Abstract
Objective
Whole-body protein metabolism is dysregulated with obesity. Our goal was to determine if activity and expression of major protein degradation pathways are compromised specifically in human skeletal muscle with obesity.
Methods
We utilized primary Human Skeletal Muscle cell (HSkM) cultures since cellular mechanisms can be studied absent of hormones and contractile activity that could independently influence metabolism. HSkM from 10 lean (BMI ≤ 26.0 kg/m2) and 8 severely obese (BMI ≥ 39.0) women were examined basally and when stimulated to atrophy (serum and amino acid starvation).
Results
HSkM from obese donors had a lower proportion of type I myosin heavy chain and slower flux through the autophagic/lysosomal pathway. During starvation, flux through the ubiquitin-proteasome system diverged according to obesity status, with a decrease in the lean and an increase in HSkM from obese subjects. HSkMC from the obese also displayed elevated proteasome activity despite no difference in proteasome content. Atrophy-related gene expression and myotube area were similar in myotubes derived from lean and obese individuals under basal and starved conditions.
Conclusions
Our data indicate that muscle cells of the lean and severely obese have innate differences in management of protein degradation, which may explain their metabolic differences.
Keywords: Protein Metabolism, Skeletal Muscle, Obesity, Human, Sarcopenia
Introduction
Obesity is a major risk factor for insulin resistance, cardiovascular disease, many cancers, and premature mortality (1). These risks are compounded by coincident low levels of muscle mass, which has been identified as “sarcopenic obesity” (2–5). Because severe obesity is accompanied by changes in circulating factors (e.g. insulin and amino acids) that influence protein synthesis and degradation (6), it is not surprising that whole-body protein metabolism is dysregulated in obese humans (7). However, it is unclear whether muscles of the obese exhibit a metabolic program, independent of neural and/or hormonal factors, where protein metabolism is compromised.
Protein degradation is a major regulator of skeletal muscle mass (total protein content) and function (regulating the amount/mass of specific proteins or organelles). Proteins are degraded mainly by the ubiquitin-proteasome system (UPS) and/or the autophagic/lysosomal system (8). Proteolysis of intracellular proteins through the UPS is dictated primarily by proteasome activity and protein polyubiquitination (9). Autophagy degrades the bulk of the remainder of cellular proteins, especially macromolecules and organelles such as mitochondria (8). Because the UPS and autophagy preferentially target different proteins, the relative flux through these systems results in an ordered loss of proteins (10, 11) and, therefore, is an important regulator of the functional characteristics (i.e. metabolic and/or contractile) of skeletal muscle.
The primary goal of this study was to determine whether skeletal muscle of the severely obese exhibit innate differences in activity and expression of the major proteolytic pathways. Accordingly, we examined primary human skeletal muscle (HSkM) cells from lean and severely obese women as 1) HSkM maintain metabolic characteristics of the donor (12–14) and 2) cell culture eliminates circulating factors (i.e. elevated free fatty and amino acids, hormonal levels) or neural input (muscular recruitment) that can differ with obesity and can confound in vivo studies (7).
Methods
Human subjects and muscle biopsies
Muscle biopsies were collected from the vastus lateralis of lean (BMI range 18.5–26.0) and severely obese (BMI range 39.0–57.3) women after an overnight fast. All subjects were free of overt disease, did not smoke, and had not participated in regular exercise in the past 6 months. Blood samples were collected during the same visit, and plasma glucose and insulin measured. Homeostatic model assessment of insulin resistance (HOMA-IR) was calculated as fasting plasma glucose × fasting plasma insulin/22.5 (15). Written, informed consent was obtained from all subjects; the study was approved by the East Carolina University and Medical Center Institutional Review Board.
Cell culture
Primary Human Skeletal Muscle (HSkM) myoblasts were isolated, expanded, and maintained as previously described (13). Upon reaching confluence, growth media (DMEM with 10% fetal bovine serum and SkGM Singlequots, Lonza) was switched to differentiation media (DMEM with 2% horse serum) to induce differentiation into multinucleated myotubes. All experiments were performed on cells passaged 4–5 times and on myotubes differentiated for 6–8 days.
For experiments using pooled myotubes (proteolytic flux, proteasome activity and proteasome content), myoblasts of each individual were cultured separately until approximately 75 % confluent, then seeded together in equal ratios in multi-well plates, allowed to adhere to the plate overnight, then switched to differentiation media to eliminate effects of differing growth rates between individual cultures. Data were collected from two independent pools (n=3 per pool), to confirm accuracy of results.
To study protein degradation responses, myotubes were starved of serum and amino acids (cultured in Hanks balanced salt solution), which dramatically accelerates protein degradation (16, 17). Alternatively, myotubes were treated with 100nM insulin-like growth factor 1 (IGF-1), which suppresses protein degradation in C2C12 myotubes (18) and increases the rate of protein synthesis in HSkM (Figure S1). Both treatments were continued for up to 30 h.
Myosin heavy chain analysis
Myosin heavy chain (MHC) isoforms were separated by high-resolution electrophoresis (19). In brief, total protein was separated overnight using 35% v/v glycerol, 8% w/v acrylamide-N,N’-methylenebisacrylamide (9:1) gels. Proteins were silver stained (Pierce Silver Stain Kit) and digital images analyzed for band intensities. Identities of specific MHC isoforms were confirmed by immunoblotting. Since this method does not reliably resolve embryonic MHC from type IIa or IIx, these have been combined in our analysis.
Protein degradation rate
Degradation rates of long-lived proteins were determined as previously described (17, 20). Proteins were radiolabeled with L-[3,5-3H]-tyrosine for 24h, chased with non-radioactive tyrosine for 2h, treatment (e.g. no serum/amino acids) started, and then 3–4 media samples collected over time. Since radioactivity release is curvilinear over 30 hours (20), we assessed degradation over two time periods, either 0–5h or 20–30h. The rates over these limited periods, calculated from the regression line of serial samples, were highly linear (R2 > 0.97) for all individuals.
To differentiate flux through the UPS and autophagic/lysosomal system, protein degradation was measured in the absence or presence of the proteasome inhibitor PS-341 (Bortezomib/Velcade, 1.0µM) or the lysosome acidification inhibitor Concanamycin A (0.1µM) (Figure S2) as previously described (17, 20). Relative flux through the pathways was determined by subtracting the inhibitor-sensitive degradation rate from the basal degradation rate (ratebasal – rateinhibited = degradation rate attributable to inhibited pathway). Differences between lean and obese myotubes were determined by subtraction (Δ Degradation rate = degradation rateobese – degradation ratelean).
Proteasome activity
Proteasomes were collected and activity of the chymotrypsin-like site was measured as described by Kisselev and Goldberg (21) using the fluorogenic peptide substrate Suc-LLVY-amc (100µM final, Bachem). Specificity was confirmed by treating extracts with the proteasome inhibitor PS-341, which eliminated over 95% of fluorescence. Results were normalized to total protein content (DC protein assay, BioRad) to control for proteasome content.
Protein content and western blots
Myotube proteins were collected in radio-immunoprecipitation assay (RIPA) buffer containing a protease inhibitor cocktail (Roche Complete). Equal amounts of total protein were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Equal loading and transfer were confirmed by MemCode Reversible Protein Stain Kit (Thermo Scientific). Membranes were incubated overnight with primary antibodies from Cell Signaling (FoxO3, #2497), Enzo Life Sciences (19S Rpt5/S6a, #PW8770; 19S Rpt6, #PW9265; 20S β1 subunit, #PW8140; 20S β5 subunit, #PW8895), Developmental Studies Hybridoma Bank (Embryonic MHC, #F1.652; type I MHC, #BA-F8), or Abcam (Fast MHC, #ab7784). Secondary antibodies, conjugated to horseradish peroxidase, were detected using an enhanced chemiluminescent substrate (Millipore). Band intensities were captured using a Bio-Rad Chemi Doc XRS and analyzed using Image Lab (BioRad) or ImageJ64 (NIH) software.
Real-time PCR
Total RNA was extracted using TRIzol Reagent (Life Technologies), reverse transcribed (Quanta Biosciences), and real-time PCR performed (SYBR green, Applied Biosystems 7900HT) (20). Fold expression of genes was calculated using the relative standard curve method with Large Ribosomal Protein (RPLPO), which was not different between groups or treatments, as the endogenous control. Amplification efficiency was near 100% for all genes. RT-PCR primers (Table 2) were generated using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and specificity confirmed by conventional PCR followed by agarose gel electrophoresis.
Table 2.
Primers used for RT-PCR
| Gene | Sequence (5' → 3’) | ||
|---|---|---|---|
| TGF-β pathway | Myostatin | Fwd | AAGACCAAAATCCCTTCTGGA |
| Rev | CTGTAACCTTCCCAGGACCA | ||
| SMAD2 | Fwd | TGTTCTTACCAAAGGCAGCA | |
| Rev | CATCGGAAGAGGAAGGAACA | ||
| SMAD3 | Fwd | GTAGCTCGTGGTGGCTGTG | |
| Rev | AACACCAAGTGCATCACCAT | ||
| Ubiquitin- proteasome system |
FoxO3 | Fwd | CTCTTGCCAGTTCCCTCATT |
| Rev | CTTCAAGGATAAGGGCGACA | ||
| Atrogin-1 | Fwd | TCAGGGATGTGAGCTGTGAC | |
| Rev | GGGGGAAGCTTTCAACAGAC | ||
| MuRF-1 | Fwd | CTTCGTGCTCCTTGCACAT | |
| Rev | ATCGTCACGGAGTGTACGG | ||
| Autophagy | LC3B | Fwd | TATCACCGGGATTTTGGTTG |
| Rev | GAGAAGACCTTCAAGCAGCG | ||
| ATG4B | Fwd | AGTATCCAAACGGGCTCTGA | |
| Rev | ACTGGGAAGATGGACGCAG | ||
| Gabarapl-1 | Fwd | TGGCTTTTGGAGCCTTCTCT | |
| Rev | CCATCCCTTTGAGTATCGGA | ||
| Control | RPLPO | Fwd | AGGCGTCCTCGTGGAAGTGACA |
| Rev | TGCTGCATCTGCTTGGAGCCC | ||
Myotube area
Myotube area was quantified by analyzing the amount of MHC covering the culture area using the method of Trendelenburg et al. (22) and ImageJ64 with a self-designed macro (Figure S3). See Supplementary Information for details (Method S1).
Statistical analysis
Statistical analysis for gene expression, degradation rate, protein content, and myotubes area were determined by two-way repeated measures ANOVA with Sidak post-test comparisons. Given our data for the protein degradation assay, a power analysis revealed that we would need 6–7 observations per group to have an 80% chance of achieving a statistically significant difference with α=0.05. Data from pooled myotubes (UPS flux, autophagic/lsysosomal flux, and proteasome activity) were determined by two-way ANOVA with Sidak post-test comparisons. Statistical analysis was performed using SPSS 19, and significance was set at α=0.05. Data is presented as mean ± SE.
Results
Subject Characteristics
Severely obese subjects were significantly heavier and had increased plasma insulin concentration and HOMA-IR than their lean counterparts (Table 1). All subjects were relatively young, but the mean age for the obese group was higher than the lean. However, protein degradation rate was not correlated with age under basal or starved conditions, indicating that age was not a confounding variable (Figure S4).
Table 1.
Subject characteristics
| Lean (n = 10) | Obese (n = 8) | |
|---|---|---|
| BMI, kg/m2 | 22.9 ± 0.8 | 46.9 ± 2.1* |
| Height, cm | 164.3 ± 2.0 | 166.9 ± 2.5 |
| Mass, kg | 61.9 ± 2.3 | 130.4 ± 9.9* |
| Age, y | 24.2 ± 1.5 | 34.1 ± 3.3* |
| Fasting glucose, (mg/dl) | 85.2 ± 2.6 | 93.7 ± 2.9 |
| Fasting insulin (µIU/ml) | 9.3 ± 1.1 | 18.2 ± 2.2* |
| HOMA-IR | 1.65 ± 0.2 | 4.27 ± 0.6* |
Data presented as mean ± SEM.
Significantly different from lean (P < 0.05).
Myosin heavy chain isoform expression is altered in myotubes of the severely obese
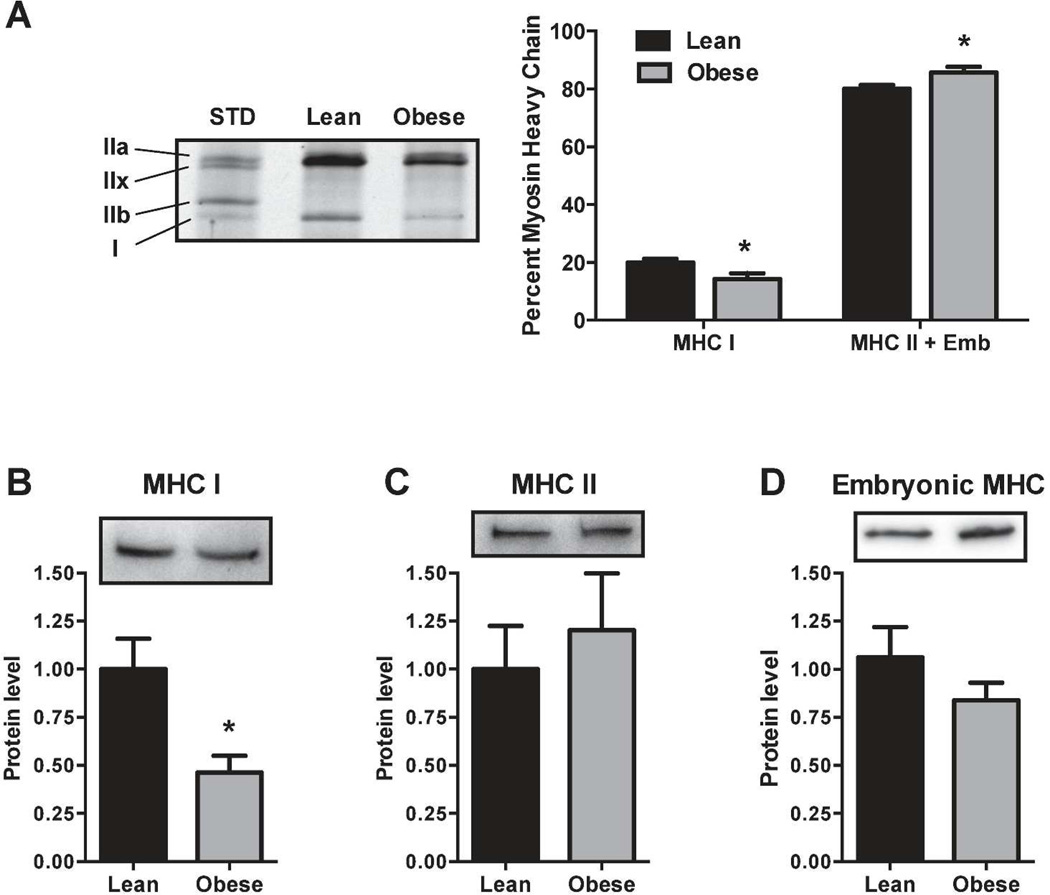
Skeletal muscle biopsies of the severely obese exhibit a lower proportion of the slow-twitch type I myosin heavy chain (MHC) compared to lean individuals (23). Therefore, to determine if this phenotypic difference is retained in cell culture, we examined MHC isoform protein expression in myotubes from lean and obese donors. Compared to the lean, the amount of type I MHC protein was lower in the severely obese when expressed either as percent of total MHC (20.0% vs 14.3%, Figure 1A) or by western blot (1.00 vs 0.46, Figure 1B). Neither type II (Figure 1C) nor embryonic MHC protein (Figure 1D) content was significantly different between the lean and obese.
Figure 1.
(a) Total protein was separated by high-resolution electrophoresis, silver stained, and MHC bands quantified. Type I MHC is a smaller fraction of total MHC in the obese than in the lean. (b) Using western blots, type I MHC protein is less abundant, but (c) type II MHC protein and (d) embryonic MHC protein are not different between lean and obese. *P < 0.05 versus lean.
Differential proteolytic flux through proteasome and autophagy/lysosome
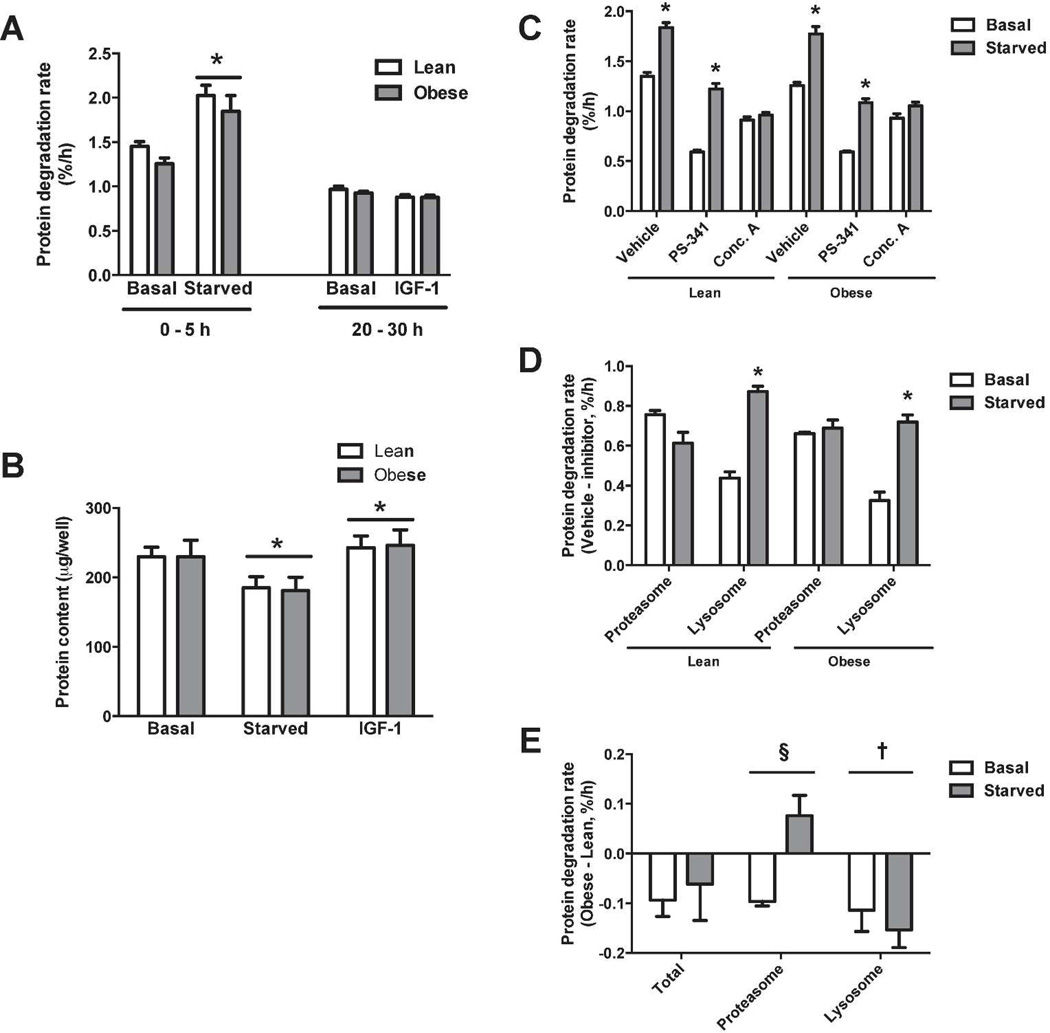
Protein degradation rates in response to hypertrophic or atrophic stimuli were determined. Starvation significantly increased protein degradation rate (~40%), and rates were similar for both groups under basal and starved conditions (Figure 2A). IGF-1 treatment had no measurable effect on degradation rates. In agreement with basal rates of degradation, myotubes from the lean and severely obese had similar rates of protein synthesis (Figure S5) and contained similar total protein under basal conditions (Figure 2B). Following starvation for 24h, protein content was reduced (~20%) in both lean and severely obese (Figure 2B). Conversely, IGF-1 increased protein content (~6%) similarly in myotubes from the lean and severely obese, presumably due to increased rate of protein synthesis (Figure S1).
Figure 2.
(a) Myotubes were subjected to starvation (removal of serum and amino acids) or IGF-1 at time 0. Protein degradation rate was calculated over the first 5 hours (starved) or from 20–30 hours (IGF-1). Each data point is the mean of four replicates. (b) Myotubes were treated for 24 h and protein content determined. (c) Protein degradation rates in the presence of inhibitors of the proteasome (PS-341) or of lysosomal acidification (Conc. A). (d) Inhibitor-sensitive proteolysis was determined by subtracting the degradation rate in the presence of inhibitors from vehicle-treated controls. (e) Differences in protein degradation rates were determined by subtracting inhibitor sensitive values of lean from obese (Δ Degradation rate = degradation rateobese – degradation ratelean). *Significant main effect of treatment (P < 0.05). †Significant main effect between groups (P < 0.05). §Significant interaction effect (P < 0.05).
Since the UPS and autophagic/lysosomal pathway preferentially target specific proteins and/or organelles for degradation (8), we sought to determine how obesity affects flux through these pathways. Proteolysis rates were measured in the presence or absence of specific inhibitors of the proteasome (PS-341) or of lysosome acidification (concanamycin A) (Figure 2C). As expected, both PS-341 and concanamycin A significantly reduced protein degradation rate compared to the vehicle control. To determine the rate of flux through each pathway, we subtracted inhibitor-sensitive rates from the vehicle-treated rate (Figure 2D). Under basal conditions, the UPS accounted for 56% (lean) and 53% (obese) of total proteolysis. During starvation, the rate of UPS-mediated protein degradation did not significantly change in either the lean or obese groups. However, during starvation, the relative contribution of the autophagic /lysosomal pathway significantly increased from 33 to 48% (lean) and 25 to 41% (obese) of total proteolysis. There was a significant group effect for myotubes of the severely obese to have slower flux (basal −26%, starved −18%) through the autophagic/lysosomal pathway. Surprisingly, starvation reduced UPS flux by 19% in myotubes of the lean myotubes, but increased UPS flux by 4% in myotubes of the severely obese, indicating a shift toward UPS-mediated protein degradation in myotubes of the severely obese during starvation. The magnitude of the difference in degradation rates between obese and lean are depicted in Figure 2E.
Proteasome specific activity is increased in skeletal muscle with obesity
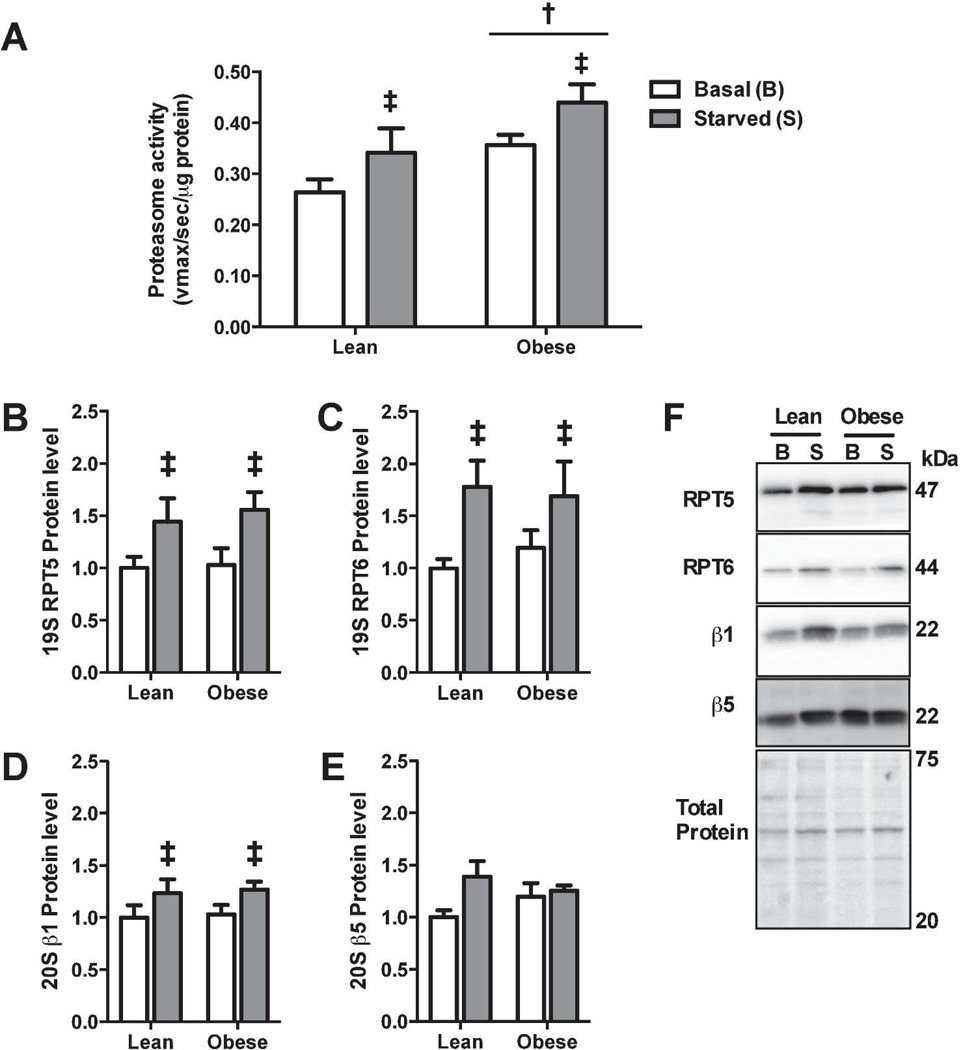
Because of the shift in the rates of protein degradation through the proteasome, we next examined how severe obesity affects proteasome activity. Proteasome activity was greater in myotubes of the obese compared to lean (Figure 3A). Furthermore, starvation increased proteasome activity in myotubes from both the lean (30%) and severely obese (23%). However, there was no significant interaction between obesity and starvation on proteasome capacity.
Figure 3.
(a) Chymotrypsin-like activity of the proteasome was assessed by the fluorogenic substrate Suc-LLVY-amc. (b–e) Western blot and protein abundance of proteasomal subunit proteins. (f) Representative blots of proteasomal subunits and membrane stained for total protein to demonstrate equal loading. ‡Significant main effect (basal versus starved, p<0.05). †Significant main effect (lean versus obese, P < 0.05).
To test whether the increase in proteasome activity was due to an increase in proteasome content, western blots for several proteasome subunits were performed. Starvation significantly (P<0.05) increased the protein levels of 19S subunits, RPT5 (Figure 3B) and RPT6 (Figure 3C), and of the 20S subunit, β1 (Figure 3D), but not the 20S subunit β5 (P=0.065) (Figure 3E). However, proteasome subunits were not differentially expressed between the lean and severely obese. Therefore, the increase in proteasome activity with obesity is likely not due to an increase in proteasome content.
Atrophy-related gene expression is similar between lean and obese
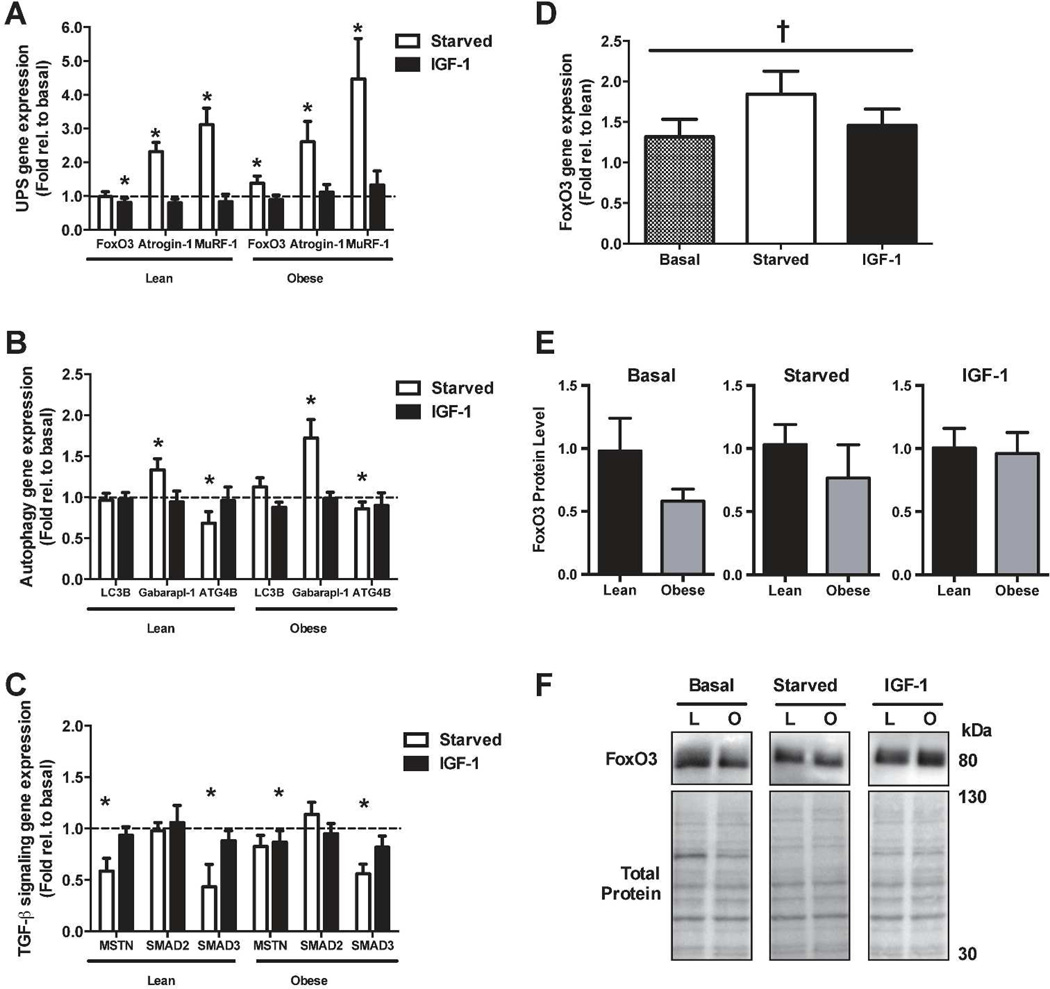
In order to determine how obesity affects the transcriptional response to stimuli that modulate protein degradation, we performed RT-PCR after 24h of starvation or IGF-1 treatment. Gene expression of the ubiquitin ligases Atrogin-1 (lean: 2.3-fold, obese: 2.6-fold) and MuRF-1 (lean: 3.1-fold and obese: 4.5-fold) increased following starvation (Figure 4A). The expression of FoxO3, a transcription factor that directly increases transcription of both Atrogin-1 (24) and MuRF-1 (16), was increased following starvation in myotubes from the severely obese (1.4-fold) but was unchanged in myotubes from lean subjects. IGF-1 treatment significantly reduced gene expression of FoxO3 (lean: 0.8 and obese: 0.9-fold), but did not affect MuRF-1 or Atrogin-1 gene expression.
Figure 4.
Real-time PCR for (a) genes involved in the ubiquitin-proteasome system (UPS), (b) the autophagic/lysosomal pathway, and (c) Transforming Growth Factor-β (TGF-β) signaling. (d) FoxO3 gene expression of obese myotubes expressed as fold relative to lean. (e) FoxO protein levels. (f) Representative blots of FoxO3 and membrane stained for total protein to demonstrate equal loading. *Significant effect versus basal (P < 0.05). †Significant main effect (obese versus lean, P < 0.05).
Starvation also significantly altered expression of genes involved in the autophagic/lysosomal pathway (Figure 4B). Specifically, mRNA content of Gabarapl-1, a gene essential for autophagosome formation, was significantly increased (lean: 1.3-fold and obese: 1.7-fold), while ATG4B, another regulator of autophagosome formation, was significantly reduced in response to starvation (lean: 0.7-fold and obese 0.9-fold). LC3B gene expression, which normally increases to sustain autophagy flux during atrophy, was unchanged in response to starvation. IGF-1 had no effect on Gabarapl-1, ATG4B, or LC3B gene expression.
Myostatin (MSTN), a muscle-specific cytokine that negatively regulates muscle mass (22, 25), can be elevated with obesity (26). Therefore, we next measured mRNA content of myostatin and its downstream transcription factors, SMAD2 and SMAD3. As shown in Figure 4C, starvation significantly reduced gene expression of MSTN (lean: 0.6 and obese: 0.8-fold) and SMAD3 (lean: 0.4 and obese 0.6-fold), but had no effect on SMAD2. IGF-1 treatment did not significantly alter expression MSTN, SMAD2, or SMAD3.
As shown in Figure 4D, there was a significant group effect (p = 0.04) for FoxO3 gene expression to be higher in myotubes from the severely obese. However, the interaction between treatments and obesity failed to reach statistical significance (p = 0.06). No other genes tested were differentially expressed between myotubes of the lean and severely obese. However, western blot analysis revealed FoxO3 protein amount was not different between groups under basal, starved, or IGF-1 treatment (Figure 4E).
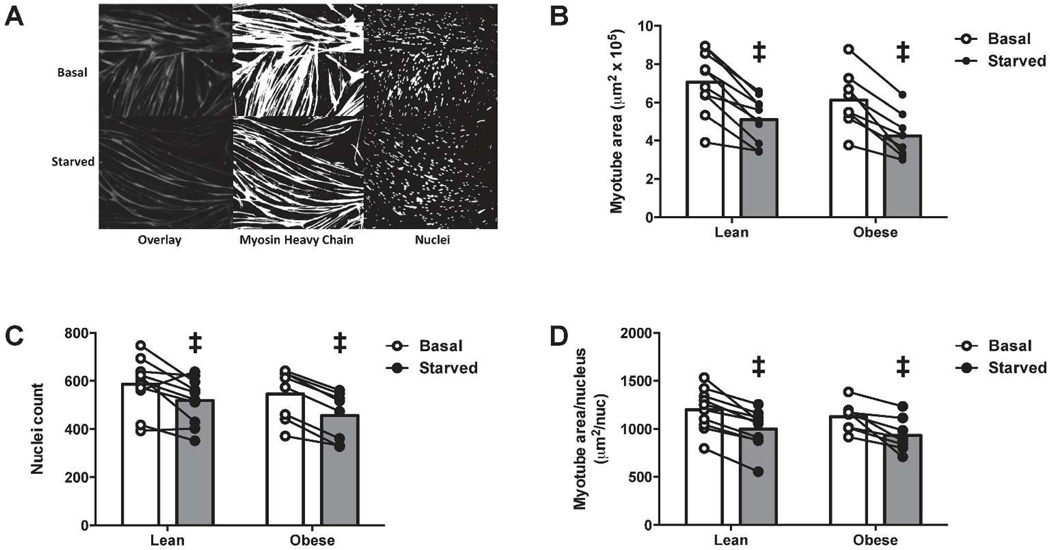
Myotubes from the lean and severely obese exhibit overall atrophy to a similar extent in response to starvation
To test whether HSkM myotubes from lean and severely obese atrophy to the same extent, myotube area (Figure 5B), nuclei number (Figure 5C) and myotube area/nucleus (Figure 5D) were determined following 24h of starvation. There was a significant main effect of starvation to decrease myotube area (~30%) and nuclei number (~15%), suggesting this treatment may induce apoptosis, as previously described in mouse myotubes (27). Despite the reduced number of nuclei, starvation significantly reduced myotube area/nucleus (~20%), demonstrating that loss of myotube size occurs to a greater extent than loss of nuclei number. There were no differences between lean and obese in myotube area, nuclei count, or myotube area/nucleus.
Figure 5.
Myotubes were kept in differentiation media (Basal) or starved of amino acids (Starved) for 24 h. (a) Representative gray-scale fluorescent images for myosin heavy chain and nuclei (Overlay), and individual components converted to threshold images representing Myosin Heavy Chain and Nuclei. (b) Myotube area. (c) Number of nuclei. (d) Myotube area per nucleus. Individual data points are presented. ‡Significant effect vs basal (P < 0.05).
Discussion
The main finding of the present study is that flux through the major protein degradation pathways is altered in human skeletal muscle with obesity. To our knowledge, this is the first study to directly measure flux through the UPS and autophagic/lysosomal pathways of human skeletal muscle. Accordingly, given the distinct roles of the UPS and the autophagic/lysosomal pathway (8), the specific proteins degraded during muscle atrophy are also likely different with obesity.
Primary Human Skeletal Muscle myotubes (HSkM) offer the opportunity to study innate characteristics of human skeletal muscle (12). Since myoblasts are purified and passaged in culture for several weeks (13), cells are subject to identical extracellular conditions; thus, any differences detected with obesity cannot be attributed to circulating factors or muscle loading. Innate metabolic dysfunction in skeletal muscle of severely obese individuals is presumably due to genetic or epigenetic differences (28). In previous studies from our group, HSkM derived from severely obese individuals are insulin resistant (29) and have impaired fatty acid oxidation (30). The present study extends these findings by demonstrating altered protein metabolism in the obese. We now also show that myotubes of the obese have a shift in MHC away from type I, which is consistent with fiber typing from muscle biopsies of obese individuals (23). The difference in MHC distribution is somewhat surprising since HSkM express both type I and type II MHC regardless of whether the source muscle fibers express exclusively type I or type II, suggesting that the MHC lineage is not pre-determined in human muscle (31, 32). We cannot discount the possibility that altered flux through the proteolytic systems observed in severe obesity is a component of the shift in muscle fiber type as characterized by MCH isoform expression. Future studies would be required to test this possibility by measuring protein degradation in HSkM cultures from subjects with wide differences of MHC isoform expression.
In relation to protein degradation, both the absolute rate of autophagic flux and its relative contribution to total proteolysis were less in myotubes with obesity. Autophagy, but not the UPS, is responsible for the degradation of entire organelles, such as mitochondria. It has been observed that severe obesity is accompanied by decreased mitochondrial content in HSkM (33) and impaired mitochondrial function in mouse skeletal muscle (34). Genetic deletion of Autoghapy-related gene 7 (Atg-7) impairs mitochondrial respiration and induces insulin resistance in mice (35). Therefore, decreased autophagic/lysosomal pathway flux may represent not only impaired protein metabolism in obese skeletal muscle, but contribute to impaired carbohydrate and lipid metabolism, possibly by slowing degradation of damaged or dysfunctional mitochondria. Likewise, in HSkM of type 2 diabetics, loss of proteasome activity by a potent chemical inhibitor impairs insulin sensitivity (14), which further demonstrates the importance of protein degradation modulating muscle phenotype. Future research should examine the cellular mechanisms by which these specific proteolytic pathways modulate insulin sensitivity.
UPS flux is determined by catabolic activity of the proteasome and protein polyubiquitination (36, 37). In the present study, we demonstrated that proteasome activity but not proteasome content is greater in HSkM with obesity. Since our experiments are not dependent on protein ubiquitination, these data indicate that specific activity of proteasomes, and thus proteolytic capacity, is inherently higher in skeletal muscle of the severely obese. It is possible that proteasome binding partners, which bind to and modify catabolic activity of the proteasome (38), are differentially expressed in skeletal muscle of the obese. However, we noted that starvation increased proteasome activity to a similar extent in myotubes regardless of obesity status. Therefore, it is unlikely that proteasome activity alone can explain the elevated UPS flux seen in response to starvation in skeletal muscle from obese individuals. Gene expression of two muscle atrophy-specific ubiquitin ligases, Atrogin-1 and MuRF-1, were also expressed to a similar extent in myotubes from the lean and severely obese under basal and starved conditions. However, it is possible that increased activity of other ubiquitin ligases may explain the increased UPS flux in myotubes of the obese in response to starvation. Alternatively, decreased expression of deubiquitinating enzymes during starvation may increase UPS flux with obesity. To date, few studies have examined the role of deubiquitinating enzymes during muscle atrophy, although several deubiquitinating enzymes are upregulated during muscle atrophy (36). Although the specific mechanism cannot be identified in the present study, the finding of differences in the UPS and MHC isoform expression with obesity in skeletal muscle provides relevant information in terms of identifying a “metabolic program” which is inherently present in human skeletal muscle with severe obesity.
Protein degradation is regulated, at least in part, at the level of gene transcription (39). Since critical atrophy-related proteins, such as LC3B, are degraded as part of overall protein degradation, increased mRNA content may be a more sensitive means to detect activation of these genes than protein content (8). We noted greater mRNA of the transcription factor FoxO3 in myotubes of the obese, but, surprisingly, expression patterns of the FoxO3-inducible genes Atrogin-1, MuRF-1, LC3B, and Gabarapl-1 were similar under basal, starved, and IGF-1 treatments in the lean and obese subjects. However, total FoxO3 protein content did not differ due to obesity status, suggesting that perhaps FoxO3 translation is inhibited or rate of FoxO3 protein degradation is specifically accelerated in skeletal muscle with obesity.
In summary, the present data indicate that flux through the major proteolytic pathways differs in human skeletal muscle with obesity. Specifically, flux through the autophagic/lysosomal pathway is decreased in skeletal muscle of the severely obese during basal and starved conditions. During starvation-induced atrophy, skeletal muscle of the severely obese experiences a relative shift toward UPS-mediated protein degradation, which may be in part due to higher specific activity of proteasomes. Therefore, muscle cells of the obese and the lean have innate differences in the management of protein degradation, which may in turn differentially regulate the disappearance of specific proteins and explain phenotypic differences in muscle with severe obesity.
Supplementary Material
What is already known about this subject
Whole body protein metabolism is dysregulated with obesity.
Protein degradation is a major regulator of skeletal muscle mass (total protein content) and skeletal muscle function (by regulating the content of specific proteins or organelles).
The major protein degradation pathways, the ubiquitin-proteasome and autophagy/lysosome, are functionally distinct; each preferentially degrades specific proteins and/or organelles.
What this study adds
Proteolytic flux through the autophagic/lysosomal pathway is slower in primary skeletal muscle cells from the severely obese.
Specific activity of the proteasome is greater in primary skeletal muscle cells of the severely obese, demonstrating increased proteolytic capacity.
Muscle cells of the lean and severely obese have innate differences in protein degradation, which further adds to the evidence of deranged metabolic profile of skeletal muscle in obesity.
Acknowledgements
Funding provided by Start-up funds from East Carolina University to JJB and CAW, NIH R00-AR05629 to CAW, NIH R01-DK56112 to JAH, and a pre-doctoral ACSM Foundation award to LMB. Monoclonal antibodies for embryonic MHC and type I MHC were obtained from Developmental Studies Hybridoma Bank at The University of Iowa. The authors thank Dr. Jill Maples for her excellent technical contributions. Current affiliation of LMB is the Department Kinesiology and Health Promotion, University of Kentucky.
References
- 1.Flegal KM, Kit BK, Orpana H, Graubard BI. Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and metaanalysis. JAMA. 2013;309:71–82. doi: 10.1001/jama.2012.113905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srikanthan P, Hevener AL, Karlamangla AS. Srikanthan P, Hevener AL, Karlamangla AS, editors. Sarcopenia exacerbates obesity-associated insulin resistance and dysglycemia: findings from the National Health and Nutrition Examination Survey III. PLoS ONE. 2010;5:e10805. doi: 10.1371/journal.pone.0010805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prado CMM, Lieffers JR, McCargar LJ, et al. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol. 2008;9:629–635. doi: 10.1016/S1470-2045(08)70153-0. [DOI] [PubMed] [Google Scholar]
- 4.Tan BHL, Birdsell LA, Martin L, Baracos VE, Fearon KCH. Sarcopenia in an overweight or obese patient is an adverse prognostic factor in pancreatic cancer. Clin Cancer Res. 2009;15:6973–6979. doi: 10.1158/1078-0432.CCR-09-1525. [DOI] [PubMed] [Google Scholar]
- 5.Stephen WC, Janssen I. Sarcopenic-obesity and cardiovascular disease risk in the elderly. J Nutr Health Aging. 2009;13:460–466. doi: 10.1007/s12603-009-0084-z. [DOI] [PubMed] [Google Scholar]
- 6.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsanos CS, Mandarino LJ. Protein metabolism in human obesity: a shift in focus from whole-body to skeletal muscle. Obesity. 2011;19:469–475. doi: 10.1038/oby.2010.290. [DOI] [PubMed] [Google Scholar]
- 8.Sandri M. Protein breakdown in muscle wasting: Role of autophagy-lysosome and ubiquitinproteasome. Int J Biochem Cell Biol. 2013;45:2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccirillo R, Demontis F, Perrimon N, Goldberg AL. Mechanisms of muscle growth and atrophy in mammals and Drosophila. Dev Dyn. 2014;243:201–215. doi: 10.1002/dvdy.24036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristensen A, Schandorff S, Hoyer-Hansen M, et al. Ordered Organelle Degradation during Starvation-induced Autophagy. Mol Cell Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Cohen S, Brault JJ, Gygi SP, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aas V, Bakke SS, Feng YZ, et al. Are cultured human myotubes far from home? Cell Tissue Res. 2013;354:671–682. doi: 10.1007/s00441-013-1655-1. [DOI] [PubMed] [Google Scholar]
- 13.Berggren JR, Tanner CJ, Houmard JA. Primary cell cultures in the study of human muscle metabolism. Exerc Sport Sci Rev. 2007;35:56–61. doi: 10.1249/JES.0b013e31803eae63. [DOI] [PubMed] [Google Scholar]
- 14.Al-Khalili L, de Castro Barbosa T, Ostling J, et al. Proteasome inhibition in skeletal muscle cells unmasks metabolic derangements in type 2 diabetes. Am J Physiol Cell Physiol. 2014;307:C774–C787. doi: 10.1152/ajpcell.00110.2014. [DOI] [PubMed] [Google Scholar]
- 15.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 16.Bollinger LM, Witczak CA, Houmard JA, Brault JJ. SMAD3 augments FoxO3-induced MuRF-1 promoter activity in a DNA-binding dependent manner. AJP: Cell Physiology. 2014;307:C278–C287. doi: 10.1152/ajpcell.00391.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–E601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 19.Mizunoya W, Wakamatsu J-I, Tatsumi R, Ikeuchi Y. Protocol for high-resolution separation of rodent myosin heavy chain isoforms in a mini-gel electrophoresis system. Anal Biochem. 2008;377:111–113. doi: 10.1016/j.ab.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Method Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 22.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol. 2009;296:C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 23.Tanner CJ, Barakat HA, Dohm GL, et al. Muscle fiber type is associated with obesity and weight loss. Am J Physiol Endocrinol Metab. 2002;282:E1191–E1196. doi: 10.1152/ajpendo.00416.2001. [DOI] [PubMed] [Google Scholar]
- 24.Sandri M, Sandri C, Gilbert A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sartori R, Milan G, Patron M, et al. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:C1248–C1257. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- 26.Hittel DS, Berggren JR, Shearer J, Boyle K, Houmard JA. Increased secretion and expression of myostatin in skeletal muscle from extremely obese women. Diabetes. 2009;58:30–38. doi: 10.2337/db08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoneich C, Dremina E, Galeva N, Sharov V. Apoptosis in differentiating C2C12 muscle cells selectively targets Bcl-2-deficient myotubes. Apoptosis. 2014;19:42–57. doi: 10.1007/s10495-013-0922-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houmard JA, Pories WJ, Dohm GL. Severe obesity: evidence for a deranged metabolic program in skeletal muscle? Exerc Sport Sci Rev. 2012;40:204–210. doi: 10.1097/JES.0b013e31825d53fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bikman BT, Zheng D, Reed MA, Hickner RC, Houmard JA, Dohm GL. Lipid-induced insulin resistance is prevented in lean and obese myotubes by AICAR treatment. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1692–R1699. doi: 10.1152/ajpregu.00190.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle KE, Zheng D, Anderson EJ, Neufer PD, Houmard JA. Mitochondrial lipid oxidation is impaired in cultured myotubes from obese humans. Int J Obes (Lond) 2012;36:1025–1031. doi: 10.1038/ijo.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaster M, Kristensen SR, Beck-Nielsen H, Schroder HD. A cellular model system of differentiated human myotubes. APMIS. 2001;109:735–744. doi: 10.1034/j.1600-0463.2001.d01-140.x. [DOI] [PubMed] [Google Scholar]
- 32.Bonavaud S, Agbulut O, Nizard R, D'honneur G, Mouly V, Butler-Browne G. A discrepancy resolved: human satellite cells are not preprogrammed to fast and slow lineages. Neuromuscul Disord. 2001;11:747–752. doi: 10.1016/s0960-8966(01)00222-x. [DOI] [PubMed] [Google Scholar]
- 33.Consitt LA, Bell JA, Koves TR, et al. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha overexpression increases lipid oxidation in myocytes from extremely obese individuals. Diabetes. 2010;59:1407–1415. doi: 10.2337/db09-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnard C, Durand A, Peyrol S, et al. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J Clin Invest. 2008;118:789–800. doi: 10.1172/JCI32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu JJ, Quijano C, Chen E, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wing SS. Deubiquitinases in skeletal muscle atrophy. Int J Biochem Cell Biol. 2013;45:2130–2135. doi: 10.1016/j.biocel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson KD. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 38.Besche HC, Sha Z, Kukushkin NV, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecker SH, Jagoe RT, Gilbert A, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.