Abstract
Dihydrofolate reductase, DHFR, is a pivotal enzyme involved in the de novo pathway for purine synthesis, and hence, represents an attractive target to disrupt systems that require rapid DNA turnover. The enzyme acquires resistance to available drugs by various molecular mechanisms, which necessitates the continuous discovery of novel antifolates. In a previous communication, we had identified a set of novel molecules that showed binding to E. coli DHFR by means of thermal shift without establishing whether they inhibited the enzyme. In this paper we show that a fraction of those molecules represent potent and novel inhibitors of DHFR activity. 7-[(4-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, AMPQD, a molecule with no reported inhibition of DHFR, potently inhibits the enzyme with a Ki value of 7.42 ± 0.92 nM by competitive displacement of the substrate dihydrofolic acid. It shows uncompetitive inhibition vis-avis NADPH indicating that the inhibitor has markedly increased affinity for the NADPH-bound form of the enzyme. Further, we demonstrate that the mode of binding of the inhibitor to the enzyme-NADPH binary complex conforms to the slow-onset, tight-binding model. On the contrary, mechanistic characterization of the parent molecule 7H-pyrrolo[3,2-f] quinazoline-1,3-diamine, PQD, shows that the lack of (4-aminophenyl)-methyl group at the 7th position abolishes the slow-onset of inhibition. This finding provides novel insights on the role of substitutions on inhibitors of E. coli DHFR and represents the first detailed kinetic investigation of a novel diaminopyrroloquinazoline derivative on a prokaryotic DHFR. Furthermore, marked differences in the potency of inhibition for the E. coli and human DHFR makes this molecule a promising candidate for development as an antibiotic.
Keywords: Mechanistic characterization; dihydrofolate reductase; drug discovery; pyrrolo[3,2-f]quinazoline-1; 3-diamine; slow-tight-binding inhibition
Introduction
Dihydrofolate reductase (E.C.1.5.1.3), DHFR, is a ubiquitous enzyme found in all kingdoms of life. The enzyme is involved in the reduction of 7,8-dihydrofolate (H2F) to 5,6,7,8-tetrahydrofolate (H4F) in which protonation of H2F on N5 precedes the hydride transfer from C4 of the NADPH cofactor to the C6 atom of the pterin ring on H2F [1]. Since DHFR is the sole source of cellular tetrahydrofolate, a metabolite essential for thymidylate and purine synthesis, its activity is indispensable. Thus, the enzyme represents an attractive target to disrupt systems that require rapid DNA turnover, e.g. proliferating cancer cells and pathogenic microbes[2]. E. coli DHFR has been extensively characterized in terms of kinetic mechanism, catalysis and structural studies[3-6]. This wealth of data makes the enzyme an attractive target to design small-molecule inhibitors as potential antibiotics. This has become all the more important given the rise in instances of nosocomial infection caused by drug-resistant E. coli[7]. However, designing inhibitors for DHFR presents considerable challenges since the enzyme acquires rapid resistance to available drugs by means of gene amplification, mutations and decreased drug uptake[8].
A lot of effort has been expended on discovering novel inhibitors for DHFR from different organisms given their potential applications to antineoplastic, anti-inflammatory and anti infective drug discovery[9-11]. Methotrexate (MTX), a 2,4-diaminopteridin, is by far the most well characterized inhibitor for DHFR, showing a slightly increased potency of inhibition for parasitic DHFR compared to either human or bacterial DHFR [2]. Other prominent antifolates include the pyrimidine-2,4-diamines pyrimethamine, which is highly specific for eukaryotic DHFRs, trimethoprim (TMP), which shows a slightly greater preference for prokaryotic DHFRs[2], metoprine (DDMP) and piritrexim (PTX). Further, to overcome the limitation imposed by the hydrophilic nature of methotrexate, which hinders its distribution across different tissues, lipophilic inhibitors like trimetrexate (TMX), a quinazoline-2,4-diamine, have been synthesized as nonclassical inhibitors of DHFRs. Pyrrolo[3,2-f]quinazoline-1,3-diamine derivatives, containing a novel tricyclic heterocycle compared to TMX, were further explored and shown to be inhibitors of parasitic DHFRs[12]. Other studies have shown 7,8-dialkyl-1,3-diaminopyrrolo[3,2-f] quinazolines compounds as high-affinity inhibitors of DHFR from Pneumocystis carinii and Candida albicans [13]. In yet another study, a high throughput screen identified twelve compounds as inhibitors of E. coli DHFR[14]. However, it should be noted that detailed kinetic studies on the inhibition brought about by these novel DHFR inhibitors is lacking.
Previously, as part of experimental validation of the virtual ligand screening algorithm FINDSITEcomb and relying on thermal shift assay methodology, we reported a set of novel molecules that showed binding to E. coli DHFR[15, 16]. In this paper, employing inhibition kinetics, we show that a fraction of those molecules represent novel inhibitors of DHFR activity and present detailed mechanistic characterization to substantiate our claims. By means of extensive steady-state and tight inhibition kinetics studies, for the first time we show that AMPQD and its parent compound, PQD, are novel tight-binding inhibitors of E. coli DHFR. These inhibitors preferentially bind to the NADPH-bound form of the enzyme at the H2F binding site. While AMPQD shows slow-onset of binding to the enzyme, PQD shows no such behavior implicating the (4-aminophenyl) methyl group as a possible origin of slow-binding behavior in E. coli DHFR. This, combined with our already reported antibacterial, antifungal and antineoplastic activity by these compounds against two different strains of E. coli (multi-drug resistant E. coli and DH5α), a strain of methicillin resistant Staphylococcus aureus, a strain of vancomycin-resistant Enterococcus faecalis, a strain of amphotericin B resistant C. albicans and HCT-116 human colon carcinoma cell line, makes these compounds potential lead candidates to target conditions arising from aberrant DHFR activity. Further, pronounced differences in the potency of inhibition and the mode of inhibitor binding for AMPQD and PQD against E. coli and human DHFR makes these molecules attractive candidates for development as novel antibiotics.
Results
Inhibition of E. coli DHFR
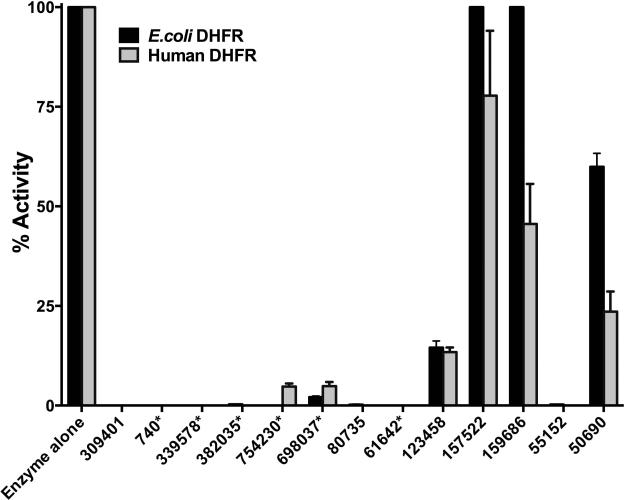
All the hits from the FINDSITEcomb experimental validation study were assessed for their ability to inhibit E. coli DHFR. The histogram in Fig 1 summarizes the results. All the reported inhibitors of DHFR from various sources viz., methotrexate (NSC740), 7H-pyrrolo(3,2-f) quinazoline-1, 3-diamine (NSC339578), methylbenzoprim (NSC382035), pralatrexate (NSC754230), pemetrexed (NSC698037) and 6,7-bis (4-aminophenyl) pteridine-2, 4-diamine (NSC61642), show unambiguous inhibition of E. coli DHFR at a 1 mM inhibitor concentration. Prior to carrying out IC-50 determination and detailed inhibition studies, the kinetic parameters for the substrate H2F and cofactor NADPH were determined and found to be in agreement with values reported in the literature within experimental error [17, 18] (Table S1 and Fig S1). For all further experiments, except when a substrate is titrated, the substrates were kept at >10 times their respective Km values. Table 1 and Fig S2A-B summarize the IC-50 values, defined as the concentration of inhibitor required to reduce the activity of the enzyme by 50%, determined for a select set of reported inhibitors of E. coli DHFR independently identified by our studies.
Fig 1.
Comparative inhibition of DHFR from E. coli and humans. Each histogram represents the activity of E. coli (black histograms) or human DHFR (gray histograms) in the presence of the inhibitor molecules tested at a fixed concentration of 1 mM. All activities are expressed as percentage activity with respect to the enzyme control for ease of comparison across the two enzymes. The numbered notations for the various inhibitor molecules represent NSC numbers. The numbers with an asterisk represent molecules that have been previously reported as having DHFR inhibitory activity from various organisms and independently “predicted” by our method.
Table 1.
IC-50 and Kiapp values for various small molecule inhibitors of E. coli and human DHFR
| Small molecule$/ NSC ID | E. coli DHFR | Human DHFR | ||
|---|---|---|---|---|
| IC-50 (nM) | Kiapp* (nM) | IC-50 (nM) | Kiapp* (nM) | |
| AMPQD/NSC309401 | 189.0 ± 1.0 | 7.8 ± 0.8 | 599.0 ± 7.2 | 19.4 ± 2.7 |
| PQD#/NSC339578 | 106.1 ± 1.2 | 4.2 ± 0.5 | 3087.1 ± 16.5 | 93.4 ± 9.0 |
| MTX#/NSC740 | 152.5 ± 1.1 | 6.7 ± 0.7 | 147.7 ± 2.3 | 5.1 ± 1.0 |
| NNCPPU/NSC80735 | 36560 ± 1100 | 2062 ± 378 | 68870 ± 1356 | 1973 ± 329 |
| NNBABD/NSC55152 | 176500 ± 1114 | 10941 ± 2156 | 254500 ± 1789 | 5161 ± 1411 |
| ISB/NSC123458 | 587800 ± 1184 | 35222 ± 5358 | ND | ND |
Kiapp was estimated by employing the Morrison equation. This equation accounts for tight binding, and hence does not assume that the free concentration of inhibitor equals the total concentration.
Reported inhibitors of DHFR from various organisms independently identified by our method as inhibitors of human and E. coli DHFR.
AMPQD, 7-[(4-aminophenyl)methyl]-7HPyrrolo[3,2-f]quinazoline-1,3-diamine; PQD, 7H-Pyrrolo(3,2-f)quinazoline-1,3-diamine; MTX, Methotrexate; NNCPPU, 1-(4-nitrophenyl)-3-[4-[4-[(4-nitrophenyl) carbamoylamino] phenoxy]phenyl]urea; NNBABD, N,N'-bis(4-aminophenyl)benzene-1,4-dicarboxamide; ISB, 2,2'-Iminostilbene. ND, Not determined since it was greater than 2 mM.
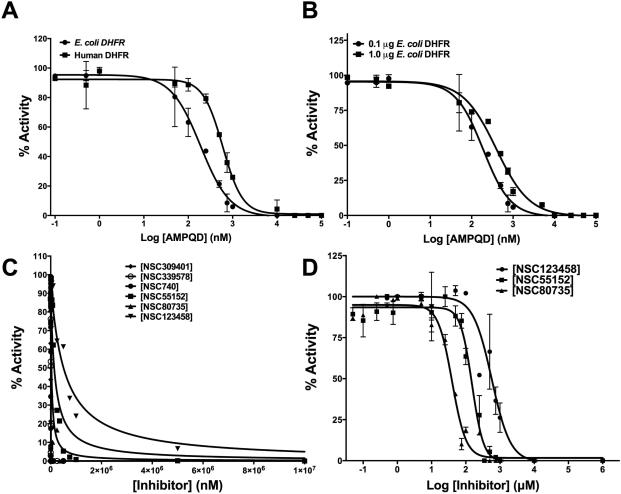
Among the nine novel binders reported by us in our previous study, seven were tested for their inhibition of E. coli DHFR[15]. Three (NSC309401, NSC80735 and NSC55152) showed almost complete inhibition while one molecule (NSC123458) showed approximately 87% inhibition at a 1mM inhibitor concentration (Fig 1). While NSC309401, AMPQD, is a substrate analogue with the quinazoline-1, 3-diamine group, NSC80735 and NSC55152 contain concatenated nitrophenyl and aminophenyl groups. To further understand their inhibition, IC-50 values were determined for the various molecules. Fig 2A shows the curve of log inhibitor concentration vs. activity for AMPQD giving an IC-50 value of 189.0 ± 1.0 nM (Table 1). This number indicates potent inhibition comparable to that shown by methotrexate, with an IC-50 value of 152.5 ± 1.1 nM. However, since it is known in the literature that IC-50 values are enzyme concentration dependent and can never get lower than [E0]/2 [19], it is highly likely that the number represents an underestimation of the actual affinity of the inhibitor for the enzyme (Fig 2B).
Fig 2.
Potency of inhibition. (A) IC-50 determination for AMPQD against E. coli and human DHFR. (B) Enzyme concentration dependence of IC-50 for the tight-binding inhibitor AMPQD for E. coli DHFR. (C) Fit of the experimental dose-response curves to Morrison's equation for tight-binding for inhibitors of E. coli DHFR. (D) IC-50 value estimates for inhibitors of E. coli DHFR not displaying tight-binding behavior (NSC80735, NSC55152 and NSC123458). On the plots, the y-axis represents % activity of the enzyme and the x-axis represents the log inhibitor concentration/inhibitor concentration. The experimental data points were fit to the respective equations using the non-linear curve-fitting algorithm of GraphPad Prism v 6.0e.
To account for this tight binding inhibition, the data were analyzed as per the methods developed by Morrison and coworkers[20, 21]. Fig 2C shows the fit of the data to the quadratic Morrison equation for tight binding and Table 1 lists the Kiapp values computed from non-linear curve fitting. As expected, the Kiapp value of 7.8 ± 0.8 nM for AMPQD is almost 25 fold lower than its IC-50. Further, compounds NSC80735, NSC55152 and NSC123458 were also titrated, and their log inhibitor concentration vs. activity were plotted to yield IC-50 values of 36.56 ± 1.1 μM, 176.5 ± 1.1 μM and 587.8 ± 1.2 μM, respectively (Fig 2D and Table 1). However, since these three compounds are sparsely soluble in water, the reported IC-50 values may, at most, represent gross approximations. Moreover, their high IC-50s and relative insolubility may lead to potential problems of bioavailability. Hence, these compounds may not represent promising lead candidates. It is worthwhile to point out that compounds NSC80735 and NSC55152 didn't show either bactericidal activity or activity against cancer cells as reported in our previous study [15]. Detailed mechanistic characterization was undertaken on AMPQD (the best hit from the FINDSITEcomb study), PQD (the parent molecule of AMPQD) and methotrexate (a well characterized DHFR inhibitor) to understand their mode of inhibition (Fig 3).
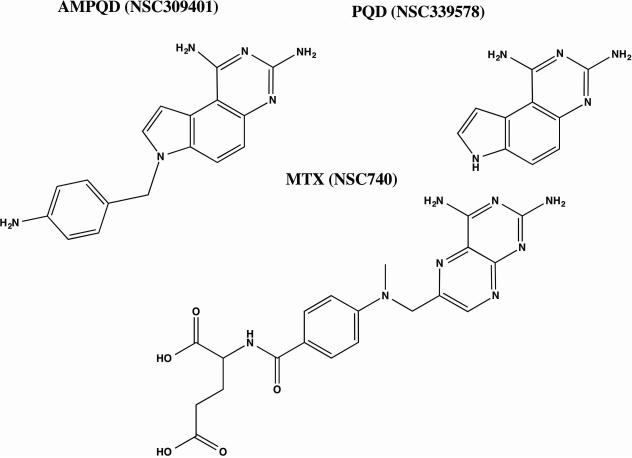
Fig 3.
Structures of (A) AMPQD (NSC309401) (B) PQD (NSC339578) and (C) MTX (NSC740). The SDF files for the structures were downloaded from PubChem database and the figures were made in ChemBioDraw 14.0.
AMPQD (NSC309401) is a competitive inhibitor of dihydrofolate binding
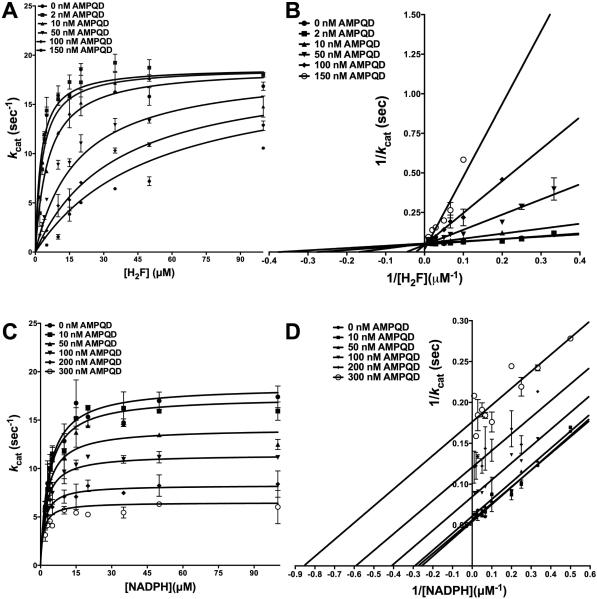
To further understand the inhibition shown by AMPQD, we resorted to detailed inhibition kinetics. Substrate dihydrofolate was titrated at several fixed concentrations of AMPQD, and the resulting curves from the primary plot, when globally fit to models for the various types of inhibition showed the best fit to the model for competitive inhibition (see Experimental Section for details) (Fig 4A) yielding a Ki, the equilibrium dissociation constant for the competitive inhibitor, of 7.42 ± 0.92 nM (Table 2). Further, for visual assessment, the data were transformed and plotted as the double-reciprocal Lineweaver-Burk plot, LB. Fig 4B shows the lines of the LB-plot intersecting on the y-axis further indicative of competitive displacement of substrate dihydrofolate by AMPQD, whereby it increases the apparent Km value for the substrate without unduly affecting the Vmax. Further, this low Ki value, similar to that obtained from fit to the Morrison equation, reinforces the fact that AMPQD is a tight-binding inhibitor, a special case where the affinity of the inhibitor for the enzyme is an order-of-magnitude lower than the minimum concentration of enzyme that can be employed in the assay mix to get reliable activity. Further, the Ki value is ~25-fold lower than the obtained IC-50. However, the Ki value for AMPQD is approximately 2-fold higher than that reported for methotrexate (the reported value is ~ 3.6 nM[22]). The above data are conclusive about AMPDQ binding to the same site as dihydrofolate and competing with the latter for high-affinity interactions with the enzyme. This competitive displacement can be ascribed to the quinazoline-1,3-diamine group shared by the two molecules (substrate and inhibitor) that might serve as the common motif responsible for binding. Though this behavior is similar to that shown by methotrexate, it is markedly different from that of a pyrimidine-2,4-diamine pyrimethamine against Plasmodium DHFR, which is a non-competitive inhibitor of the latter [23].
Fig 4.
Inhibition kinetics of AMPQD (NSC309401) for E. coli DHFR (A) Fit of the primary data to the competitive inhibition model for H2F titration at several fixed concentrations of AMPQD. (B) Double reciprocal Lineweaver-Burk plot of H2F titration at several fixed concentrations of AMPQD. (C) Fit of the primary data to the uncompetitive inhibition model for NADPH titration at several fixed concentrations of AMPQD (D) Double reciprocal Lineweaver-Burk plot of NADPH titration at several fixed concentrations of AMPQD. The y-axis shows the kcat value. The experimental data points were fit to the respective models using the non-linear curve-fitting algorithm of GraphPad Prism v 6.0e.
Table 2.
Parameters from inhibition kinetics and time-dependent inactivation of E. coli DHFR
| Inhibitors | Substrate | Inhibition# | Ki/αKi (nM)* | koff (min1) | kon (nM−1min−1) | Kd (nM) |
|---|---|---|---|---|---|---|
| AMPQD | H2F | C | 7.42 ± 0.92 | 0.118 ± 0.017 | 0.008 ± 0.001 | 14.57 ± 2.10 |
| NADPH | U | 162.70 ± 9.06 | NA | NA | NA | |
| PQD | H2F | C | 3.18 ± 0.51 | 0.094 ± 0.010 | 0.021 ± 0.002 | 4.48 ± 0.63 |
| NADPH | U | 72.17 ± 4.23 | NA | NA | NA | |
| MTX | H2F | C | 3.6£ | 0.223 ± 0.078 | 0.013 ± 0.004 | 17.15 ± 2.92 |
| NADPH | U | 111.00 ± 7.32 | NA | NA | NA |
C: competitive inhibition, U: uncompetitive inhibition
The Ki reported is for competitive inhibition while αKi is reported for uncompetitive inhibition; KD represents koff/ kon; NA, Not applicable.
The value reported is from the study [21]
AMPQD (NSC309401) is an uncompetitive inhibitor of NADPH binding
To understand AMPQD's effect on the cofactor NADPH binding, the latter was titrated at several fixed concentrations of AMPQD, and the resulting curves from the primary plot, when globally fit to models for the various types of inhibition showed the best fit to the model for uncompetitive inhibition (Fig 4C) yielding an αKi, the equilibrium dissociation constant for the uncompetitive inhibitor, of 162.9 ± 9.1 nM (Table 2). This higher αKi value shows that AMPQD binding site is fully formed only when the enzyme is bound to NADPH. It is worthwhile to point out that AMPQD Ki was ~7.4 nM at saturating NADPH (see previous section). Further, for visual assessment, the resulting data were transformed and plotted as the double-reciprocal LB plot. Fig 4D shows parallel lines on the LB-plot confirming the fit of primary data to model for uncompetitive inhibition. The data on competition of AMPQD with NADPH is strongly indicative of an ordered binding event whereby NADPH binding facilitates inhibitor binding. It should be noted that this pattern of uncompetitive inhibition against NADPH is similar to the way MTX behaves (Fig S3A-B).
Though, in principle, E. coli DHFR can bind to both NADPH and H2F randomly as shown by some studies[6], productive catalysis proceeds through an ordered ternary complex formation with NADPH binding prior to dihydrofolate. Furthermore, the pattern is also consistent with the pH-independent and pH-dependent models for E. coli DHFR kinetic mechanism proposed by Fierke et al that shows that dihydrofolate always binds to the NADPH-bound form of the enzyme [4]. In a previous study, we have shown that AMPQD independently binds to the enzyme in the absence of NADPH. Hence, the type of inhibition should ideally be either noncompetitive or linear mixed-type. However, the difference between Ki and αKi is large. Hence, for all practical purposes, this inhibition can be considered as uncompetitive. Thermal shift assay measurements carried out from 0 nM - 500 nM AMPQD in the absence of NADPH were unsuccessful in stabilizing the protein and showed preferential binding to the denatured form of the protein. However, at a high concentration of 1mM, the inhibitor showed binding to the enzyme, even in the absence of added NADPH, as seen in the thermal stability profile (Fig S4). This further proves that AMPQD binding in the nM concentration range is absolutely conditional upon NADPH binding to the enzyme.
7H-Pyrrolo(3,2-f) quinazoline-1, 3-diamine (PQD) inhibition kinetics
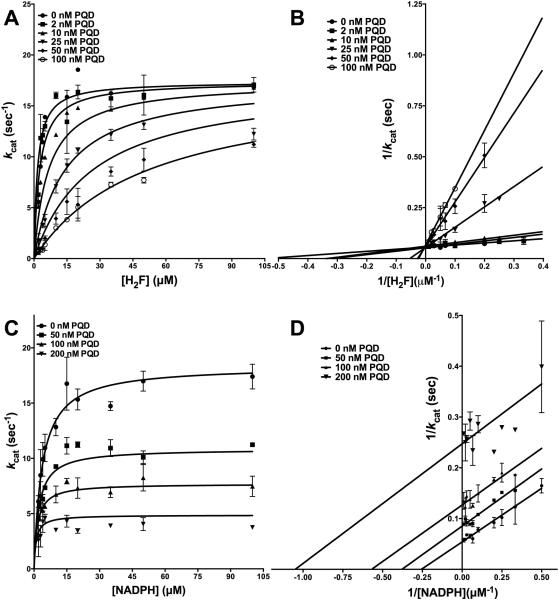
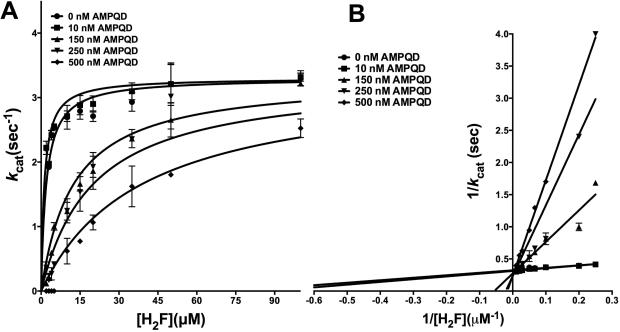
PQD has been shown to possess inhibitory activity against DHFRs from eukaryotic sources, inhibiting fungal DHFRs [13]. In order to assess its inhibition of a prokaryotic enzyme, it was tested against E. coli DHFR (Fig 1 and Fig S2A). PQD is the parent molecule for AMPQD and lacking the latter's (4-aminophenyl) methyl group. Figs 5A-B show the primary curves for H2F titration at several different concentrations of PQD fit to the model of competitive inhibition and the double-reciprocal LB plot. Further, Fig 5C-D shows the primary curves for NADPH titration at several different concentrations of PQD fit to the model for uncompetitive inhibition and the double reciprocal LB plot. These patterns show that PQD occupies the H2F binding site and preferentially binds to the NADPH-bound form of the enzyme, mirroring the behavior shown by its derivative AMPQD. However, the Ki value of 3.18 ± 0.51 nM for PQD is approximately half of that shown by AMPQD indicative of tighter binding (Table 2).
Fig 5.
Inhibition kinetics of PQD (NSC339578) for E. coli DHFR (A) Fit of the primary data to the competitive inhibition model for H2F titration at several fixed concentrations of PQD. (B) Double reciprocal Lineweaver-Burk plot of H2F titration at several fixed concentrations of PQD. (C) Fit of the primary data to the uncompetitive inhibition model for NADPH titration at several fixed concentrations of PQD (D) Double reciprocal Lineweaver-Burk plot of NADPH titration at several fixed concentrations of PQD. The y-axis shows the kcat value. The experimental data points were fit to the respective models using the non-linear curve-fitting algorithm of GraphPad Prism v 6.0e.
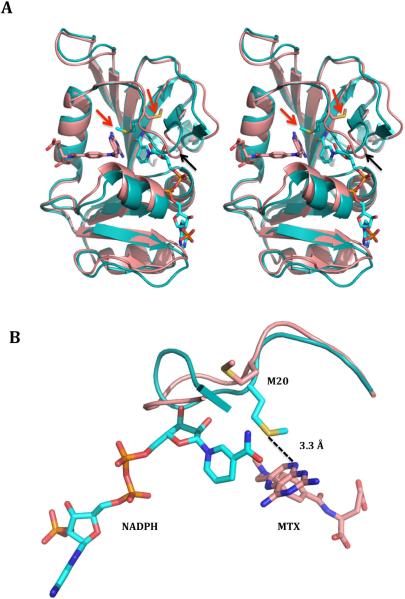
In an attempt to rationalize the ordered binding behavior, whereby all three inhibitors preferably bind to the NADPH-bound binary complex of the enzyme, structures of E. coli DHFRs in complex with NADPH (PDB ID: 1RX1) and methotrexate (PDB ID: 3DRC) were analyzed. When the structures were superimposed with their respective ligands, maximum change was noticed in the M20 loop that covers the active site where the hydride transfer reaction happens from NADPH to H2F (Fig 6A). Furthermore, it became clear that the dramatic change in the orientation of M20 from the MTX-bound form to NADPH-bound form of the enzyme might be the principal reason of why the inhibitors prefer the NADPH-form (Fig 6A and 6B). This shift in the orientation between the two structures makes the thio group of methionine come within hydrogen bonding distance of the N8 group on MTX in the NADPH-bound structure. Since AMPQD shares this substructure with MTX, it is highly likely that this interaction with M20 increases the affinity of the inhibitor for the enzyme manifold. Further, apart from this principal interaction, the whole M20 loop with several charged and bulky residues undergoes a change between the two structures. Other residues that can have possible roles in this preferential binding of inhibitor to NADPH-bound form are M16 and E17.
Fig 6.
(A) Stereo image of the superimposed cartoon representation for NADPH-bound (PDB ID: 1RX1) and methotrexate-bound (PDB ID: 3DRC) E. coli DHFR highlighting the movement of M20 loop. The NADPH-bound structure is shown in teal color, the methotrexate-bound structure is shown in salmon color, and the ligands are shown in a stick representation in the respective colors. The red arrows indicate the position of the flipped methionine 20 and the black arrows show the movement of M20 loop upon NADPH binding. (B) Zoomed in representation highlighting the almost 180° flip of the methionine side chain in NADPH-bound E. coli DHFR that brings the thio group of methionine within hydrogen bonding distance of the N8 group of methotrexate. The hydrogen bonds are shown in dotted representation. The figures were generated with MacPyMOL.
Slow onset tight binding inhibition: comparative study between AMPQD and PQD
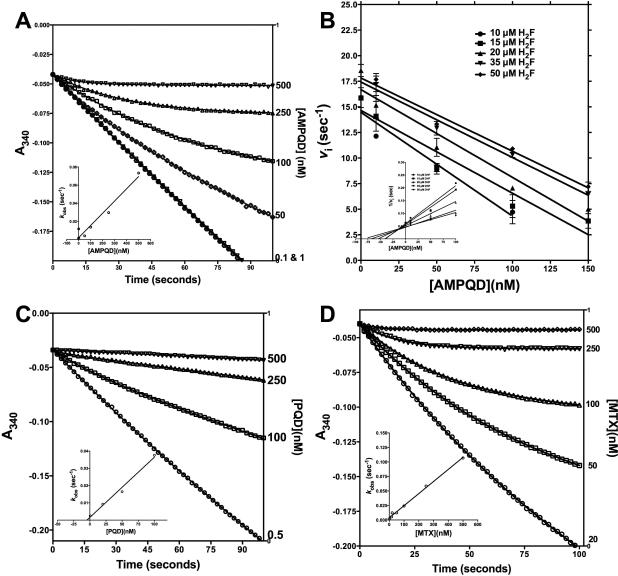
AMPQD and PQD bind to E. coli DHFR with the same kinetic behavior as the well-characterized inhibitor methotrexate. Both are competitive with respect to H2F and show preferential binding to the NADPH-bound form. Since it is known that methotrexate shows a slow-onset tight binding mode to the enzyme[22], we wanted to ascertain whether this also holds true for AMPQD and PQD. The Kiapp values obtained for both AMPQD and PQD using steady state methods show that the inhibitors are tight-binding inhibitors (Table 1). Progress curve analysis was used to determine whether the inhibitors showed slow-onset of tight binding in inhibiting E. coli DHFR. Upon addition of AMPQD, the rate of product formation decreased exponentially with time from an initial velocity (vi) to a steady state velocity (vs) (Fig 7A). Additionally, both vi, vs and the time required to reach vs decreased with increasing concentrations of the inhibitor, whereas kobs increased (Fig. 7A inset). This non-linear behavior in product formation in the presence of inhibitor complies with both the simple reversible slow-onset tight-binding inhibition model and inhibitor binding followed by isomerization model. However, upon assessing the effect of preincubation time with inhibitor on the steady state velocity of the reaction, whereby v/vi was plotted against time at various fixed inhibitor concentrations, the behavior conformed to the classic reversible slow onset inhibition in which no isomerization happens after the rapid formation of the initial E-I complex (data not shown). A simple reversible equilibrium between the enzyme and inhibitor with association and dissociation rate constants k3 and k-3, respectively, defines this model aptly as shown in Scheme I. This behavior is similar to that of any reversible inhibitor, except that the values of k3 and/or k-3 are smaller, leading to the slow-onset of inhibition. Further, as can be seen in the inset of Fig 7A, increase in kobs is linear with respect to inhibitor concentration conforming to the mechanism of reversible slow binding with slope equal to k3 and y-intercept equal to k-3. However, it should be noted that the measured value of k3 is apparent, since this rate constant is substrate concentration dependent as is seen in the vi vs. AMPQD plot at several fixed concentrations of H2F (Fig 7B). Hence, the apparent value of Ki (Kiapp) for an inhibitor of this type can be calculated from the ratio of k-3/k3app, which is equivalent to the ratio of the y-intercept/slope from the linear fit of the data plotted as in Fig 7A inset. The above analysis unequivocally proves that inhibition by AMPQD conforms to the classical slow-onset tight binding reversible inhibition.
Fig 7.
(A) Time-dependent inactivation of E. coli DHFR by 0–500 nM AMPQD. Inset shows the kobs plotted as a function of [AMPQD] (B) Direct plot for the effect of AMPQD on the initial velocity of H2F reduction of DHFR at various substrate concentrations. Inset shows the linearized plot of the data in (B). (C) Time-dependent inactivation of E. coli DHFR by 0–500 nM PQD. Inset shows the kobs plotted as a function of [PQD] (D) Time-dependent inactivation of E. coli DHFR by 0–500 nM MTX. Inset shows the kobs plotted as a function of [MTX]. The solid curves represent the best fit of the data to Eq. 7 for slow binding inhibition using of GraphPad Prism v 6.0e.
SCHEME I.
When the time-dependent inhibition data for PQD was analyzed, it was clear that the inhibition can be merely classified as classical tight-binding reversible inhibition since the curves show no hint of biphasic non-linearity even at high concentration of the inhibitor (Fig 7C). Since PQD is the parent molecule of AMPQD, the slow onset behavior of the latter compound implicates the 4-aminophenyl methyl substitution at the 7th position. It should also be noted that, unlike the inhibition mode of PQD, AMPQD inhibition of the enzyme is similar to that shown by methotrexate (Fig 7D). However, the physical basis for the slow-onset behavior of both AMPQD and methotrexate in DHFRs remains unexplained, and we propose that substitutions on the 7H-Pyrrolo[3,2-f] quinazoline-1,3-diamine for AMPQD and 2,4-Diaminopteridin on MTX might be the principal determinant of the slow-onset behavior.
Differential inhibition of human and E. coli DHFR
E. coli and human DHFR shares 28 % sequence identity and are structurally highly conserved (Fig S5). It is well demonstrated that inhibitors designed against prokaryotic DHFRs inhibit the activity of DHFRs from eukaryotic sources, given the high sequence and structural similarity of this protein across different evolutionary lineages[2]. There are several examples of such broad inhibition with the most prominent example being the commonly employed antifolate methotrexate, which is known to inhibit DHFRs from E. coli, rat and Plasmodium species [2]. However, differences in potency and mode of inhibition and the fact that antifolates target only rapidly proliferating cells like pathogenic microbes and tumors enables selective employment of them for specific treatment goals. With this aim, the hits obtained from the FINDSITEcomb study[15] were assessed for their inhibitory activity on human DHFR. Fig 1 shows the comparative histogram of inhibition demonstrating that both human and E. coli DHFR are inhibited to similar extent at 1mM inhibitor concentrations. To understand the inhibition further, the IC-50 values for the various molecules were estimated (Table 1 , Fig 2A and Fig S2). Marked differences in the potency of AMPQD and PQD were seen in their inhibition of the homologous proteins from humans and E. coli. AMPQD, the novel hit from our study, showed an IC-50 of 599.0 ± 7.2 nM for the human DHFR, which is about 3-fold less potent than the IC-50 for E. coli DHFR (Fig 2A and Table 1). The most dramatic difference was PQD's inhibition of human DHFR with an IC-50 value of 3.09 ± 0.17 μM, representing a 30-fold reduction in the potency of inhibition vis-à-vis E. coli DHFR (Fig S2A and Table 1). However, the IC-50 values for inhibition of human and E. coli DHFR by methotrexate were comparable, with a value of ~148 nM for the former and ~152 nM for the latter, respectively. For the rest of the molecules, the parameters are summarized in Table 1 and Fig S2B&C.
In order to further assess the effect of AMPQD on the human enzyme, competition experiments were performed by titrating H2F at several fixed concentration of AMPQD. The curves thus obtained were fit to various inhibition models with the best fit obtained for competitive inhibition (Fig 8A). Further, the double reciprocal LB plot shows intersection of the lines on the y-axis reinforcing the competitive inhibition (Fig 8B). This indicates that AMPQD inhibits the human enzyme by competitive displacement of substrate H2F, similar to its mode of action against the E. coli enzyme. However, the Ki value of 22.47 ± 3.66 nM for the human enzyme is almost 3-fold higher than that obtained for the E. coli enzyme (7.4 nM), indicative of poorer inhibition of the former.
Fig 8.
Inhibition kinetics of AMPQD (NSC309401) for human DHFR (A) Fit of the primary data to the competitive inhibition model for H2F titration at several fixed concentrations of AMPQD. (B) Double reciprocal Lineweaver-Burk plot of H2F titration at several fixed concentrations of AMPQD. The y-axis shows the kcat value. The experimental data points were fit to the model using the non-linear curve-fitting algorithm of GraphPad Prism v 6.0e.
Furthermore, upon analyzing the time-dependence of NADPH depletion in the presence of AMPQD for the human enzyme, no non-linearity was evident. This is indicative of neither slow onset nor slow dissociation of the inhibitor molecule to the human enzyme (Fig S6A). This is unlike the behavior displayed by the inhibitor AMPQD on the E. coli enzyme where prominent non-linearity was evident from the time-course of NADPH depletion in the presence of the inhibitor (Fig 7A). Likewise, PQD also didn't show any slow-onset of inhibition on the human enzyme exactly mirroring its behavior on E. coli DHFR (Fig S6B and 7B). The differences in the mode of inhibition by AMPQD of the human and E. coli enzyme is strongly indicative of differences in the binding site microenvironment between the two homologs. In fact, a recent study by Bhabha et al[24] has shown that despite high structural similarity, the dynamics of the active site loop movements varies substantially between human and E. coli DHFR. This, they hypothesize, results in markedly different inhibition by the product NADP+ of the two homologs (IC-50 of ~620 μM for human DHFR versus ~5 mM for E. coli DHFR). However, inhibition of human DHFR by the known antifolate methotrexate shows signs of pronounced non-linearity in the time-course curves of NADPH consumption indicating that the inhibitor retains its slow-onset behavior as seen with E. coli DHFR (Fig S6C). Moreover, the plot of kobs versus MTX concentration is hyperbolic, indicative of isomerization after inhibitor binding (Fig S6D). This is yet another behavior seen in the inhibition of the human enzyme that is markedly different from that shown for its E. coli counterpart whereby, in the case of the latter, there was no isomerization whatsoever as seen in the linear kobs versus [MTX] plots (insets of Fig 7D).
Discussion
Since DHFR is a pivotal enzyme in the synthesis of precursors of DNA, it has been the target for both anticancer and antibacterial drugs[2]. There has been a plethora of folate analogues that have been synthesized and tested for potential inhibitory activity against DHFRs from various sources[2, 25]. Principal among which are methotrexate, used prevalently as an anticancer drug, and trimethoprim, used as an antibacterial drug. In spite of multiple inhibitors designed against DHFRs from various organisms, detailed mechanistic characterization is available only for a few of these molecules. However, detailed kinetic characterization of an inhibitor is essential for designing efficient inhibitors with greater potency against the intended target, for determining the proper dose for testing on cellular/animal disease models and for understanding the pharmacodynamics. Further, since DHFR acquires rapid resistance to newly discovered antifolates, it is necessary to keep discovering novel small-molecules that inhibit this enzyme especially given the rise in instances of nosocomial E. coli infections in hospitalized patients. Several reports in literature highlight the fact that the incidence of E. coli mediated infections in hospitalized patients are on the rise, with one study showing multidrug resistant E. coli as the causative agent of UTI responsible for 40-50% of total nosocomial infections[7, 26-28]. Our study shows that AMPQD, a novel 7HPyrrolo[3,2-f] quinazoline-1, 3-diamine, is a potent inhibitor of the bacterial enzyme. Further, it also shows that compounds NSC80735, NSC55152 and NSC123458 show reasonable inhibition with μM IC-50 values and represent scaffolds amenable to modifications for development of novel DHFR inhibitors. While NSC80735 and NSC55152 contain concatenated nitrophenyl and aminophenyl groups, NSC123458 is a dibenzazepine that is a common structural scaffold in many antidepressants and analgesics.
In the current study, detailed steady state inhibition experiments on E. coli DHFR have shown that the small-molecules AMPQD and PQD bind to the H2F binding site and prefer the NADPH-bound binary form of the enzyme. This hints at sequential binding of the substrates NADPH followed by H2F, given that the small-molecules are structural analogues of H2F. This order of substrate and substrate analogue binding is in conformity with those proposed in the literature[4]. Further, it is well documented in literature that NADPH exerts a synergistic effect on folate analogues binding to DHFR [29]. NADPH enhances the binding of all the classical (phenyltriazine, DADMP) and slow-binding (MTX, TMP) inhibitors of DHFR, while it has no effect on either pyrimethamine or folate binding to DHFR. It has been reported that the degree of NADPH synergism can vary as much as 1000 fold for the binding among the different folate analogues[29]. We posit that since NADP and NADPH are present in equal concentrations in the prokaryotic cytosol (as against the eukaryotic cytosol where NADP is no more than 1% of the concentration of NADPH)[30]), for an inhibitor to be an effective drug, it might be advantageous for it to inhibit the product-forming ternary form of enzyme. Hence, inhibitors showing greater synergy may be preferable as potential lead candidates.
Tight-binding inhibitors are an important class where the affinity of the inhibitor for its cognate enzyme is so high that the equilibrium assumptions employed to compute the affinity of the inhibitor for the enzyme no longer remain valid. Further, a lot of well-known inhibitors extensively employed as drugs also display the property of slow-onset of inhibition because of the time-dependent establishment of equilibrium between the enzyme-bound form and free inhibitor. A few examples are captopril[31], an angiotensin inhibitor, Dup697[32], a COX2 inhibitor, and methotrexate [22, 33], a DHFR inhibitor. Methotrexate inhibits DHFR in a time-dependent manner involving the rapid formation of enzyme-NADPH-MTX complex that undergoes relatively slow, reversible isomerization to form a thermodynamically stable ternary enzyme complex resulting in enhanced inhibition. Hence, the MTX concentration required for inhibition is comparable to the enzyme concentration employed in steady-state kinetic studies. This type of inhibition is categorized as slow-on/slow-off tight binding inhibition[22]. Other folate analogues that exhibit slow-onset, tight-binding inhibition are 5-deazamethotrexate, aminopterin and trimethoprim, though the extent to which they bring about isomerization for slow dissociation differs significantly. Here, we demonstrate that AMPQD shows a slow-onset tight-binding behavior similar to that shown by methotrexate. However, upon analyzing the time-dependent inhibition curves, it becomes obvious that there is no pronounced isomerization, if any, which could be detected. Mainly, the kobs vs. inhibitor plots are linear both for AMPQD and MTX (Fig 7A and D) even at high inhibitor concentrations. Further, we could convincingly rule out the possibility that there is isomerization of the enzyme between two different forms with the inhibitor preferably binding to one form by the fact that kobs increased (rather than decreased) with increasing inhibitor. For DHFR from E. coli, it has been reported that all folate analogues that act as tight binding inhibitors exhibit slow binding characteristics. However slow-onset does occur without tight binding as observed with 5-deazafolate. To the best of our knowledge, for the first time, we show that PQD is a tight-binding inhibitor of DHFR not displaying any slow-binding behavior, as is evident from the linear time-course curves (Fig 7B).
Further, pronounced differences in the potency of inhibition of AMPQD and PQD for the human and E. coli homologue, with an order-of-magnitude higher affinity for the latter makes these molecules good candidates for development as effective antibiotics. It has been shown in the literature that recombinantly expressed human DHFR cannot complement DHFR-deficient E. coli cells, and this was ascribed to the differences in the dynamics and conformational plasticity of the active site loops across the two different DHFRs [24]. This conjecture is further validated by our studies showing similar site of binding on both human and E. coli DHFR for the inhibitor AMPQD (H2F binding site) (see Fig 4A, Fig 8A) albeit with different modes (slow-onset tight binding for the E. coli enzyme, while merely tight-binding for the human homologue as in Fig 7A and Fig S6A). Further, the isomerization of MTX after initial binding on the human enzyme, as evident in the nonlinearity of kobs versus MTX plot (Fig S6D), is completely absent from its binding to the E. coli DHFR (Fig 7D inset).
In conclusion, employing detailed inhibition kinetics, this study reports a set of novel and potent inhibitors of prokaryotic DHFR that have the potential to be developed as antibiotics for amelioration of conditions arising from bacterial infections. This, combined with the already reported antibacterial activity of AMPQD against two different strains of E. coli (Multi-drug resistant E. coli and DH5α), a strain of methicillin resistant S. aureus and a strain of vancomycin-resistant E. faecalis, makes this molecule a potential candidate for development as an antibiotic. Further, detailed kinetic assessment of the inhibition brought about by this molecule shows that it is competitive with respect to H2F and uncompetitive with respect to NADPH, with a preference of the inhibitor molecule for the NADPH-bound form of the enzyme. Furthermore, we show that while AMPQD is a slow-onset tight binding inhibitor of E. coli DHFR, similar to methotrexate, PQD is merely a tight-binding inhibitor with no slow-onset behavior. This detailed kinetic characterization of the inhibitors, by means of providing additional insight on the structure-activity relationship, paves the way for development of better antifolates.
Experimental Procedures
Reagents
All reagents and chemicals, unless mentioned otherwise, were of high quality and were procured from Sigma-Aldrich Co., USA, Amresco, or Fisher Scientific. E. coli dihydrofolate reductase was provided by Prof. Eugene Shakhnovich, Harvard University. The small molecule 7-[(4-aminophenyl) methyl]-7H-Pyrrolo[3,2-f]quinazoline-1,3-diamine (NSC309401), methotrexate (NSC740), 7H-Pyrrolo(3,2-f) quinazoline-1, 3-diamine (NSC339578), methylbezoprim (NSC382035), pralatrexate (NSC754230), pemetrexed (NSC698037), 6,7-bis(4-aminophenyl) pteridine-2,4-diamine (NSC61642), 2,2’-iminostilbene (NSC123458), benzoylpas (NSC159686), cibanaphthol RPH (NSC50690), NSC80735, NSC157522 and NSC55152 were provided by the Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI), National Institutes of Health (NIH). Dihydrofolate reductase assay kit (CS0340) was obtained from Sigma (Sigma-Aldrich, St. Louis, MO) and contained 0.1 units of human DHFR (D6566), dihydrofolic acid, methotrexate and NADPH.
Dihydrofolate reductase assay
The stock solutions of H2F and NADPH were reconstituted as per the manufacturer's instructions. DHFR catalyzes the transfer of a hydride from NADPH to H2F with an accompanying protonation to produce H4F. Overall, H2F is reduced to H4F and NADPH is oxidized to NADP+, resulting in a net decrease in the absorbance of NADPH at 340 nm. To understand the kinetics of E. coli DHFR and human DHFR, the rate of reduction of H2F to H4F was monitored by the decrease in absorbance at 340 nm for 100 seconds. The amount of product formed from the slope of initial velocity curves was computed using a molar extinction coefficient (ε) of 6.2 × 103 M-1cm-1 for β-NADPH at 340 nm[34]. The non-enzymatic hydrolysis of NADPH was normalized by monitoring the reaction in a double beam Hitachi U-2010 UV-Vis spectrophotometer (Hitachi High Technologies America, Inc., San Jose, CA, USA) with an appropriate blank. All the assays were carried out in the linear range of enzyme concentration. Assays were initiated with the addition of enzyme to the sample cuvette after zeroing the absorbance reading with respect to the reference cuvette. The initial velocities, where product formation was less than 5%, were measured for reaction mixtures containing 100 mM HEPES pH 7.3 at room temperature (~ 22 ° C).
To determine the Km and Vmax for H2F and NADPH, the respective substrate was titrated at fixed saturating concentration of the other, and the resultant velocities were plotted against substrate concentration and fit to equation 1 for one-site binding hyperbola
| (1) |
where d[P]/dt is the rate of product formation, Vmax is the maximum velocity, [S] is the substrate concentration and Km is the Michaelis-Menten constant for the substrate assayed.
All the measurements were performed in duplicate, and the error values indicated are standard deviations (S.D.). The concentration of E. coli DHFR used was 16.7 nM (see below for protocol of enzyme concentration estimation). Unless mentioned otherwise, all the data were fit using non-linear curve fitting subroutines of GraphPad Prism, version 4.0 (GraphPad Software, Inc., San Diego, CA).
Velocity-titration curves for enzyme concentration estimation
To interpret inhibition data without errors (see next section), an accurate estimate of catalytically active [Et] is essential. Methods suggested in the paper by Williams et al., 1979 [33], whereby velocity measurements after preincubation with a ligand (where enzyme-ligand complex is inactive and dissociates slowly) were followed to estimate the catalytically-active total enzyme concentration. Briefly, 0.1μg of E. coli DHFR and 0.16 μg of human DHFR was preincubated with various concentrations of MTX in 100 mM HEPES pH 7.3 and 60 μM NADPH for 300 seconds. The reaction was initiated with 50 μM H2F, and the resultant velocity was plotted as a function of MTX concentration (Fig S7A-D). The total concentration of the catalytically active enzyme is given by the intercept of the curve with the abscissa, which corresponds to 16.7 nM for of E. coli DHFR and 12.4 nM for human DHFR, respectively. The experiment was repeated with 600 seconds preincubation time giving identical results. The concentration was also estimated employing the molar extinction coefficient of 33460 M−1 cm−1 for the E. coli DHFR and 25440 M−1 cm−1 for human enzyme at 280 nm, respectively.
Inhibition kinetics
Various inhibitors were assessed for their inhibitory effect on the dihydrofolic acid reducing ability of E. coli DHFR and human DHFR. Initial inhibition was assessed in a reaction mixture containing 100 mM HEPES pH 7.3, 60 μM NADPH, 50 μM H2F and 1 mM of each inhibitor. 16.7 nM of E. coli DHFR and 12.4 nM of human DHFR were used in the assay mix. Subsequently, both the potency of the inhibitor and its affinity for the enzyme were computed by experimental IC-50 determination and competition assays to determine its Ki. IC-50 determination assays were carried out in 100 mM HEPES pH 7.3, 60 μM NADPH, 50 μM H2F and variable concentration of each inhibitor. The enzyme concentration was as specified above. The curves were fit to equation (2), where I is the inhibitor concentration, and y is the percentage of activity.
| (2) |
Furthermore, Kiapp were computed from the IC-50 curves by fitting them to the quadratic Morrison equation (3) for tight binding inhibition. This equation accounts for tight binding by doing away with the assumption that the free concentration of inhibitor equals the total concentration.
| (3) |
where vi represents velocity in the presence of inhibitor, v0 represents velocity in the absence of inhibitor, [E]T represents total enzyme, [I]T represents total inhibitor and K appi represents apparent Ki.
Experimental Ki value determinations were carried out by titrating the substrates H2F and NADPH, around their respective Km values, at various fixed concentrations of the inhibitors. The resulting [substrate] vs. velocity curves were fit to the models of competitive inhibition (equation 4), non-competitive inhibition (equation 5) and uncompetitive inhibition (equation 6) in order to discriminate between the different types of inhibition and to estimate the various inhibition constants (Ki).
Competitive:
| (4) |
Non-competitive:
| (5) |
Uncompetitive:
| (6) |
where v is the velocity of the reaction, Vmax is the maximum velocity, [S] is the substrate concentration, and [I] is the inhibitor concentration. Km is the Michaelis-Menten constant, and Ki is the inhibition constant. Visual assessment of the type of inhibition was undertaken by plotting the double reciprocal Lineweaver-Burk plot from experimental data points constituting the primary plot.
Progress Curve Analysis: Slow tight binding
The slow-onset inhibition of E. coli and human DHFR brought about by AMPQD, PQD and MTX was monitored by initiating the reaction with 16.7 nM of E. coli DHFR and 12.4 nM of human DHFR in assay mixtures containing 100 mM HEPES pH 7.3, 50 μM H2F, 60 μM NADPH and varying concentration of inhibitor (0-25 μM). The reactions were allowed to proceed until the progress curve became linear, indicating that the enzyme has attained steady state. To ensure that substrate depletion does not significantly affect the reaction rate, substrate concentrations greater than 10 times the respective Km values were used. Progress curves were analyzed as described previously[35]. Briefly, the resulting progress curves were fit to the integrated rate Eq. 7 for slow binding inhibition by nonlinear regression analysis.
| (7) |
Where, At and A0 are the absorbance at time t and time 0, kobs is the pseudo-first order rate constant for approach to the steady state, whereas νi and νs correspond to the initial and final slopes of the progress curve. Values for νi, νs, and kobs were obtained at each inhibitor concentration. Progress curves were approximately linear in the absence of added inhibitor. The values of kobs, vi , and vs obtained from the fit to equation 7 were replotted to obtain the koff and kon for inhibitor binding for a classical single-step inhibition mechanism in which rapid reversible binding of the inhibitor occurs to the enzyme.
Further, the patterns were also analyzed for possible two-step inhibition mechanism. In this case, which signifies a second slow step of isomerization after inhibitor binding to form the final enzyme-inhibitor complex, the following equation was used to fit the replot of kobs vs inhibitor to gauge its non-linearity.
| (8) |
where k4 and k-4 represent the forward and reverse rate constants for the isomerization step.
Thermal shift assay methodology
High throughput thermal shift assays were carried out following established guidelines [36, 37]. Briefly, thermal melt curves for proteins were obtained from samples aliquoted in 96-well plates using a RealPlex quantitative PCR instrument from Eppendorf (Eppendorf, NY, USA) with 5 X Sypro orange dye as the fluorescent probe (Invitrogen). (λexcitation is 465 nm and λemission is 580 nm). A heating gradient of 1 °C/min from 25 °C to 75 °C was used. The thermal melt experiments were done in 100 mM HEPES pH 7.3 and 150 mM NaCl with 20 μl total volume of the reaction mix. The concentration of AMPQD was varied from 0 nM-500 nM at 5 μM of E. coli DHFR. All experiments were done in duplicate, with the mean value considered for further analysis.
The curves were fit to Boltzmann's equation (Eq. 9) for estimating the Tm (melting temperature) from the observed intensity of fluorescence, I.
| (9) |
Imin and Imax are the minimum and maximum intensities; a denotes the slope of the curve at the transition midpoint temperate, Tm [36]. Thermodynamic parameters were estimated as specified in the previous literature [15].
Supplementary Material
Acknowledgements
This project was funded by GM-37408 and GM-48835 of the Division of General Medical Sciences of the NIH. The authors wish to thank Prof. Eugene Shakhnovich, Harvard University, for providing purified E. coli DHFR protein. We would also like to thank the Developmental Therapeutics Program of the National Cancer Institute for providing the small molecules used in this study.
Abbreviations
- DHFR
dihydrofolate reductase
- AMPQD
7-[(4-aminophenyl) methyl]-7H-pyrrolo[3,2-f] quinazoline-1, 3-diamine
- PQD
7H-pyrrolo[3,2-f] quinazoline-1, 3-diamine
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced
- H2F
dihydrofolate
- MTX
methotrexate
Footnotes
Author Contributions
BS conceived of the study, participated in its design, carried out the experiments, analyzed and interpreted the results, and drafted the manuscript. JS conceived of the study, participated in its design and coordination, provided appropriate resources, helped analyze the data, and was involved in drafting and critically reviewing the manuscript. All authors read and approved the final manuscript.
References
- 1.Liu CT, Francis K, Layfield JP, Huang X, Hammes-Schiffer S, Kohen A, Benkovic SJ. Escherichia coli dihydrofolate reductase catalyzed proton and hydride transfers: Temporal order and the roles of Asp27 and Tyr100. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18231–6. doi: 10.1073/pnas.1415940111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schweitzer BI, Dicker AP, Bertino JR. Dihydrofolate reductase as a therapeutic target. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1990;4:2441–52. doi: 10.1096/fasebj.4.8.2185970. [DOI] [PubMed] [Google Scholar]
- 3.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 4.Fierke CA, Johnson KA, Benkovic SJ. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987;26:4085–92. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 5.Cayley PJ, Dunn SM, King RW. Kinetics of substrate, coenzyme, and inhibitor binding to Escherichia coli dihydrofolate reductase. Biochemistry. 1981;20:874–9. doi: 10.1021/bi00507a034. [DOI] [PubMed] [Google Scholar]
- 6.Stone SR, Morrison JF. Kinetic mechanism of the reaction catalyzed by dihydrofolate reductase from Escherichia coli. Biochemistry. 1982;21:3757–65. doi: 10.1021/bi00259a006. [DOI] [PubMed] [Google Scholar]
- 7.Bean DC, Krahe D, Wareham DW. Antimicrobial resistance in community and nosocomial Escherichia coli urinary tract isolates, London 2005- 2006. Annals of clinical microbiology and antimicrobials. 2008;7:13. doi: 10.1186/1476-0711-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst P, Ouellette M. New mechanisms of drug resistance in parasitic protozoa. Annual review of microbiology. 1995;49:427–60. doi: 10.1146/annurev.mi.49.100195.002235. [DOI] [PubMed] [Google Scholar]
- 9.Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat. 2012;15:183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Hagner N, Joerger M. Cancer chemotherapy: targeting folic acid synthesis. Cancer Manag Res. 2010;2:293–301. doi: 10.2147/CMR.S10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chio LC, Queener SF. Identification of highly potent and selective inhibitors of Toxoplasma gondii dihydrofolate reductase. Antimicrob Agents Chemother. 1993;37:1914–23. doi: 10.1128/aac.37.9.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan J, Zhang Q, O'Neil M, Obaldia N, 3rd, Ager A, Gerena L, Lin AJ. Antimalarial activities of new pyrrolo[3,2-f]quinazoline-1,3-diamine derivatives. Antimicrob Agents Chemother. 2005;49:4928–33. doi: 10.1128/AAC.49.12.4928-4933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuyper LF, Baccanari DP, Jones ML, Hunter RN, Tansik RL, Joyner SS, Boytos CM, Rudolph SK, Knick V, Wilson HR, Caddell JM, Friedman HS, Comley JC, Stables JN. High-affinity inhibitors of dihydrofolate reductase: antimicrobial and anticancer activities of 7,8-dialkyl-1,3-diaminopyrrolo[3,2-f]quinazolines with small molecular size. J Med Chem. 1996;39:892–903. doi: 10.1021/jm9505122. [DOI] [PubMed] [Google Scholar]
- 14.Zolli-Juran M, Cechetto JD, Hartlen R, Daigle DM, Brown ED. High throughput screening identifies novel inhibitors of Escherichia coli dihydrofolate reductase that are competitive with dihydrofolate. Bioorg Med Chem Lett. 2003;13:2493–6. doi: 10.1016/s0960-894x(03)00480-3. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan B, Zhou H, Kubanek J, Skolnick J. Experimental validation of FINDSITE(comb) virtual ligand screening results for eight proteins yields novel nanomolar and micromolar binders. J Cheminform. 2014;6:16. doi: 10.1186/1758-2946-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan B, Skolnick J, Zhou H. Molecules with potent DHFR binding affinity and antibacterial activity in (USPTO, ed) Georgia Tech Research Corporation; (Atlanta, GA, US), United States: 2014. [Google Scholar]
- 17.Murakami C, Ohmae E, Tate S, Gekko K, Nakasone K, Kato C. Cloning and characterization of dihydrofolate reductases from deep-sea bacteria. Journal of biochemistry. 2010;147:591–9. doi: 10.1093/jb/mvp206. [DOI] [PubMed] [Google Scholar]
- 18.Stone SR, Morrison JF. Dihydrofolate reductase from Escherichia coli: the kinetic mechanism with NADPH and reduced acetylpyridine adenine dinucleotide phosphate as substrates. Biochemistry. 1988;27:5493–9. doi: 10.1021/bi00415a016. [DOI] [PubMed] [Google Scholar]
- 19.Cha S. Tight-binding inhibitors-I. Kinetic behavior. Biochemical pharmacology. 1975;24:2177–85. doi: 10.1016/0006-2952(75)90050-7. [DOI] [PubMed] [Google Scholar]
- 20.Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochimica et biophysica acta. 1969;185:269–86. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 21.Williams JW, Morrison JF. The kinetics of reversible tight-binding inhibition. Methods in enzymology. 1979;63:437–67. doi: 10.1016/0076-6879(79)63019-7. [DOI] [PubMed] [Google Scholar]
- 22.Stone SR, Montgomery JA, Morrison JF. Inhibition of dihydrofolate reductase from bacterial and vertebrate sources by folate, aminopterin, methotrexate and their 5-deaza analogues. Biochemical pharmacology. 1984;33:175–9. doi: 10.1016/0006-2952(84)90472-6. [DOI] [PubMed] [Google Scholar]
- 23.Tahar R, de Pecoulas PE, Basco LK, Chiadmi M, Mazabraud A. Kinetic properties of dihydrofolate reductase from wild-type and mutant Plasmodium vivax expressed in Escherichia coli. Molecular and biochemical parasitology. 2001;113:241–9. doi: 10.1016/s0166-6851(01)00230-4. [DOI] [PubMed] [Google Scholar]
- 24.Bhabha G, Ekiert DC, Jennewein M, Zmasek CM, Tuttle LM, Kroon G, Dyson HJ, Godzik A, Wilson IA, Wright PE. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nature structural & molecular biology. 2013;20:1243–9. doi: 10.1038/nsmb.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abraham A, McGuire JJ, Galivan J, Nimec Z, Kisliuk RL, Gaumont Y, Nair MG. Folate analogues. 34 Synthesis and antitumor activity of nonpolyglutamylatable inhibitors of dihydrofolate reductase. Journal of medicinal chemistry. 1991;34:222–7. doi: 10.1021/jm00105a035. [DOI] [PubMed] [Google Scholar]
- 26.Schaberg DR, Culver DH, Gaynes RP. Major trends in the microbial etiology of nosocomial infection. The American journal of medicine. 1991;91:72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 27.Caini S, Hajdu A, Kurcz A, Borocz K. Hospital-acquired infections due to multidrug-resistant organisms in Hungary, 2005-2010. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2013;18 [PubMed] [Google Scholar]
- 28.Hassan SA, Jamal SA, Kamal M. Occurence of multidrug resistant and ESBL producing E coli causing urinary tract infections. journal of basic and applied sciences. 2011;7:39–43. [Google Scholar]
- 29.Stone SR, Morrison JF. Mechanism of inhibition of dihydrofolate reductases from bacterial and vertebrate sources by various classes of folate analogues. Biochimica et biophysica acta. 1986;869:275–85. doi: 10.1016/0167-4838(86)90067-1. [DOI] [PubMed] [Google Scholar]
- 30.Appleman JR, Beard WA, Delcamp TJ, Prendergast NJ, Freisheim JH, Blakley RL. Unusual transient- and steady-state kinetic behavior is predicted by the kinetic scheme operational for recombinant human dihydrofolate reductase. The Journal of biological chemistry. 1990;265:2740–8. [PubMed] [Google Scholar]
- 31.Baudin B, Beneteau-Burnat B. Mixed-type inhibition of pulmonary angiotensin I-converting enzyme by captopril, enalaprilat and ramiprilat. Journal of enzyme inhibition. 1999;14:447–56. doi: 10.3109/14756369909030335. [DOI] [PubMed] [Google Scholar]
- 32.Dannhardt G, Kiefer W. Cyclooxygenase inhibitors--current status and future prospects. European journal of medicinal chemistry. 2001;36:109–26. doi: 10.1016/s0223-5234(01)01197-7. [DOI] [PubMed] [Google Scholar]
- 33.Williams JW, Morrison JF, Duggleby RG. Methotrexate, a highaffinity pseudosubstrate of dihydrofolate reductase. Biochemistry. 1979;18:2567–73. doi: 10.1021/bi00579a021. [DOI] [PubMed] [Google Scholar]
- 34.Horecker BL, Kornberg A. The extinction coefficients of the reduced band of pyridine nucleotides. The Journal of biological chemistry. 1948;175:385–90. [PubMed] [Google Scholar]
- 35.Rawat R, Whitty A, Tonge PJ. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13881–6. doi: 10.1073/pnas.2235848100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 37.Crowther GJ, He P, Rodenbough PP, Thomas AP, Kovzun KV, Leibly DJ, Bhandari J, Castaneda LJ, Hol WG, Gelb MH, Napuli AJ, Van Voorhis WC. Use of thermal melt curves to assess the quality of enzyme preparations. Analytical biochemistry. 2010;399:268–75. doi: 10.1016/j.ab.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.