prior to analysis, saving time and cost. As a result, chemists are likely to spend more time thinking about issues such as biostability and intracellular delivery. Getting synthetic nucleic acids to pass through the cellular membrane is a long-standing problem; we suspect that the next ten years will see strong advances in that direction.
A second, and related, major trend for the next ten years will be the rapid development of synthetic biology. For chemists, this means increasing development of replacements for cellular components, and inserting them into functioning biological pathways. In the nucleic acids, we will see more studies of modified backbones and bases in vivo, and we will see tests of whether altered cells can take up nucleic acid components from the medium. Pathways involving synthetically altered ribozymes, riboswitches, and other biologically active RNAs will be explored. Many applications might arise, including the ability to encode many more amino acids than nature currently allows. The tools of chemistry will work intimately with those of molecular biology to make this happen.
Acknowledgments
ETK thanks the U.S. National Institutes of Health for support of his laboratory’s research in modified nucleic acids over the past decade through grants GM063587, GM072705, GM067201, and GM068122. OK acknowledges the California HIV/AIDS Research Program at the University of California for support of his postdoctoral training through fellowship F08-ST-220.
Biographies

Omid Khakshoor is a postdoctoral fellow at Stanford. He received his BSc and MSc from Sharif University of Technology, Tehran, Iran, and his PhD from University of California, Irvine (UCI). He has been awarded the 2006 ACS Division of Medicinal Chemistry Predoctoral Fellowship and the 2009–2011 California HIV/AIDS Research Program (CHRP) Postdoctoral Fellowship. His research interest includes organic chemistry, chemical biology, and biochemistry.

Eric Kool is the George and Hilda Daubert Professor of Chemistry at Stanford University. His research interests include the organic chemistry, biochemistry, biophysics and synthetic biology of modified nucleic acids.
Notes and references
- 1.Guarnieri DJ, Dileone RJ. Ann Med. 2008;40:197. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 2.(a) Mattick JS, Makunin IV. Hum Mol Genet. 2006;15:R17. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]; (b) Carthew RW, Sontheimer EJ. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wilusz JE, Sunwoo H, Spector DL. Genes Dev. 2009;23:1494. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Amarzguioui M, Rossi JJ, Kim D. FEBS Lett. 2005;579:5974. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]; (b) Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 4.(a) Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Röhl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Nature. 2004;432:173. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]; (b) Dykxhoorn DM, Palliser D, Lieberman J. Gene Ther. 2006;13:541. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 5.(a) Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Angew Chem, Int Ed. 2007;46:1212. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]; (b) Link KH, Breaker RR. Gene Ther. 2009;16:1189. doi: 10.1038/gt.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Blouin S, Mulhbacher J, Penedo JC, Lafontaine DA. ChemBioChem. 2009;10:400. doi: 10.1002/cbic.200800593. [DOI] [PubMed] [Google Scholar]
- 6.(a) Seela F, Sirivolu VR. Chem Biodiversity. 2006;3:509. doi: 10.1002/cbdv.200690054. [DOI] [PubMed] [Google Scholar]; (b) Weisbrod SH, Marx A. Chem Commun. 2008:5675. doi: 10.1039/b809528k. [DOI] [PubMed] [Google Scholar]; (c) Raindlová V, Pohl R, Sanda M, Hocek M. Angew Chem, Int Ed. 2010;49:1064. doi: 10.1002/anie.200905556. [DOI] [PubMed] [Google Scholar]
- 7.Lutz JF. Angew Chem, Int Ed. 2007;46:1018. doi: 10.1002/anie.200604050. [DOI] [PubMed] [Google Scholar]
- 8.(a) Amblard F, Cho JH, Schinazi RF. Chem Rev. 2009;109:4207. doi: 10.1021/cr9001462. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) El-Sagheer AH, Brown T. Chem Soc Rev. 2010;39:1388. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 9.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc Natl Acad Sci U S A. 2007;104:16793. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg MM. Org Biomol Chem. 2007;5:18. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 11.(a) Haraguchi K, Greenberg MM. J Am Chem Soc. 2001;123:8636. doi: 10.1021/ja0160952. [DOI] [PubMed] [Google Scholar]; (b) Iwai S. Nucleosides, Nucleotides Nucleic Acids. 2006;25:561. doi: 10.1080/15257770600685826. [DOI] [PubMed] [Google Scholar]; (c) Küpfer PA, Leumann CJ. Nucleic Acids Res. 2007;35:58. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summerer D, Marx A. Angew Chem, Int Ed. 2001;40:3693. doi: 10.1002/1521-3773(20011001)40:19<3693::aid-anie3693>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 13.Scaringe SA, Wincott FE, Caruthers MH. J Am Chem Soc. 1998;120:11820. [Google Scholar]
- 14.Jepsen JS, Sørensen MD, Wengel J. Oligonucleotides. 2004;14:130. doi: 10.1089/1545457041526317. [DOI] [PubMed] [Google Scholar]
- 15.(a) Luisier S, Leumann CJ. ChemBioChem. 2008;9:2244–2253. doi: 10.1002/cbic.200800322. [DOI] [PubMed] [Google Scholar]; (b) Lebreton J, Escudier JM, Arzel L, Len C. Chem Rev. 2010;110:3371. doi: 10.1021/cr800465j. [DOI] [PubMed] [Google Scholar]; (c) Obika S, Rahman SMA, Fujisaka A, Kawada Y, Baba T, Imanishi T. Heterocycles. 2010;81:1347. [Google Scholar]
- 16.Schöning KU, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A. Science. 2000;290:1347. doi: 10.1126/science.290.5495.1347. [DOI] [PubMed] [Google Scholar]
- 17.Meggers E, Zhang L. Acc Chem Res. 2010;43:1092. doi: 10.1021/ar900292q. [DOI] [PubMed] [Google Scholar]
- 18.Krueger AT, Lu HG, Lee AHF, Kool ET. Acc Chem Res. 2007;40:141. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur H, Babu BR, Maiti S. Chem Rev. 2007;107:4672. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- 20.Chaput JC, Ichida JK, Szostak JW. J Am Chem Soc. 2003;125:856. doi: 10.1021/ja028589k. [DOI] [PubMed] [Google Scholar]
- 21.Schlegel MK, Meggers E. J Org Chem. 2009;74:4615. doi: 10.1021/jo900365a. [DOI] [PubMed] [Google Scholar]
- 22.Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO, Hunkapiller M. Science. 1998;280:1540. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- 23.(a) Ju J, Kim DH, Bi L, Meng Q, Bai X, Li Z, Li X, Marma MS, Shi S, Wu J, Edwards JR, Romu A, Turro NJ. Proc Natl Acad Sci U S A. 2006;103:19635. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Morozova O, Marra MA. Genomics. 2008;92:255. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]; (c) Mardis ER. Trends Genet. 2008;24:133. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Kanai Y. Cancer Sci. 2010;101:36. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Proc Natl Acad Sci U S A. 1996;93:9821. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto A. Org Biomol Chem. 2009;7:21. doi: 10.1039/b813595a. [DOI] [PubMed] [Google Scholar]
- 27.(a) Kriaucionis S, Heintz N. Science. 2009;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T. Angew Chem, Int Ed. 2010;49:5375. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 28.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(a) Ranasinghe RT, Brown T. Chem Commun. 2005:5487. doi: 10.1039/b509522k. [DOI] [PubMed] [Google Scholar]; (b) Silverman AP, Kool ET. Chem Rev. 2006;106:3775. doi: 10.1021/cr050057+. [DOI] [PubMed] [Google Scholar]; (c) Martí AA, Jockusch S, Stevens N, Ju JY, Turro NJ. Acc Chem Res. 2007;40:402. doi: 10.1021/ar600013q. [DOI] [PubMed] [Google Scholar]
- 30.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 31.Wang KM, Tang ZW, Yang CYJ, Kim YM, Fang XH, Li W, Wu YR, Medley CD, Cao ZH, Li J, Colon P, Lin H, Tan WH. Angew Chem, Int Ed. 2009;48:856. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]; (b) Inouye M, Ikeda R, Takase M, Tsuri T, Chiba J. Proc Natl Acad Sci U S A. 2005;102:11606. doi: 10.1073/pnas.0502078102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lubin AA, Plaxco KW. Acc Chem Res. 2010;43:496. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(a) Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; (b) Xu K, Huang JR, Ye ZZ, Ying YB, Li YB. Sensors. 2009;9:5534. doi: 10.3390/s90705534. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Agasti SS, Rana S, Park MH, Kim CK, You CC, Rotello VM. Adv Drug Delivery Rev. 2010;62:316. doi: 10.1016/j.addr.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.(a) Grossmann TN, Strohbach A, Seitz O. ChemBioChem. 2008;9:2185. doi: 10.1002/cbic.200800290. [DOI] [PubMed] [Google Scholar]; (b) Kolpashchikov DM. Chem Rev. 2010;110:4709. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]
- 35.Bao G, Rhee WJ, Tsourkas A. Annu Rev Biomed Eng. 2009;11:25. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.(a) Li XY, Liu DR. Angew Chem Int Ed. 2004;43:4848. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]; (b) Wrenn SJ, Harbury PB. Annu Rev Biochem. 2007;76:331. doi: 10.1146/annurev.biochem.76.062205.122741. [DOI] [PubMed] [Google Scholar]; (c) Berger J, Oberhuber M. Chem Biodiversity. 2010;7:2581. doi: 10.1002/cbdv.201000108. [DOI] [PubMed] [Google Scholar]; (d) Scheuermann J, Neri D. ChemBioChem. 2010;11:931. doi: 10.1002/cbic.201000066. [DOI] [PubMed] [Google Scholar]
- 37.Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Nature. 2004;431:545. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.(a) Benner SA, Sismour AM. Nat Rev Genet. 2005;6:533. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Keasling JD. ACS Chem Biol. 2008;3:64. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]; (c) Purnick PEM, Weiss R. Nat Rev Mol Cell Biol. 2009;10:410. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 39.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, Smith HO, Venter JC. Science. 2010;329:52. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 40.(a) Benner SA. Acc Chem Res. 2004;37:784. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]; (b) Yang ZY, Hutter D, Sheng PP, Sismour AM, Benner SA. Nucleic Acids Res. 2006;34:6095. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.(a) Zimmermann N, Meggers E, Schultz PG. J Am Chem Soc. 2002;124:13684. doi: 10.1021/ja0279951. [DOI] [PubMed] [Google Scholar]; (b) Zimmermann N, Meggers E, Schultz PG. Bioorg Chem. 2004;32:13. doi: 10.1016/j.bioorg.2003.09.001. [DOI] [PubMed] [Google Scholar]; (c) Clever GH, Kaul C, Carell T. Angew Chem, Int Ed. 2007;46:6226. doi: 10.1002/anie.200701185. [DOI] [PubMed] [Google Scholar]; (d) Clever GH, Shionoya M. Coord Chem Rev. 2010;254:2391. [Google Scholar]
- 42.(a) Kool ET. Acc Chem Res. 2002;35:936. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]; (b) Leconte AM, Matsuda S, Romesberg FE. J Am Chem Soc. 2006;128:6780. doi: 10.1021/ja060853c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. Nat Methods. 2006;3:729. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 43.(a) Hirao I, Mitsui T, Kimoto M, Yokoyama S. J Am Chem Soc. 2007;129:15549. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]; (b) Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2009;131:14620. doi: 10.1021/ja906186f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim TW, Delaney JC, Essigmann JM, Kool ET. Proc Natl Acad Sci U S A. 2005;102:15803. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.(a) Liu HB, Gao JM, Lynch SR, Saito YD, Maynard L, Kool ET. Science. 2003;302:868. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]; (b) Gao JM, Liu HB, Kool ET. Angew Chem, Int Ed. 2005;44:3118. doi: 10.1002/anie.200500069. [DOI] [PubMed] [Google Scholar]; (c) Krueger AT, Kool ET. J Am Chem Soc. 2008;130:3989. doi: 10.1021/ja0782347. [DOI] [PubMed] [Google Scholar]
- 46.Minakawa N, Kojima N, Hikishima S, Sasaki T, Kiyosue A, Atsumi N, Ueno Y, Matsuda A. J Am Chem Soc. 2003;125:9970. doi: 10.1021/ja0347686. [DOI] [PubMed] [Google Scholar]
- 47.Doi Y, Chiba J, Morikawa T, Inouye M. J Am Chem Soc. 2008;130:8762. doi: 10.1021/ja801058h. [DOI] [PubMed] [Google Scholar]
- 48.Lu HG, Krueger AT, Gao JM, Liu HB, Kool ET. Org Biomol Chem. 2010;8:2704. doi: 10.1039/c002766a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaney JC, Gao JM, Liu HB, Shrivastav N, Essigmann JM, Kool ET. Angew Chem, Int Ed. 2009;48:4524. doi: 10.1002/anie.200805683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeman NC. J Theor Biol. 1982;99:237. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 51.(a) Winfree E, Liu FR, Wenzler LA, Seeman NC. Nature. 1998;394:539. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]; (b) Lin CX, Liu Y, Rinker S, Yan H. ChemPhysChem. 2006;7:1641. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 52.Zhang YW, Seeman NC. J Am Chem Soc. 1994;116:1661. [Google Scholar]
- 53.Gothelf KV, LaBean TH. Org Biomol Chem. 2005;3:4023. doi: 10.1039/b510551j. [DOI] [PubMed] [Google Scholar]
- 54.(a) Tanaka K, Shionoya M. Coord Chem Rev. 2007;251:2732. [Google Scholar]; (b) Yang H, Metera KL, Sleiman HF. Coord Chem Rev. 2010;254:2403. [Google Scholar]
- 55.(a) Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. Nature. 2008;451:553. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]; (b) Nykypanchuk D, Maye MM, van der Lelie D, Gang O. Nature. 2008;451:549. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]; (c) Mastroianni AJ, Claridge SA, Alivisatos AP. J Am Chem Soc. 2009;131:8455. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.(a) Liu D, Park SH, Reif JH, LaBean TH. Proc Natl Acad Sci U S A. 2004;101:717. doi: 10.1073/pnas.0305860101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rothemund PWK, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E. J Am Chem Soc. 2004;126:16344. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]; (c) Mitchell JC, Harris JR, Malo J, Bath J, Turberfield AJ. J Am Chem Soc. 2004;126:16342. doi: 10.1021/ja043890h. [DOI] [PubMed] [Google Scholar]
- 57.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao CD. Nature. 2008;452:198. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 58.Rothemund PWK. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 59.Nangreave J, Han DR, Liu Y, Yan H. Curr Opin Chem Biol. 2010;14:608. doi: 10.1016/j.cbpa.2010.06.182. [DOI] [PubMed] [Google Scholar]
- 60.Liu H, Liu DS. Chem Commun. 2009:2625. doi: 10.1039/b822719e. [DOI] [PubMed] [Google Scholar]
- 61.(a) Gao JM, Strässler C, Tahmassebi D, Kool ET. J Am Chem Soc. 2002;124:11590. doi: 10.1021/ja027197a. [DOI] [PubMed] [Google Scholar]; (b) Hrdlicka PJ, Babu BR, Sørensen MD, Harrit N, Wengel J. J Am Chem Soc. 2005;127:13293. doi: 10.1021/ja052887a. [DOI] [PubMed] [Google Scholar]; (c) Wilson JN, Kool ET. Org Biomol Chem. 2006;4:4265. doi: 10.1039/b612284c. [DOI] [PubMed] [Google Scholar]; (d) Mayer-Enthart E, Wagenknecht HA. Angew Chem, Int Ed. 2006;45:3372. doi: 10.1002/anie.200504210. [DOI] [PubMed] [Google Scholar]; (e) Chiba J, Takeshima S, Mishima K, Maeda H, Nanai Y, Mizuno K, Inouye M. Chem–Eur J. 2007;13:8124. doi: 10.1002/chem.200700559. [DOI] [PubMed] [Google Scholar]; (f) Samain F, Malinovskii VL, Langenegger SM, Häner R. Bioorg Med Chem. 2008;16:27. doi: 10.1016/j.bmc.2007.04.052. [DOI] [PubMed] [Google Scholar]; (g) Varghese R, Wagenknecht HA. Chem Commun. 2009:2615. doi: 10.1039/b821728a. [DOI] [PubMed] [Google Scholar]; (h) Grigorenko NA, Leumann CJ. Chem–Eur J. 2009;15:639. doi: 10.1002/chem.200801135. [DOI] [PubMed] [Google Scholar]
- 62.Wilson JN, Teo YN, Kool ET. J Am Chem Soc. 2007;129:15426. doi: 10.1021/ja075968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varghese R, Wagenknecht HA. Chem–Eur J. 2009;15:9307. doi: 10.1002/chem.200901147. [DOI] [PubMed] [Google Scholar]
- 64.(a) Heilemann M, Tinnefeld P, Mosteiro GS, Parajo MG, Van Hulst NF, Sauer M. J Am Chem Soc. 2004;126:6514. doi: 10.1021/ja049351u. [DOI] [PubMed] [Google Scholar]; (b) Heilemann M, Kasper R, Tinnefeld P, Sauer M. J Am Chem Soc. 2006;128:16864. doi: 10.1021/ja065585x. [DOI] [PubMed] [Google Scholar]; (c) Hannestad JK, Sandin P, Albinsson B. J Am Chem Soc. 2008;130:15889. doi: 10.1021/ja803407t. [DOI] [PubMed] [Google Scholar]
- 65.Dai N, Teo YN, Kool ET. Chem Commun. 2010;46:1221. doi: 10.1039/b926338a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teo YN, Wilson JN, Kool ET. J Am Chem Soc. 2009;131:3923. doi: 10.1021/ja805502k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samain F, Ghosh S, Teo YN, Kool ET. Angew Chem, Int Ed. 2010;49:7025. doi: 10.1002/anie.201002701. [DOI] [PMC free article] [PubMed] [Google Scholar]