Abstract
Targeting synthetic lethal interactions is a promising new therapeutic approach to exploit specific changes that occur within cancer cells. Multiple approaches to investigate these interactions have been developed and successfully implemented, including chemical, siRNA, shRNA, and CRISPR library screens. Genome-wide computational approaches, such as DAISY, also have been successful in predicting synthetic lethal interactions from both cancer cell lines and patient samples. Each approach has its advantages and disadvantages that need to be considered depending on the cancer type and its molecular alterations. This review discusses these approaches and examines case studies that highlight their use.
Keywords: synthetic lethality, synthetic lethal interactions, drug discovery, drug screening assays, antitumor, cancer, neoplasms, library screens, siRNA screen, siRNA library, shRNA screen, shRNA library, CRISPR screen, CRISPR library, computational screen, DAISY
Introduction
A major challenge in developing new anti-cancer therapies is to identify compounds that have sufficient therapeutic windows comprised of the concentrations causing tumor regression with minimal normal tissue toxicity. Most cancer therapies suffer from narrow therapeutic windows causing numerous side effects that greatly reduce a patient’s quality of life. Further elucidation of the unique differences between cancer and normal tissue allows for the development of targeted therapies with reduced side effects. During malignant progression, cancer cells acquire multiple mutations, including the activation of proto-oncogenes, the inactivation of tumor suppressors, and other additional genetic or epigenetic alterations resulting in a drastically transformed genome [1,2]. Indeed, most of the hallmarks of cancer are associated with changes to the cancer cell’s genetic makeup in order to enable it to endlessly proliferate [3]. These changes can be exploited to specifically target and kill cancer cells while sparing normal cells [2,4]. Traditional chemotherapy, including irradiation or DNA damaging agents, relies on this principle. Most cancer cells have defects in their cell cycle checkpoints where DNA would normally be repaired before replication [5,6]. Thus, radiotherapy and DNA damaging agents target DNA in the continuously dividing cancer cells, causing cell death. Normal cells, with their intact cell cycle checkpoints, repair the damage before dividing [5,6]. However, these therapies often have serious side effects and greatly reduce a patient’s quality of life [7]. Synthetic lethality represents a more targeted approach that exploits the specific changes within the cancer at a single gene level that separate it from healthy tissue.
Synthetic Lethality
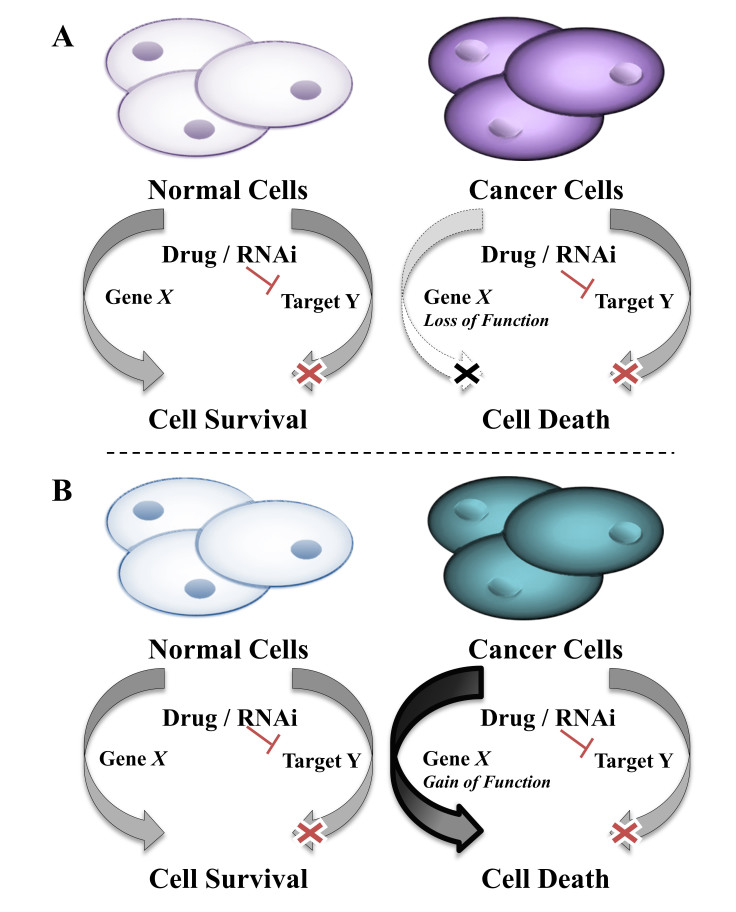
Synthetic lethality is defined as the interaction between two co-essential genes such that inhibiting the function of either gene separately results in cell survival, but inhibiting the function of both genes results in cell death (Figure 1) [8-10]. Inhibition of a gene may be achieved by chemical or genetic means. For instance, the genetic inhibition of a gene’s function may occur through RNAi, mutation, deletion, epigenetic changes, or perturbations of upstream regulators. Chemical inhibition of the gene’s function may be achieved by treatment with a chemical compound. The inhibition of co-essential genes can occur at any level, and the loss of function does not have to occur in the same manner for both co-essential genes. For example, one gene can be lost due to deletion and the other inhibited by a chemical compound. While synthetic lethality is best known in the context of loss-of-function mutants [9], other perturbations including gene overexpression [11,12], epigenetic changes [13], and cell extrinsic differences [14,15] can cause these interactions to occur [10] (Figure 1). In identifying genes that are synthetically lethal within each cancer type, we gain an understanding of the molecular biology of those cancers and how it can be specifically exploited. There are multiple approaches that have been successfully used to identify synthetic lethal interactions in cancer.
Figure 1.
Synthetic lethal interactions spare normal cells while selectively killing cancer cells. A) In the loss of function, phenotype cancer cells have lost the function of gene X due to genetic loss, epigenetic changes, cell extrinsic changes, and more. When cells that express gene X are treated to inhibit gene X’s synthetic lethal partner gene Y, they remain viable, but cancer cells lacking gene X die. B) In the gain of function phenotype, also called synthetic dosage lethal (SDL), cancer cells have an overexpression or overactivation of gene X due to oncogenic mutation, generation of fusion proteins, changes in upstream regulators, epigenetic changes, cell extrinsic changes, and more. When cells with wild type (WT) gene X are treated to inhibit gene X’s synthetic lethal partner gene Y they survive, but cancer cells with the gene X gain of function die.
Different Approaches to Identify Synthetic Lethal Interactions
Hypothesis-Driven Approach
It is possible to identify synthetic lethal interactions using a candidate or hypothesis-driven approach if the alterations to the molecular pathways in the cancer are well established. For instance, if a tumor suppressor is frequently lost and the resulting changes to the gene expression profile are known, then it may be possible to use RNA interference (RNAi) or chemical compounds to inhibit the genes that are expected to be upregulated to compensate for the loss. This may result in a synthetic lethal interaction. Mutations in the breast cancer early onset (BRCA) 1 and BRCA2 genes are known to occur frequently in breast and ovarian cancers. Knowing that BRCA1/2 are involved in homologous recombination (HR) and DNA double-strand break repair, both Byant et al. [16] and Farmer et al. [17] hypothesized that inhibition of Poly (ADP-Ribose) Polymerase (PARP), responsible for DNA single-strand break repair, would be synthetically lethal with loss of BRCA1/2. This hypothesis is based on the knowledge that PARP deficiency results in spontaneous single-strand breaks at the DNA replication fork, which require HR for repair. Cells deficient in BRCA1/2 are unable to provide HR for repair of DNA single- and double-strand breaks caused by chemical or genetic PARP inhibition, causing synthetic lethality. The synthetic lethal interaction was extended to in vivo studies, followed by clinical studies where treatment of BRCA1/2-deficient tumors with a DNA damaging agent combined with a PARP inhibitor extended patient survival over chemotherapy alone [18,19].
Synthetic lethal interactions also may explain why particular chemical compounds have increased efficacy in cancers characterized with specific genetic alterations. While clinical trials for PARP inhibitors in ovarian cancer were successful in increasing progression free survival by 62 percent overall [20], in a follow-up analysis of the same study, it found that the BRCA1/2-deficient group had a much higher rate of 82 percent [21]. Another study by Leibowitz et al. [22] hypothesized that non-steroidal anti-inflammatory drugs (NSAIDs) are highly effective in preventing colorectal tumorigenesis due to a synthetic lethal interaction with the loss of the adenomatous polyposis coli (APC) gene. While NSAIDs were able to activate cell death pathways in both cancer and normal cells, APC deficiency triggered the activation of BH3 interacting-domain (BID), a death agonist in extrinsic apoptotic pathway, resulting in synthetic lethality [22]. In situations in which putative targets cannot be identified, a screening-based approach is necessary.
Screening-Based Approaches
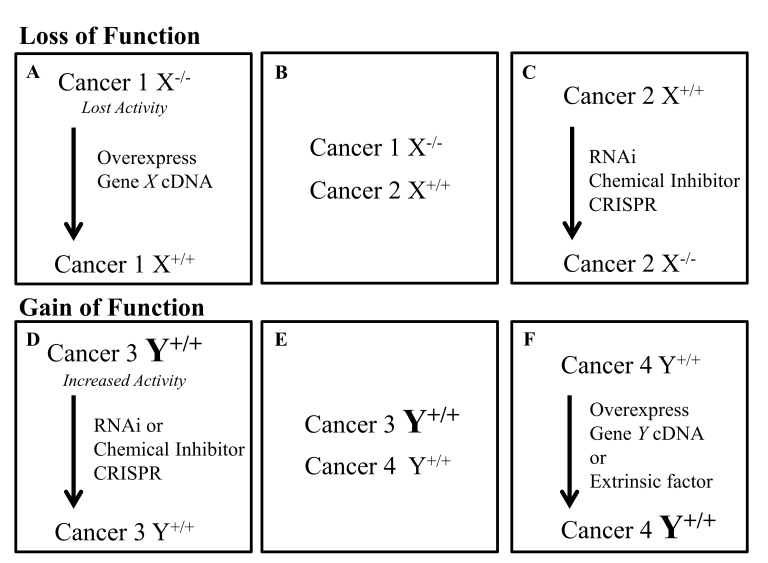
Most large scale approaches investigating synthetic lethal interactions in cancer rely on the comparison of drug or RNAi treatment in “matched” cell lines (Figure 2). Matched cell lines are generated so that their only difference is in the expression/activation status of the gene of interest. In studying a loss-of-function phenotype (Figure 1A), the parental cancer cell line may have lost the expression of a gene, have inactivating mutations, or have been treated with an extrinsic factor (like a chemical compound) such that the activity of the gene is lost. In this model, a matched cell line could be generated from a parent cancer cell line deficient in the gene by overexpressing it (Figure 2A). Next, multiple cell lines with and without expression of the gene could be compared (Figure 2B). Finally, one could inactivate the gene in a cell line expressing it (Figure 2C). In studying a gain-of-function phenotype (Figure 1B), the parental cancer cell line may have acquired a new gene fusion, oncogenic mutation resulting in constitutive activation, overexpression of the gene, or have been treated with an extrinsic factor (like a receptor ligand) such that the gene’s activity would increase. In this model, a matched cell line could be generated from a parent cancer cell line with an overactive gene by inactivating it (Figure 2D). Next, multiple cancer cell lines with and without the increase in the gene’s activity might be investigated (Figure 2E). Additionally, the gene could be overexpressed in the parent cell line, expressing it at a normal level (Figure 2F). Finally, a cell extrinsic factor like a receptor ligand could be used to treat the cells with low receptor activity such that activity of the receptor would increase (Figure 2F). These matched cell lines often differ in the activity of a single gene; one of them represents a cancer cell line characterized by a specific gene alteration, and the other represents a cancer cell line characterized by the absence of that alteration. In this respect, the latter simulates the normal tissue where genetic alterations are absent. Once the matched cell lines have been generated, they can be used in high-throughput screens. These screens can be separated into two categories: chemical libraries and genome-wide interference. Chemical libraries include both annotated and non-annotated libraries where the targets of the chemical compounds are known and unknown respectively. Genome-wide interference screens have been conducted successfully using siRNA, shRNA, and CRISPR.
Figure 2.
Approaches to generating matched cell lines for synthetic lethality screens. There are multiple approaches to generate matched cell lines if a loss of function (A-C) or a gain of function (D-F) of a specific gene is studied. For studying synthetic lethal interactions in cancer cells that have lost expression of a gene: A) the cDNA for the gene can be re-expressed in the deficient cell line; B) multiple cancer lines both expressing and deficient in the gene can be investigated; or C) cancer lines that express the gene can be treated to inactivate the gene. For studying synthetic lethal interactions in cancer cells with oncogenic mutations that increase activity of the gene or create a new gene fusion (like BCR-ABL), multiple approaches can be used: D) cells can be treated with RNAi or a chemical inhibitor in order to reduce the expression back to or below normal levels; E) multiple cancer lines both with and without the mutation could be investigated; or F) the activating mutation can be introduced or the gene can be overexpressed within a cancer line that has normal expression of WT gene.
Many chemical library screens rely on the use of chemical compounds with unknown molecular targets. These screens usually contain tens of thousands of different compounds, which greatly increase the chances of getting a “hit.” A hit is a compound that is synthetically lethal within cancer cells containing the gene alteration being studied. The difficulty of these screens is in identifying the molecular target of the hit. Genome-wide interference screens rely on the use of siRNA, shRNA, or CRISPR libraries in order to inhibit genes involved in every molecular pathway within cells. Smaller libraries can be useful if specific pathways are established. Genome-wide interference screens suffer from the need to identify chemical compounds that inhibit the molecular targets identified before the interaction can be utilized in clinical trials. If the compounds are not available, their development can be long and arduous. The use of an annotated chemical library can offer both a small molecule compound and the molecular target assuming the hit is acting through the predicted target. This review will investigate multiple case studies for each approach, the screens involved, the methods of validation the studies used, and the synthetic lethal interactions identified. Our goal is to present the merits of each approach and the context in which they have been successfully applied.
Chemical Library Screens
The use of large non-annotated chemical libraries (Figure 3A), wherein the molecular targets of the drugs are unknown, have been an effective method for discovering synthetically lethal interactions in a number of different cancers including renal cell carcinomas (RCC) [23,24] and ovarian cancer [25]. Most chemical library screens rely on fluorescently labeled matched cell lines and measure fluorescence intensity as a surrogate for cell count as a high-throughput viability assay. Following the identification of hits, an investigation of the actual drug targets and mechanism of action is required. Often an additional screen is necessary to accomplish this. If the drug class has been previously investigated and its targets have been established, then confirmation that the drug is acting “on-target” can be performed using RNAi to knock down the putative target. Annotated chemical libraries, in which the targets of the drugs are known, can be useful tools for identifying synthetic lethal interactions. Since annotated libraries are often small (hundreds of chemical compounds) in comparison to non-annotated libraries (tens of thousands of compounds), the benefit from knowing the molecular target of a compound may be offset by the reduced chance in identifying hits from a screen. Likely for this reason, the use of non-annotated chemical library screens appears to be more frequently described in the literature. Below, we discuss two papers in which screens using non-annotated chemical libraries were employed.
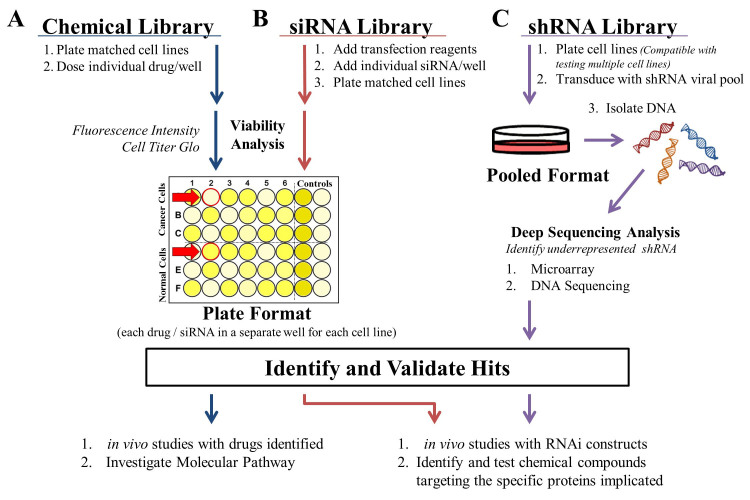
Figure 3.
Approaches to investigating synthetic lethal interactions. A) Chemical library screens are conducted using matched cell lines in a plate format wherein each drug is dosed separately to both lines. Fluorescence intensity from GFP or EYFP labeled cell lines can be used as a surrogate for cell count to make the assay high-throughput. B) siRNA library screens are also conducted in a plate format. Libraries are often custom ordered with individual siRNAs or siRNA pools and transfection reagent contained in each well. The matched cell lines can then be plated and following incubation analyzed in parallel using a high-throughput method like Cell Titer-Glo. Hits from screens performed using the plate format (A and B) are identified by comparing the normalized values of each well between the two matched cell lines. A hit, indicated by the red arrows, kills cancer cells while sparing the matched cell line (simulated normal tissue). C) The shRNA library approach is most often performed by transducing matched cell lines with a shRNA viral pool, although plated approaches can be conducted similar to the siRNA library approach. The pooled approach is more conducive for studying multiple cell lines and relies on deep sequencing analysis to identify hits. A hit is identified by identifying shRNAs that are lost after library transduction in cancer cell lines and present in matched (normal) cell lines, since synthetic lethal interactions will result in the cells death. Once hits have been identified and validated in each screen, the approaches A, B, C split. With a chemical library screen (A) in vivo studies can be readily initiated while the investigation into the molecular pathway is conducted. For siRNA and shRNA screens (B, C), in vivo studies can also be initiated using RNAi constructs while small molecules are developed.
The first paper exploited the fact that inactivation of the tumor suppressor Von Hippel Lindau (VHL) has been shown to occur in more than 70 percent of RCC [26]. The inactivation of VHL results in abrogation of its E3 ubiquitin ligase activity and overexpression of its targets. Turcotte et al. [23] utilized a library of 64,000 chemical compounds to identify synthetic lethal interactions in VHL-deficient RCC (Figure 1A). The screen used a VHL-deficient RCC4 parent cell line and matched RCC4-VHL cell line obtained by overexpression of VHL cDNA (Figure 2A). Both matched lines stably expressed Enhanced Yellow Fluorescent Protein (EYFP), and fluorescence was used to monitor cell number. The matched lines were treated in parallel with the chemical library in a plate format (Figure 3A). Clonogenic assays, in which the cells were plated at low densities and allowed to form colonies, were used to confirm their hits. The authors observed that VHL-deficient RCC cells treated with the hit STF-62247 accumulated intracytoplasmic vesicles more readily than RCC-VHL cells, causing RCC cell death. The synthetic lethal effect of STF-62247 also was confirmed in vivo using multiple matched RCC lines. Since the molecular target of STF-62247 was unknown, the authors utilized a yeast deletion pool to identify the target. This additional screen confirmed that STF-62247 was disrupting the trans-golgi network stimulating the maturation of autophagosomes to autolysosomes selectively in the VHL-deficient RCC.
The second paper was aimed at the identification of chemical compounds that would re-sensitize cancer cells to DNA damaging agents such as ionizing radiation (IR) or Cisplatin. These agents cause DNA intrastrand crosslinks and are effective in treating a wide variety of cancers with perturbed DNA repair pathways, including ovarian cancer [27]. Eventually, the cancer cells evolve to acquire drug resistance through various mechanisms, including activation of DNA repair pathways like the Fanconi Anemia (FA) pathway [28]. For example, DNA damaging agents are initially efficacious in ovarian cancer, but most patients relapse with resistant disease that is no longer sensitive to the treatment [29]. Jacquemont et al. [25] utilized a library of 16,000 chemical compounds to identify hits that would inhibit the overactive FA pathway, thus re-sensitizing cancer cells to DNA damaging agents (Figure 1B). The parent PD20 fibroblast cell line is Fanconi Anemia Group D2 (FANCD2)-deficient, causing FA pathway disfunction. The PD20-EGFP-FANCD2 cell line utilized for the screen was obtained by overexpression of Enhanced Green Fluorescent Protein (EGFP) labeled version of the wild type (WT) FANCD2 cDNA. This overexpression made the PD20-EGFP-FANCD2 cell line resistant to DNA damaging agents. The chemical compound screen was performed using 96-well plates seeded with PD20-EGFP-FANCD2 cells treated and untreated with IR (Figure 2F, extrinsic factor). Cells were dosed with the ICCB bioactives and Commercial Diversity Set 1, Chembridge DiverSet, and NINDS II chemical compound libraries. The plates were imaged using EGFP microscopy to identify hits that selectively reduced EGFP-FANCD2 foci formation by greater than 50 percent in IR treated cells, which indicates that the hit reduces DNA repair events, causing restoration of sensitivity to the IR treatment. From the 43 hits obtained, 26 hits also successfully reduced EGFP-FANCD2 foci formation in cells treated with Cisplatin. Fifteen hits were then validated in multiple ovarian cancer cell lines. Since the FA pathway is known to be involved in DNA repair and homologous recombination (HR), in a follow-up study, the HR efficiency was tested using a GFP-based reporter system, where GFP expression was correlated with HR events. The majority of the drugs acted by inhibiting the HR process (like the compound Bortezomib) or by directly inhibiting FANCD2 foci formation.
Taken together, non-annotated chemical library screens, as described above, can be an effective means to identify synthetic lethal interactions within individual matched cell lines. This method is not suited to studying a large number of cell lines in parallel since it relies on the plate format. One advantage that chemical library screens have in comparison to RNAi or CRISPR-based screens (see below) is that chemical compounds tend to inhibit families of targets, whereas RNAi is specific for individual members within the family. Different interactions can be identified with both approaches since some genes within the same family might be redundant and inhibiting a single gene would not cause synthetic lethality in an RNAi screen. In contrast, inhibiting the whole family by a chemical compound might be too cytotoxic in a chemical library screen, wherein inhibiting a single member may have been synthetically lethal. Overall, this approach is suited for studying specific cancer lines, generates a large number of hits, and provides compounds that can be used in vivo, but requires establishment of molecular targets.
siRNA Library Screens
The siRNA library screen relies on a plate format wherein each siRNA is transfected separately in its own well (Figure 3B), which makes it very similar to the format described above for the chemical library (Figure 3A). In some screens, multiple siRNAs targeting a gene are pooled into the same well (called a siRNA pool, usually comprised of four individual siRNAs). Cell Titer-Glo luminescent cell viability assay from Promega is often used to assess viability and identify hits. Both studies investigated below rely on the addition of extrinsic factors to generate matched cell lines. Once hits have been identified, in vivo studies can be conducted using RNAi constructs and chemical compounds for the established targets can be investigated.
In the first study, Kranz et al. [32] performed a genome-wide siRNA screen to sensitize the human glioblastoma cell line U251MG treated with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) to apoptosis (Figure 1B). TRAIL is an important ligand in the death receptor-mediated apoptosis pathway, which is often inactivated in cancers, including glioblastoma [30,31]. This screen was performed in a 384-well format using matched U251MG cells both with and without TRAIL treatment (Figure 2F, extrinsic factor) and Dharmacon’s SMART-pool siRNA library targeting 5,000 genes with four pooled siRNAs per target in each well (Figure 3B). Plates were analyzed using the Cell Titer-Glo viability assay to identify hits. Validation of the hits from the screen was performed, using the same system, by transfecting individual siRNAs for every gene hit instead of transfecting siRNA pools. FAT1 was chosen for further investigation since all four individual siRNAs resulted in decreased survival in response to TRAIL. This synthetic lethal interaction was also identified in five additional cell lines. The authors found that FAT1 is linked to the extrinsic apoptosis pathway and that it impedes Caspase-8 recruitment to the Death-Inducing Signaling Complex (DISC) in an ubiquitin-independent mechanism. Synthetic lethality of FAT1 with TRAIL treatment was confirmed by knocking out FAT1 using CRISPR/Cas9-mediated genome engineering.
In the second study, Josse et al. [35] performed a siRNA library screen to identify genes that are synthetically lethal with TOPoisomerase I (TOP1) inhibition in the breast cancer line MDA-MB-231 (Figure 1A). TOPoisomerases (TOPs) are enzymes that reverse the supercoiling of DNA that occurs during replication and transcription [33]. Inhibition of TOPs has been shown to be an effective means for killing constitutively proliferating cancer cells [34]. Inhibitors trap the TOP enzyme on the DNA and result in stalled replication forks and transcriptional complexes, causing DNA double strand breaks. Importantly, TOP1 inhibitors are in clinical trials or approved for ovarian cancer, colorectal cancer, breast cancer, and more [33,34]. The siRNA screen was performed using parallel 384-well plates of matched MDA-MB-231 cells treated with the TOP1 inhibitor Camptothecin (CPT) or vehicle control (Figure 2C, chemical compound). The Qiagene human druggable genome library version 4.1, targeting 7,000 genes with four individual siRNAs per gene, was transfected into cells. Cell Titer-Glo was used to assess viability and identify hits (Figure 3B). Forty-two hits were identified that selectively killed CPT-treated MDA-MB-231 while sparing vehicle-treated cells. To validate the top hits, the effects of the siRNAs were investigated with varying CPT concentrations and additional siRNAs for more than 40 of the candidate genes. One of the top hits was Ataxia Telangiectasia Mutated (ATM). Since ATM and Rad3-related protein kinase (ATR) function in parallel, and ATR inhibitors already have been developed and are in clinical trials, the authors confirmed that ATR inhibitors are synthetically lethal with TOP1 treatment. The authors found that ATR and its downstream target Chk1 are crucial factors in repairing DNA lesions that occur when TOP1 is inhibited. This synthetic lethal interaction was confirmed in vivo in mice with COLO205 colon cancer xenografts, and combination treatment with ATR and TOP1 inhibitors was more effective than individual treatment for either drug.
In summary, the siRNA approach, like the chemical library approach, is limited by the plate format since each siRNA or siRNA pool must be transfected into an individual well (Figure 3B, plate format) as compared to the shRNA library approach (discussed below) where shRNAs are transduced in a viral pool (Figure 3C, pooled format). Thus, the plate format limits the number of cancer types that can be investigated concurrently, but the screen can be performed using a high-throughput viability assay like Cell Titer-Glo. Hits can later be validated in additional cell lines. When performing secondary siRNA screens, each gene must be targeted with multiple non-pooled individual siRNAs to avoid false positives from off-target effects. The study by Josse et al. [35] investigated more than seven individual siRNAs for their top hits in order to identify the best candidates for further analysis. Overall, the siRNA approach is well suited to investigating individual matched cell lines where understanding the specific genes involved within the synthetic lethal interaction is important.
shRNA Library Screens
The use of shRNA libraries (Figure 3C) can aid in identifying the key molecular pathways that a tumor is dependent on for survival. There are a number of common strategies when performing shRNA screens. First, most screens consist of shRNA virus pools, which makes the approach applicable for studying multiple cancer lines in parallel, although a plate format with individual shRNAs per well can also be used [36], similar to siRNA screens (Figure 3B). The readout from the pooled screen is often in the form of deep sequencing [37] or a microarray [38]. The hits from the screens are then validated by performing viability assays using the individual shRNAs. Viability assays are often conducted using a kit like Cell Titer-Glo or clonogenic assays. The process of identifying the mechanism of action and chemical compounds targeting the gene hits is then different for each study.
In the first example, Etemadmoghadam et al. [38] performed shRNA library screening on 102 cancer cell lines including ovarian, colon, pancreas, lung, and other cancer types to investigate synthetic lethality with Cycline E1 (CCNE1) amplification (Figure 1B). Overexpression of the CCNE1 oncogene occurs in multiple cancers and is associated with poor prognosis in high-grade serous ovarian cancer [39,40]. CCNE1 overexpression results in increased proliferation through decreased regulation of the S phase cell cycle check point [41]. The authors obtained microarray data from shRNA experiments from the Integrative Genomics Portal, and the data was analyzed using the GenePattern module ScorebyClassComp and GENE-E software. The shRNA library contained a pool of 54,020 shRNAs targeting 11,194 genes. All cancer cell lines were divided into CCNE1 amplified/overexpressing or normal CCNE1 copy number/expression representing matched cell lines (Figure 2E). Each cancer type was analyzed separately. The relative abundance of each shRNA sequence in the matched cell lines was compared since shRNAs causing synthetic lethality should be eliminated from CCNE1 amplified/overexpressing cells. The screen identified 835 genes that were essential for survival in CCNE1-amplified or overexpressing cancer cell lines and 25 high confidence hits. The authors validated the results of the shRNA screen by performing an siRNA screen for their top 115 hits plus an additional 27 candidate genes. For the siRNA screen SK-OV-3 (CCNE1-unamplified cells) and OVCAR-3 (CCNE1-amplified cells) (Figure 2E) were plated in parallel, and viability was measured using the Cell Titer-Glo assay in a plate format (similar to Figure 3B). Among the best hits were cyclin-dependent kinase 2, BRCA1, and other genes involved in DNA damage repair (including HR) and cell division. Based on this screen and the chemical library screen by Jacquemont et al. [25] (see above) the authors used the chemical compound Bortezomib to target the FA pathway and disrupt HR. Bortezomib was synthetically lethal with CCNE1 amplification in multiple ovarian cancer cell lines.
In the second example, Hoffman et al. [37] identified synthetic lethal interactions in Brahma Related Gene 1 (BRG1)-deficient cancers (Figure 1A) using an epigenome-focused shRNA screen to identify epigenetic cancer dependencies. Epigenetic disregulation caused by alterations in the chromatin remodeling complex SWItch/Sucrose Non Fermentable (SWI/SNF) have been shown to be important in tumorigenesis [42]. Inactivation of BRG1, a DNA-dependent ATPase and part of the SWI/SNF complex, is associated with increased cell proliferation, dysregulation of cell cycle checkpoints, and poor clinical prognosis [43]. Matched lines for BRG1 were obtained from the Cancer Cell Line Encyclopedia and consisted of a panel of 58 human cancer cell lines differing in their BRG1 status (Figure 2B). In order to study epigenetics-based synthetic lethality, the authors constructed a Deep Coverage Design shRNA library (DECODER) containing 6,500 shRNAs, with 17 shRNAs per gene, to specifically target enzymes involved in epigenetic regulation. The DECODER lentiviral pool was transduced into each cell line in duplicate. The relative abandance of each shRNA was measured by Illumina GA2X-based next generation sequencing of each shRNA’s barcode in BRG1-deficient and expressing cell lines. Hits were shRNAs that were selectively eliminated from BRG1-deficient cells. From this screen, the gene with the strongest synthetic lethal effect was BRahMa (BRM), a catalytic subunit of the SWI/SNF chromatin remodeling complex and paralog of BRG1. The authors hypothesized that BRM compensates for BRG1 loss in the deficient cancer cell lines. To confirm this, the authors then showed that cancer lines with homozygous loss of BRG1 are more dependent on BRM than those with heterozygous loss. To further confirm the synthetic lethal interaction between BRG1 and BRM, the authors introduced an inducible shRNA targeting BRM into several BRG1-deficient and WT cell lines. BRM knockdown in BRG1-deficient cells caused irreversible growth arrest and significant induction of repressive H3K9me3 histone methylation marks. The synthetic lethal interaction was confirmed in vivo using an inducible shRNA for BRM in BRG1-deficent lung cancer xenografts of NCI-H1299.
In the third example, Scholl et al. [36] identified a synthetic lethal interaction in Kirsten rat sarcoma (KRAS) oncogene-dependent cancer cells using a subset of the Broad Institute TRC shRNA Library that consists of 5,024 shRNAs (Figure 1B). Oncogenic KRAS mutations occur within most human cancers [44-46] and drive tumorigenesis by increasing proliferation, anabolism, evasion of apoptosis and the immune system and by stimulating metastasis [44-46]. The authors hypothesized that oncogenic KRAS mutations cause secondary dependencies on genes that then can be exploited for causing synthetic lethal interactions. The shRNA screen was conducted in the acute myeloid leukemia (AML) cell lines NOMO-1 (mutant KRAS), THP-1 (WT KRAS), fibroblasts (WT KRAS), and human mammary epithelial cells (WT KRAS) (Figure 2E). Similar to the siRNA plate format approach (Figure 3B), the screen was conducted in a 384-well plate format in which each well was transduced with a single shRNA. Cell Titer-Glo was used to determine cell viability for each well. The top candidate gene STK33, a member of the calcium/calmodulin-dependent protein kinase subfamily of serine/threonine protein kinases, was then validated in additional matched AML lines (Figure 2E). The authors then tested the consequence of STK33 knockdown in multiple non-AML cancer types. The synthetic lethal interaction was only observed in cancers that are dependent on mutant KRAS for viability and proliferation. The interaction was confirmed in vivo using inducible shRNA targeting STK33 in pancreatic, breast, lung, and colon cancer xenografts. Further investigation has shown that in mutant KRAS cells, STK33 acts through S6K1-induced inactivation of Bcl-2-associated death promoter to suppress the mitochondia-mediated apoptosis and promote survival.
The shRNA approach requires redundancy in order to rule out false positive hits, since each individual shRNA has varying levels of efficacy in different cell types due to target transcript variation, shRNA design, and varying degrees of off-target effects. Overall, this approach allows for the investigation of synthetic lethality in a large number of cancer cell lines in parallel through pooled viral libraries and generates a large number of hits. These methods require the use of microarray or DNA sequencing, which can be cost prohibitive; small molecule compounds targeting the identified genes may not be readily available.
CRISPR Library Screens
A new approach to genome editing was discovered in bacteria and archaea through the identification of clustered repeat sequences in 1987 that was later named CRISPR, or clustered regularly interspaced short palindromic repeats, by Jansen et al. [47] in 2002. CRISPR serves as a bacterial immune system against bacteriophages. Invading viral DNA is incorporated into the bacterial host genome between the CRISPR repeat sequences, which can then be used to specifically target the viral DNA for degradation [48]. The CRISPR system is now a powerful tool for specifically editing the genome [49] and can be applied to identifying synthetic lethal interactions (Figure 4).
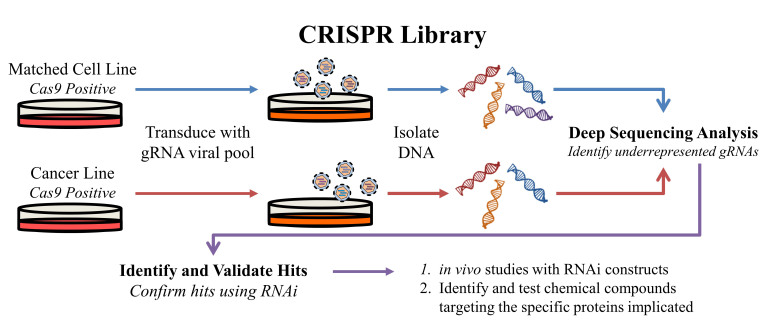
Figure 4.
An overview of the CRISPR approach for synthetic lethality screens. Matched cell lines stably expressing Cas9 are transduced with the gRNA library in a pooled virus format to produce a library of single cell mutants. The mutant library for each line is then screened for synthetic lethal interactions by isolating DNA and identifying underrepresented gRNA sequences in the cancer cell line, which are present in the matched (normal) cells. Interactions identified from the screen can be investigated and validated using RNAi. Small molecules interacting with the targets of interest can be developed and RNAi constructs may be used for in vivo studies.
Hiroko et al. [50] performed a genome-wide CRISPR screen in mouse cells to identify genes that confer susceptibility to Clostridium septicum alpha-toxin. The aim of this study was to identify genes that when targeted would confer resistance to the toxin. While not technically identifying synthetic lethal interactions, since the screen identified genes that resulted in survival when targeted by guide RNAs (gRNAs) (reverse of Figure 1B), this study exemplifies how a CRISPR-based approach could be conducted for studying synthetic lethal interactions in cancer. The CRISPR system generates bi-allelic mutations in each targeted gene resulting in a loss of function phenotype. Lentiviral CRISPR-Cas9 constructs that target 19,150 mouse protein-coding genes were generated using 87,897 gRNAs. The gRNA library covered 94.3 percent of the genome with at least two gRNAs per gene. Mouse embryonic stem cells (mESC) were transduced with the lentiviral CRISPR pool and treated with or without alpha-toxin for 5 days (Figure 2F). The surviving cells were analyzed by deep sequencing to detect which genes had been lost. In a synthetic lethality screen, the gRNA could be barcoded, allowing for hits to be identified by detecting underrepresented gRNA in the cancer population that are still present in the matched (WT) cell line (Figure 4). The authors identified 13 genes associated with alpha-toxin resistance and validated genes that were targeted by at least two independent gRNAs. Four validated hits conferring resistance to alpha-toxin were B4galt7, 1700016K19Rik, Cstf3, and Ext2. The authors verified B4galt7 and Ext2 by re-introducing the full length cDNA into the corresponding gene knockout cell lines, which resulted in sensitivity to the toxin being restored. No further molecular analysis was conducted, and the results were not pursued yet with in vivo studies or chemical compound studies.
One major advantage to using a CRISPR library is that the system provides for greater efficiency with fewer off-target effects when compared to an RNAi screen. As such, the need for redundant targeting of the same gene is reduced. CRISPR screens, like shRNA library screens, are well-suited for studying multiple cell lines in parallel since a pooled viral library can be employed. This is also a disadvantage, since the screen will then rely on costly microarray or deep sequencing to identify hits. Since CRISPR is a relatively new system, it is likely that synthetic lethal screens using this approach will become more frequent.
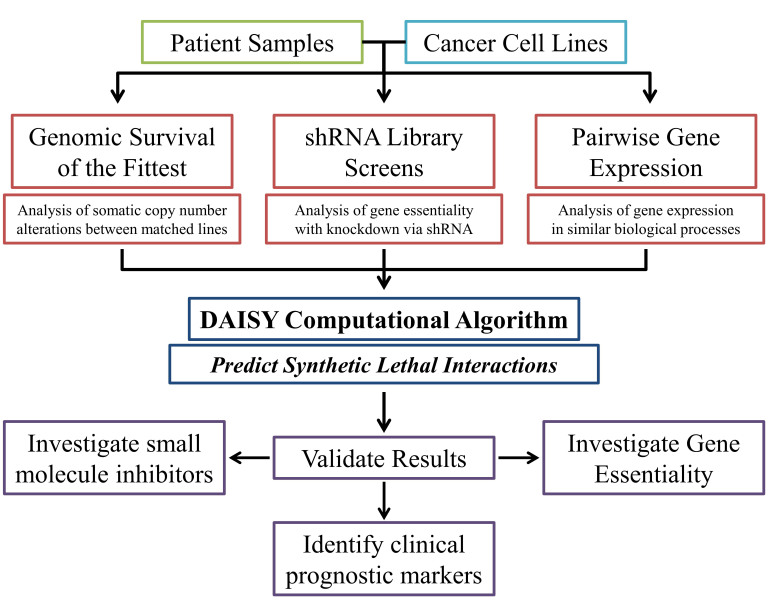
The DAISY Approach
Most computational approaches to identifying synthetic lethal interactions on a global level have been performed in yeast [9,51] and then applied to human cancer by investigating the effect in their human orthologs. Combining multiple approaches into a single screen can be an effective way to computationally identify synthetic lethal interactions on a global scale in human cancer. The data mining synthetic lethality pipeline, known as DAISY, is an approach published by Arnon et al. [52]. It consists of a data-driven computational screen for identifying synthetic lethal pairs through the analysis of cancer genomic data (Figure 1A). DAISY is a combination of three inference strategies: survival of the fittest (SoF), shRNA functional examination, and pairwise gene co-expression (Figure 5). The authors also analyzed synthetic dosage lethality (SDL), which is the overexpression of a gene that leads to the essentiality of a second gene, making a SDL pair (Figure 1B). DAISY statistically infers synthetic lethal interactions from data sets generated from cancer cell lines and patient samples (Figure 2B,E).
Figure 5.
An overview of the DAISY computational approach. DAISY is a computational program that scans large genomic data sets from cell lines and patient samples to identify synthetic lethal interactions. DAISY uses three approaches that independently interpret multiple data sets. The first approach is called survival of the fittest (SoF), which examines genomic DNA data sets for gene co-inactivation and somatic copy number alterations (SCNA). The second approach analyzes the essentiality of a potential synthetic lethal gene pair using databases from shRNA screens. The last approach uses gene expression data sets to search for genes that are expressed in the similar biological processes by analyzing pairwise gene expression. Predicted synthetic lethality pairs are then tested and validated using in vitro gene essentiality assays. If drugs targeting the identified synthetic lethal gene pairs are available then in vitro drug screens can be performed. The expression pattern of the synthetic lethal gene pairs can also be assessed for potential use as biomarkers for clinical prognosis.
The first approach, SoF, is based on the assumption that cells with reduced expression of synthetic lethal gene pairs will not survive in a heterogeneous cell population. Since cancer cells with the inactivation of two synthetically lethal genes will not survive, by studying the somatic copy number alteration (SCNA) of a specific gene from the pooled populations of multiple cell lines and comparing it to the average change in all other genes, a synthetic lethal interaction can be inferred. The co-inactivation of synthetically lethal genes should occur less frequently than the average. This is similar to the shRNA screen approach, wherein microarray or sequencing is used to identify which shRNAs have been lost from the pooled population with the exception that it is on a much larger scope: each gene investigated is being compared to every other gene across multiple cell lines and patient samples. The second approach is a shRNA functional examination and relies on three separate shRNA library screens in 46, nine, and 92 cancer cell lines to identify genes that become essential due to the knockdown of another gene. Each separate screen comprised an individual data set including the SCNA and gene expression profiles for each cancer cell line examined within the screen. This relies on the hypothesis that synthetically lethal genes will be simultaneously lost from the viral pool in multiple cancer cell lines. The third approach is pairwise gene co-expression analysis, which is based on the assumption that synthetic lethal pairs are involved in closely related biological processes. The combined analysis of the three approaches analyzed more than 535 million gene pairs. A hit from the DAISY screen is defined as two genes in which the inactivation of one renders the other essential for survival.
DAISY’s effectiveness was evaluated against previously identified synthetic lethal gene pairs, including the partners of PARP1, VHL, MutS protein Homolog 2 (MSH2), and KRAS. The validation was performed on 7,276 gene pairs that were tested in six large screens. PARP1 and BRCA1/BRCA2, as well as MSH2 and Dihydrofolate reductase, were successfully confirmed as synthetic lethal pairs. Daisy’s predictive ability was also used to identify genes that become essential with VHL deficiency. Forty-four genes were predicted, and a small siRNA screen was performed to test the validity of the prediction. DAISY identified 3.83 times more synthetically lethal genes than the follow-up siRNA screen. This indicates that additional validation is needed for predicted pairs to eliminate false positives.
The predictive ability of the combined DAISY approach was then used to generate genome-wide networks of synthetic lethal and SDL interactions in cancer. More than 2,000 synthetic lethal genes and more than 3,000 SDL genes were identified in both normal and cancer cells. The identified gene pairs were also investigated in a large study based on clinical samples and cancer cell lines to test gene essentiality, clinical prognosis potential, and drug efficacy.
DAISY does have its limitations. It can only be applied to cancer cells since it relies on large genomic mutation data sets, which at times can be hard to work with because of their inaccuracy. It does not account for epigenetic or posttranslational regulation that can alter synthetic lethal and SDL interactions. DAISY’s predictive power can be utilized to identify novel co-essential gene pairs, which may lead to new and improved therapeutics for treating cancer.
Conclusions and Outlook
Harnessing synthetic lethal interactions to create novel cancer-specific therapies is a rapidly growing part of cancer biology. The specificity of these interactions will allow for improved overall survival while also increasing patient quality of life. Synthetic lethality allows for the exploitation of cancer specific mutations that are not “druggable” by identifying interactions with other co-essential genes. It provides an explanation as to why some drugs have increased efficacy in specific cancer types and may even provide new clinically relevant diagnostic markers. With the discovery and implementation of CRISPR, it is very likely that studies will be conducted using CRISPR libraries. Future computational based genome-wide inference studies like DAISY could be a powerful tool for mining the myriad public data sets available to identify synthetic lethal interactions in specific cancer types. Other screens could identify synthetic lethal interactions using a microRNA library approach. As technology advances and sophisticated approaches become easier to conduct, it might be possible for synthetic lethal screens to be extended to personalized medicine.
Abbreviations
- AML
acute myeloid luekemia
- APC
adenomatous polyposis coli
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein kinase
- BID
BH3 interacting-domain death agonist
- BRCA
breast cancer early onset
- BRG1
Brahma related gene 1
- BRM
Brahma
- CCNE1
cycline E1
- CPT
camptothecin
- DECODER
Deep Coverage Design shRNA Library
- DISC
death-inducing signaling complex
- FA
Fanconi anemia
- FANCD2
Fanconi anemia group D2
- gRNA
guide RNA
- HR
homologous recombination
- IR
ionizing radiation
- KRAS
Kirsten rat sarcoma
- mESC
mouse embryonic stem cells
- MSH2
MutS protein homolog 2
- NSAIDs
nonsteroidal anti-inflammatory drugs
- PARP1
Poly [ADP-ribose] Polymerase 1
- RCC
renal cell carcinomas
- RNAi
RNA interference
- SCNA
somatic copy number alteration
- SDL
synthetic dosage lethality
- SoF
survival of the fittest
- SWI/SNF
switch/sucrose NonFermentable
- TOP
topoisomerase
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- VHL
Von Hippel Lindau
Author contributions
JMT: Wrote the paper and compiled Figures 1, 2, and 3; QHN: wrote the CRISPR section and compiled Figure 4; MS: wrote the DAISY section and compiled Figure 5; OVR: Outlined the review content and helped to finalize the text and figures. QHN and MS contributed equally to the work. All authors contributed to literature search and revising the article. This review was supported by ACS fund IRG 98-27907 to OVR.
References
- Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21(24):3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3(8):448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Reddy A, Kaelin WG Jr.. Using cancer genetics to guide the selection of anticancer drug targets. Curr Opin Pharmacol. 2002;2(4):366–373. doi: 10.1016/s1471-4892(02)00178-9. [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- Hosoya N, Miyagawa K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014;105(4):370–388. doi: 10.1111/cas.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block KI, Koch AC, Mead MN, Tothy PK, Newman RA, Gyllenhaal C. Impact of antioxidant supplementation on chemotherapeutic toxicity: a systematic review of the evidence from randomized controlled trials. Int J Cancer. 2008;123(6):1227–1239. doi: 10.1002/ijc.23754. [DOI] [PubMed] [Google Scholar]
- Hartman J, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;1001(291):1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8(6):437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov. 2011;10(5):351–364. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel M, Molina-Arcas M, Weigelt B, Marani M, Warne PH, Kuznetsov H. et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012;22(8):1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair B, Kubicek S, Nijman SMB. Exploiting epigenetic vulnerabilities for cancer therapeutics. Trends Pharmacol Sci. 2014;35(3):136–145. doi: 10.1016/j.tips.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N. et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70(20):8045–8054. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PM, Eppink B, Essers J, Stap J, Rodermond H, Odijk H. et al. Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc Natl Acad Sci USA. 2011;108(24):9851–9856. doi: 10.1073/pnas.1101053108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H, Schultz N, Thomas HD. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Farmer H, Mccabe N, Lord CJ. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Ellisen L. PARP inhibitors in cancer therapy: promise, progress, and puzzles. Cancer Cell. 2011;19(2):165–167. doi: 10.1016/j.ccr.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luvero D, Milani A, Ledermann JA. Treatment options in recurrent ovarian cancer: latest evidence and clinical potential. Ther Adv Med Oncol. 2014;6(5):229–239. doi: 10.1177/1758834014544121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C. et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G. et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- Leibowitz B, Qiu W, Buchanan ME, Zou F, Vernon P, Moyer MP. et al. BID mediates selective killing of APC-deficient cells in intestinal tumor suppression by nonsteroidal antiinflammatory drugs. Proc Natl Acad Sci USA. 2014;111(46):16520–16525. doi: 10.1073/pnas.1415178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14(1):90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Nguyen P, Turcotte S, Lai EW, Banh A. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3(94):94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont C, Simon JA, D’Andrea AD, Taniguchi T. Non-specific chemical inhibition of the Fanconi anemia pathway sensitizes cancer cells to cisplatin. Mol Cancer. 2012;11:26. doi: 10.1186/1476-4598-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan J, Lal P, Dondeti V, Letrero R. HIF-α Effects on c-Myc Distinguish Two Subtypes of Sporadic VHL-Deficient Clear Cell Renal Carcinoma. Cancer Cell. 2008;14(6):435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabik C, Dolan M. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33(1):9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon K, Swisher E, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102(4):663–669. doi: 10.1111/j.1349-7006.2010.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3(7):502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2(4):277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5(11):876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- Fesik D, Boutros M. A synthetic lethal screen identifies FAT 1 as an antagonist of caspase-8 in extrinsic apoptosis. EMBO J. 2014;33(3):181–197. doi: 10.1002/embj.201385686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6(10):789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8(1):82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse R, Martin SE, Guha R, Ormanoglu P, Pfister TD, Reaper PM. et al. The ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase I inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014;74(23):6968–6979. doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Fröhling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY. et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111(8):3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J. et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci USA. 2013;110(48):19489–19494. doi: 10.1073/pnas.1314302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr D, Kanitz V, Anderegg B, Luthardt B, Engel J, Löhrs U. et al. Analysis of Gene Amplification and Prognostic Markers in Ovarian Cancer Using Comparative Genomic Hybridization for Microarrays and Immunohistochemical Analysis for Tissue Microarrays. Am J Clin Pathol. 2006;126(1):101–109. doi: 10.1309/n6x5mb24bp42kp20. [DOI] [PubMed] [Google Scholar]
- Etemadmoghadam D, Beroukhim R. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15(4):1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akli S, Keyomarsi K. Cyclin E and its low molecular weight forms in human cancer and as targets for cancer therapy. Cancer Biol Ther. 2003;2(4 Suppl 1):S38–S47. [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Bai J, Mei P, Zhang C, Chen F, Li C, Pan Z. et al. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One. 2013;8(3):e59772. doi: 10.1371/journal.pone.0059772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Szappanos B, Kovács K, Szamecz B, Honti F, Costanzo M, Baryshnikova A. et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet. 2011;43(7):656–662. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E. et al. Predicting Cancer-Specific Vulnerability via Data-Driven Detection of Synthetic Lethality. Cell. 2014;158(5):1199–1209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]