Abstract
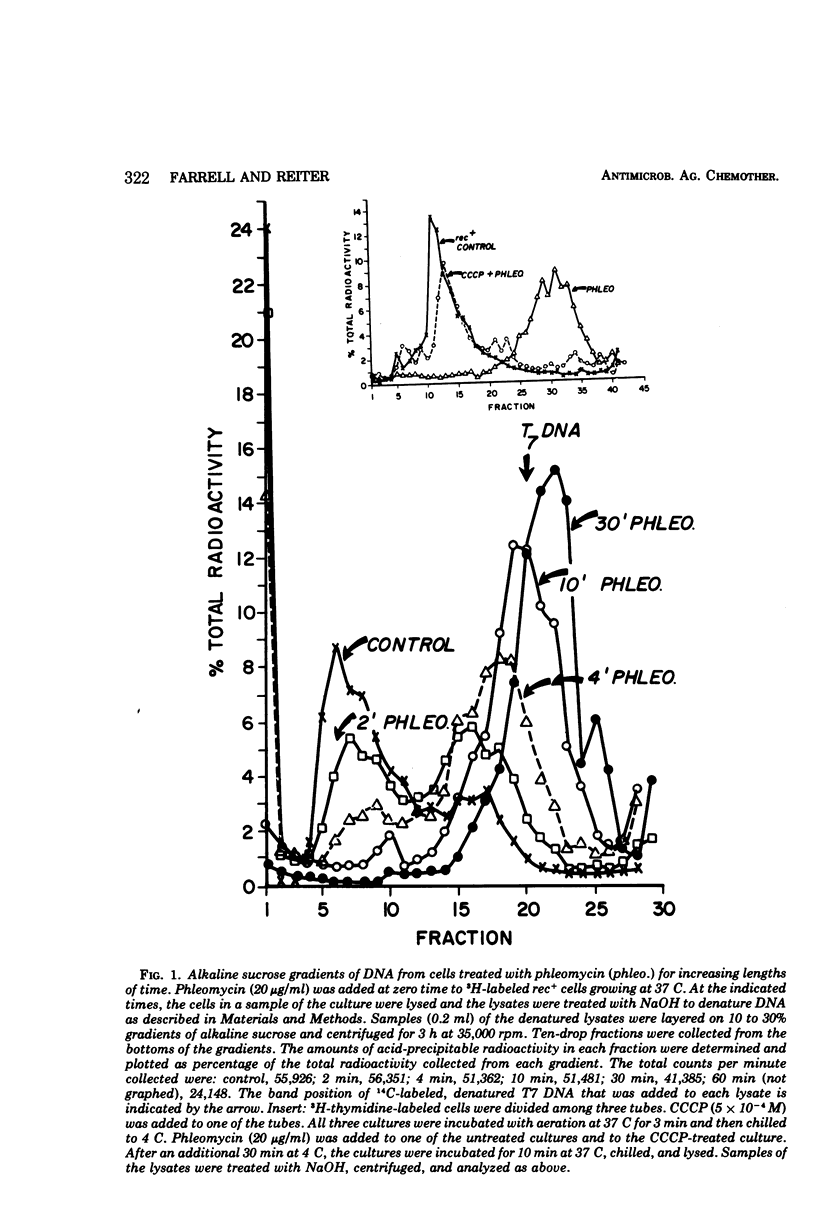
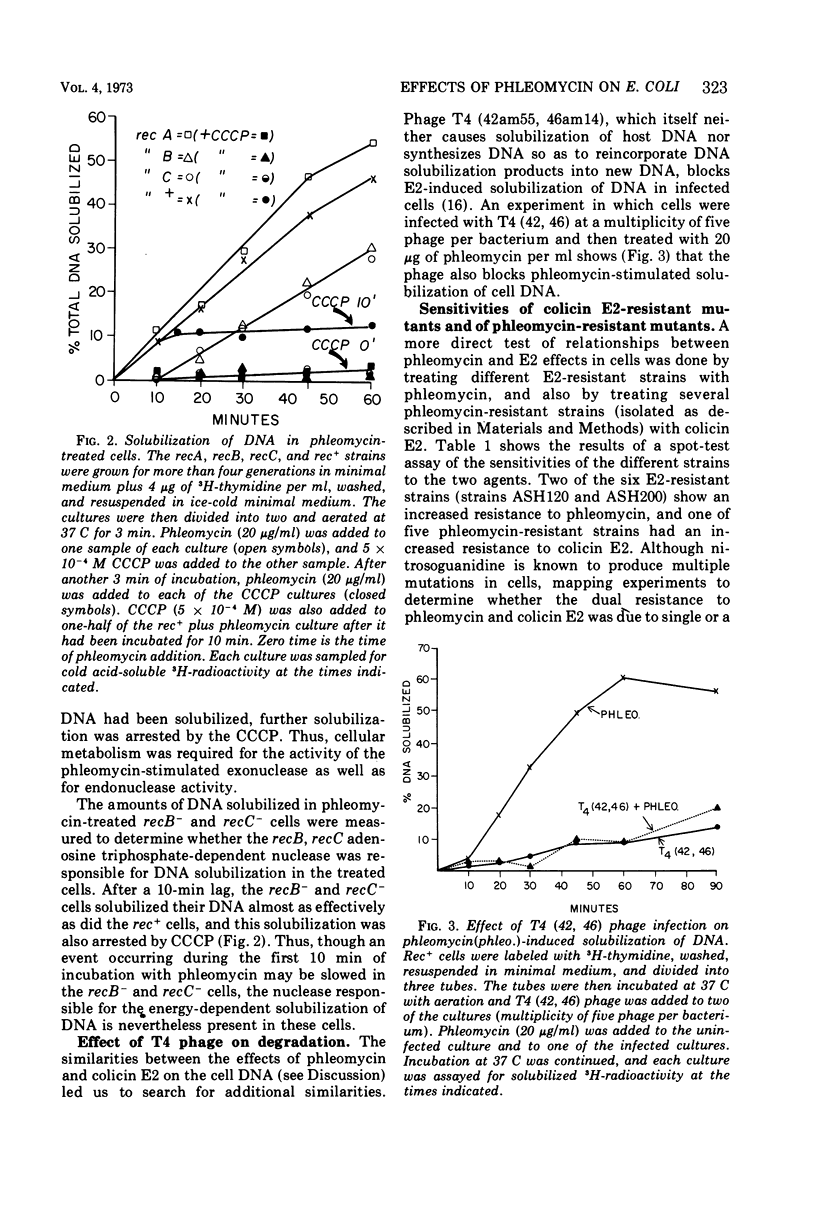
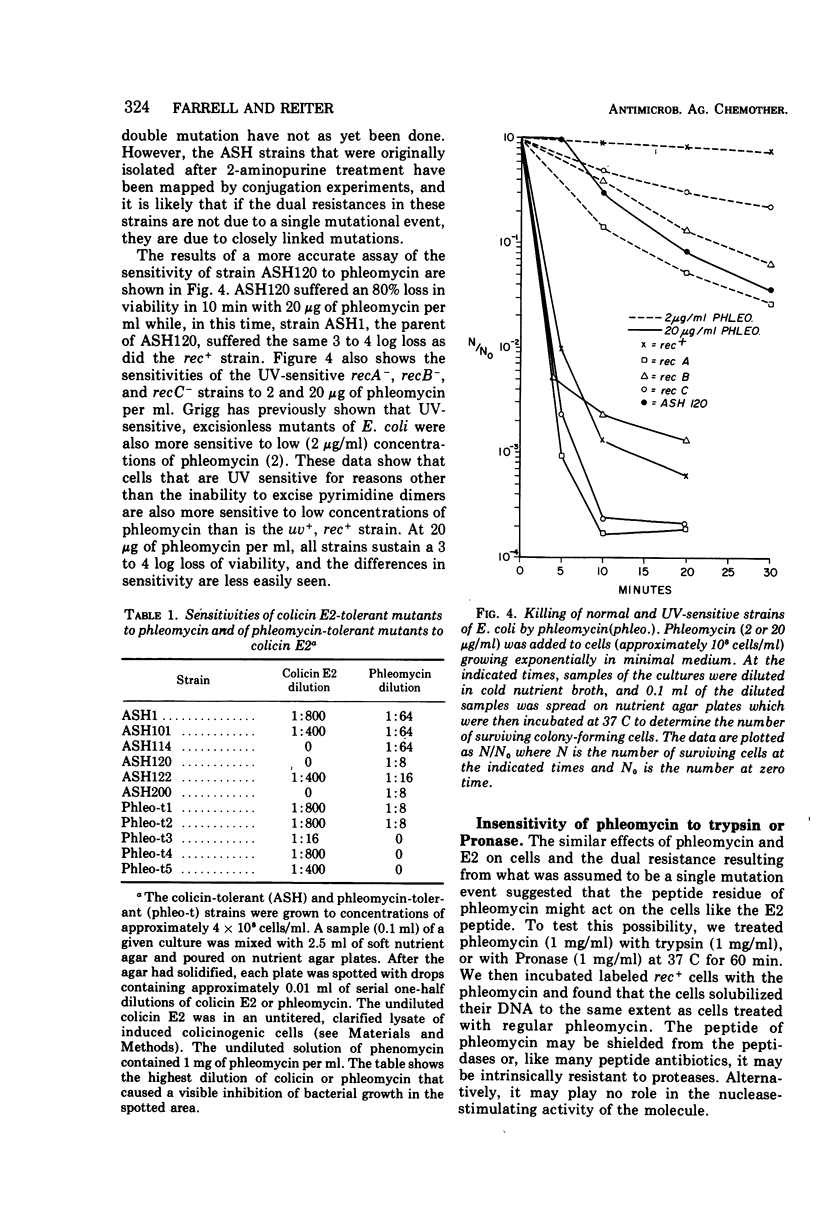
Phleomycin stimulates the degradation of DNA by energy-dependent endonuclease and exonuclease reactions in Escherichia coli rec+ cells and in recB− and recC− cells that lack an adenosine triphosphate-dependent nuclease functioning in the repair of ultraviolet (UV) lesions. Exonuclease activity is blocked in T4 phage-infected cells. The endonuclease reaction produces 107-dalton segments resembling those produced in colicin E2-treated cells. These differ from the random-sized segments produced in UV-irradiated cells, or the 106-dalton segments made in T4 phage-infected cells. A mutant selected for phleomycin tolerance is cross-tolerant to colicin E2, and some mutants selected for colicin E2 tolerance are cross-tolerant to phleomycin. On the basis of these cross-tolerances and the similarities between the effects of phleomycin and E2-stimulated nucleases, the suggestion is made that both agents may stimulate the same nuclease reactions in E. coli cells.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beppu T., Arima K. Dissociating activity of purified colicin E2 on the isolated membrane complex of Escherichia coli. Biochim Biophys Acta. 1970 Dec 1;219(2):512–514. doi: 10.1016/0005-2736(70)90235-x. [DOI] [PubMed] [Google Scholar]
- Grigg G. W. Induction of DNA breakdown and death in Escherichia coli by phleomycin. Its association with dark-repair processes. Mol Gen Genet. 1969;104(1):1–11. doi: 10.1007/BF00277357. [DOI] [PubMed] [Google Scholar]
- HEYTLER P. G., PRICHARD W. W. A new class of uncoupling agents--carbonyl cyanide phenylhydrazones. Biochem Biophys Res Commun. 1962 May 4;7:272–275. doi: 10.1016/0006-291x(62)90189-4. [DOI] [PubMed] [Google Scholar]
- Holland E. M., Holland I. B. Induction of DNA breakdown and inhibition of cell division by colicin E2. Nature of some early steps in the process and properties of the E-2-specific nuclease system. J Gen Microbiol. 1970 Dec;64(2):223–239. doi: 10.1099/00221287-64-2-223. [DOI] [PubMed] [Google Scholar]
- Holland I. B., Threlfall E. J., Holland E. M., Darby V., Samson A. C. Mutants of Escherichia coli with altered surface properties which are refractory to colicin E2, sensitive to ultraviolet light and which can also show recombination deficiency, abortive growth of bacteriophage lambda and filament formation. J Gen Microbiol. 1970 Aug;62(3):371–382. doi: 10.1099/00221287-62-3-371. [DOI] [PubMed] [Google Scholar]
- Kutter E. M., Wiberg J. S. Degradation of cytosin-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4D wild type and with mutants defective in genes 46, 47 and 56. J Mol Biol. 1968 Dec;38(3):395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- NOMURA M. MECHANISM OF ACTION OF COLICINES. Proc Natl Acad Sci U S A. 1964 Dec;52:1514–1521. doi: 10.1073/pnas.52.6.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Yamaki H., Suzuki H., Tanaka N., Umezawa H. The combined effects of bleomycin and sulfhydryl compounds on the thermal denaturation of DNA. Biochim Biophys Acta. 1969 Mar 18;179(1):165–171. doi: 10.1016/0005-2787(69)90132-4. [DOI] [PubMed] [Google Scholar]
- Pietsch P., Garrett H. Primary site of reaction in the in vitro complex of phleomycin in DNA. Nature. 1968 Aug 3;219(5153):488–489. doi: 10.1038/219488a0. [DOI] [PubMed] [Google Scholar]
- Reiter H., Milewskiy M., Kelley P. Mode of action of phleomycin on Bacillus subtilis. J Bacteriol. 1972 Aug;111(2):586–592. doi: 10.1128/jb.111.2.586-592.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose P. S. Interaction between colicin E2 and DNA in vitro. FEBS Lett. 1972 Jun 15;23(2):241–243. doi: 10.1016/0014-5793(72)80351-x. [DOI] [PubMed] [Google Scholar]
- Ringrose P. Sedimentation analysis of DNA degradation products resulting from the action of colicin E2 on Escherichia coli. Biochim Biophys Acta. 1970 Aug 8;213(2):320–334. doi: 10.1016/0005-2787(70)90040-7. [DOI] [PubMed] [Google Scholar]
- Rupp W. D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968 Jan 28;31(2):291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- Strauss B., Searashi T., Robbins M. Repair of DNA studied with a nuclease specific for UV-induced lesions. Proc Natl Acad Sci U S A. 1966 Sep;56(3):932–939. doi: 10.1073/pnas.56.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift R. L., Wiberg J. S. Bacteriophage T4 inhibits colicin E2-induced degradation of Escherichia coli deoxyribonucleic acid. I. Protein synthesis-dependent inhibition. J Virol. 1971 Sep;8(3):303–310. doi: 10.1128/jvi.8.3.303-310.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKITA T. Studies on purification and properties of phleomycin. J Antibiot (Tokyo) 1959 Nov;12:285–289. [PubMed] [Google Scholar]
- TANAKA N. EFFECT OF PHLEOMYCIN ON DNA POLYMERASE OF TUMOR ORIGIN. J Antibiot (Tokyo) 1965 Mar;18:111–111. [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]



