Abstract
Background
Small non-coding RNA molecules (miRNAs) play a pivotal role in regulating gene expression in development. miRNAs regulate key processes at the cellular level and thereby influence organismal and tissue development including kidney morphogenesis. A miRNA molecule is initially synthesized as a longer hairneedle-shaped RNA transcript and then processed through an enzymatic complex that contains the RNA-processing enzyme Drosha and its essential interactor Dgcr8. Resulting pre-miRNAs are then cleaved by Dicer. Recent data showed that loss of Dicer resulted in severe developmental kidney phenotypes. However, as Dicer has multiple miRNA-independent functions, it was not entirely clear whether the observed renal phenotypes could be exclusively attributed to a lack of miRNA expression.
Methods
We analyzed the role of miRNAs in kidney development by conditional gene deletion of Dgcr8 in the developing kidney using a transgenic mouse line that expresses Cre recombinase in the distal nephron and derivatives of the ureteric bud in kidney development.
Results
Animals with a gene deletion of Dgcr8 in these tissues developed severe hydronephrosis, kidney cysts, progressive renal failure and premature death within the first two months after birth, a phenotype strongly resembling Dicer deletion.
Conclusions
Here we show that conditional gene deletion of the essential miRNA-processing enzyme Dgcr8 in the developing renal tubular system results in severe developmental defects and kidney failure. These data confirm earlier findings obtained in Dicer knock-out animals and clearly illustrate the essential role of miRNAs in kidney development. The data suggests that miRNA dysregulation may play an important, yet ill-defined role in the pathogenesis of inborn defects of the genitourinary system and indicate that miRNA defects may be causative in the development of human disease.
Electronic supplementary material
The online version of this article (doi:10.1186/s12882-015-0053-1) contains supplementary material, which is available to authorized users.
Keywords: CAKUT, Dgcr8, Dicer, Hydronephrosis, Kidney, miRNA
Background
MicroRNAs are important regulators of gene expression and have been shown to be crucial to developmental processes in many different tissues [1]. To study the role of miRNAs in the kidney several publications have addressed this question using a conditional knockout of Dicer, the RNAse III enzyme catalyzing the maturation from pre-miRNA to mature microRNAs [2-9]. This strategy revealed major defects in both tubular and glomerular development and maintenance. The loss of Dicer in derivatives of the ureteric bud and in the tubular system lead to a severe hydronephrosis coupled with cystic kidneys and loss of functional parenchyme [2-4,6]. These phenotypes strongly resemble congenital anomalies of the kidney and urinary tract (CAKUT) in the clinical setting. However, whether this is truly due to loss of microRNAs has remained elusive since Dicer fulfills several other important functions which may well be involved in renal development [10]. Among these are its role as a DNAse in genomic DNA fragmentation during apoptosis, the processing of endogenous siRNA and the detoxification of repeat elements [11-13]. In an elegant study elucidating the role of microRNAs in skin development this issue has been addressed using the conditional knockout of genes involved in different steps of miRNA processing displaying a phenotypic overlap [14,15]. As to the kidney this has been done successfully regarding the effects of podocyte-specific loss of microRNAs using a conditional knockout allele of Drosha [16], which confirmed previous studies based on Dicer knockout in podocytes [7-9].
Consequently, we set out to confirm the role of microRNAs in renal development using a conditional allele of Dgcr8. Dgcr8 interacts with Drosha and is essential for its role in processing pri-microRNAs in the nucleus [17].
Methods
Mice
Dgcr8 fl/fl animals were described before [18] and generously provided by Elaine Fuchs (Rockefeller University, NYC, USA). To generate a kidney tubulus specific Dgcr8 knockout these mice were crossed to a KspCre transgenic line (contributed by Peter Igarashi, UT Southwestern Medical Center, Dallas, USA) that expresses the Cre recombinase under the control of a ksp-cadherin promotor resulting in Cre expression in the developing genitourinary tract and kidney tubulus system was performed as described before [19]. Animals were housed in standardized specific pathogen-free conditions in the animal facility of the CMMC (University of Cologne).
All animal procedures were performed according to European (EU directive 86/609/EEC), national (TierSchG), and institutional guidelines and were approved by local governmental authorities (LANUV NRW).
Histology
The kidneys were fixed in formalin, embedded in paraffin and stained with PAS according to standard protocols. To analyse the expression of Ki-67, slides of fixed and paraffin-embedded mouse kidneys were de-paraffinized using Xylol and descending concentrations of ethanol. Antigen retrieval was carried out by warming kidney slides in citrate buffer (10 mM, pH6) for 10 min using a microwave. After blocking with 3% H2O2 and Avidin and Biotin (Vector Laboratories, Inc.) for 15 min each, slides were sequentially incubated with the Ki-67 antibody (rabbit Ki-67 ab16667, abcam, 1:500 dilution, over night at 4°C) and after washing with PBS with biotinylated anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA, USA; 1 h at room temperature). Kidney slides were labelled with ABC kit (Vector Laboratories, Inc.), and development was carried out using diaminobenzidine solution (Sigma Aldrich). Slides were counterstained with hematoxylin (Sigma-Aldrich), dehydrated and afterwards mounted with Histomount (National Diagnostics). Stained slides were scanned using a Slidescanner (Leica) and analyzed using the ImageScope software (version 12.0.1.5030, Aperio).
Laboratory medicine
Heparinized blood was obtained by cardiac puncture. Plasma was prepared by centrifugation at 3000 rpm for 10 min. Urea was measured in the central laboratory medicine unit of the University Hospital of Cologne using the kinetic UV test (Roche Diagnostics). Significance was calculated using a two-tailed Student’s t test for all measurements (urea, body weight of mice).
qPCR
RNA was extracted from whole mouse kidneys using acid guanidinium thiocyanate-phenol-chloroform extraction [20]. RT reactions were performed using the Taqman microRNA Reverse Transcription Kit (ABI). Expression of mir-192 (assay ID 000491), and miR-200b (assay ID 4426961) was analyzed using Taqman assays (ABI), and snoRNA135 (assay ID 001230) served as endogenous control. All qPCR experiments were performed on the ABI 7900HT System. All data points were generated using the number of biological replicates indicated in the figure. Data analysis and statistics were performed using the Expression Suite v1.3 software package applying the comparative Ct method and using one standard deviation for error bar calculation (LifeTechnologies).
TUNEL assay
To analyse apoptosis in the kidneys of Dgcr8 knockout and littermate control mice we utilized the Promega DeadEnd Fluorometric Kit according to the manufacturers protocol. Pictures were taken with an inverted microscope (Axiovert200, equipped with an ApoTome system and an AxioCam MRm camera. Objective used: Plan Apochromat 20×/0.8 NA. Carl Zeiss) using Axiovision 4.8 (Carl Zeiss).
Results
To analyze the role of Dgcr8 and thereby miRNAs in the developing tubular system independent of a Dicer mouse model, we crossed a Dgcr8 fl/fl mouse line [18] with KspCre mice [19], resulting in a conditional knockout of Dgcr8 in the developing urogenital tract and tubulus system.
Dgcr8 fl/fl; KspCre positive mice (afterwards named Dgcr8 knockout) showed an obvious delay in growth (Figure 1A) in comparison to the control littermates and a significantly reduced body weight (Figure 1B). This phenotype was most likely caused by the developing renal failure in these mice, since they showed a marked elevation in serum urea (Figure 1C). The phenotype became apparent in the first weeks of life, several mice died during the weaning period leading to a significantly reduced number of Dgcr8 knockout animals after weaning in comparison to the control genotypes (Additional file 1: Figure S1A). None of the Dgcr8 knockout mice analyzed so far survived longer than 8 weeks, most likely due to development of end stage renal disease. In order to confirm that loss of Dgcr8 abrogates miRNA biogenesis we quantified two miRNAs that had been shown before to be primarily expressed in renal tubular cells and to be depleted by KspCre driven loss of Dicer [3,6,21]. Both expression of miR-192 and miR-200b were greatly reduced in the conditional Dgcr8 knockout mouse line (Figure 2).
Figure 1.
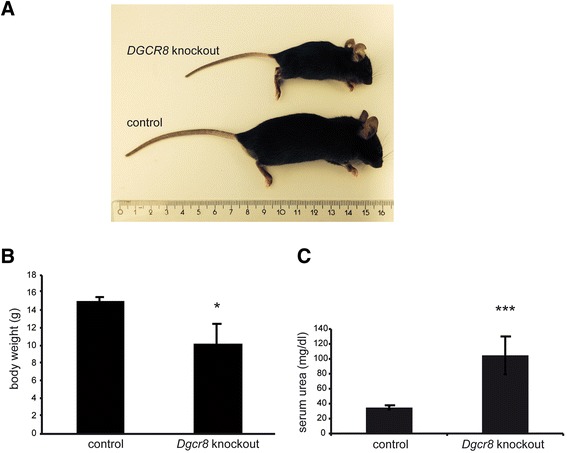
Kidney-specific knockout of Dgcr8 results in end stage renal disease. A Conditional Dgcr8 fl/fl; KspCre knockout mice are markedly smaller than their control littermates (7 week old animals) B Dgcr8 knockout mice have a significant lower body weight in comparison to their control littermates (4 week old animals; * = p < 0.05; error bars represent SEM; knockout: n = 3; control: n = 5) C Dgcr8 knockout mice develop end stage renal disease as displayed by elevated serum urea levels in comparison to control animals (4–7 week old animals; *** = p < 0.001; error bars represent SEM; knockout: n = 6; control: n = 13).
Figure 2.
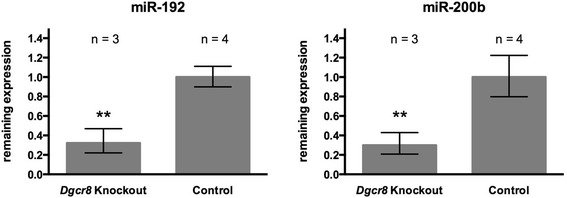
KspCre-mediated loss of Dgcr8 induces depletion of tubulus-specific miRNAs. Both expression of miR-192 and miR-200b is strongly reduced in Dgcr8 knockout kidneys when compared to WT littermates (4-7 week old animals; ** = p < 0.01; error bars represent SEM).
Further examination of the kidneys of the Dgcr8 knockout mice showed the macroscopic picture of severe hydronephrosis with a dilated ureter and kidney pelvis (Figure 3A). There were no signs for a complete obstruction of the ureter, since the bladder of the knockout animals was filled with urine (data not shown).
Figure 3.
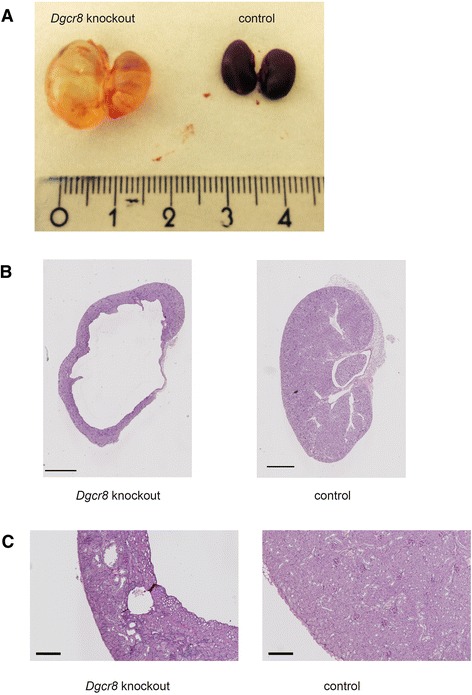
Hydronephrosis and cystic kidneys of Dgcr8 knockout animals. A Conditional knockout of Dgcr8 in the renal tubulus system leads to hydronephrosis. B + C Histological analysis confirms hydronephrosis with severe loss of kidney parenchyma especially in the medulla region, a thinned cortex and kidney cysts (bar = 1000 μm (B) and 200 μm (C)).
Histopathological analyses of the kidneys confirmed the diagnosis of hydronephrosis and obstructive nephropathy. In most affected animals nearly the entire medulla was missing, and the cortex was very thin (Figure 3B). In addition, some kidneys displayed dilated tubuli and cysts were observed (Figure 3C). As described for Dicer knockout mice before [3] the Dgcr8 knockout kidneys showed a reduced glomerular density pointing towards a branching defect (Additional file 1: Figure 1B). To further analyse the cellular basis to this phenotype we performed TUNEL assays and Ki-67 stainings. These revealed a strong induction of apoptosis and cellular proliferation in tubular cells of Dgcr8 knockout animals (Figure 4).
Figure 4.
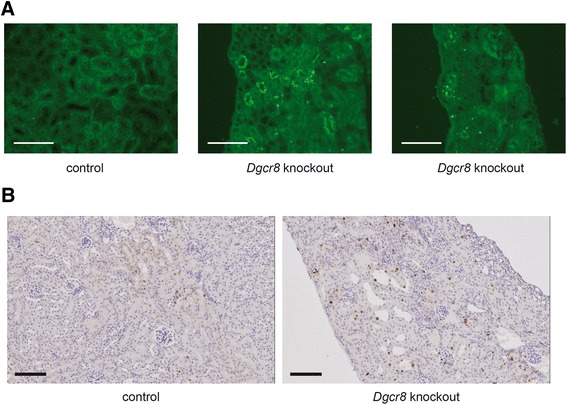
Loss of Dgcr8 induces apoptosis and proliferation. A TUNEL staining reveals a dramatic increase in apoptosis in Dgcr8 deficient kidneys (representative images of renal cortex, bar = 100 μm). B Staining for Ki-67 reveals an increase in proliferation in Dgcr8 knockout kidneys (bar = 100 μm).
Discussion
The loss of Dgcr8 specifically in renal epithelial cells nicely resembled the phenotype observed earlier when knocking out Dicer in the same tissue using the same cre mouse line. Both Dicer and Dgcr8 are involved in a number of different processes primarily regarding the processing of nucleic acids [10-13,22,23]. Since the known overlapping function of these two genes is microRNA processing our study strongly supports the conclusion that the severe phenotype in both mouse models including hydronephrosis and renal failure is caused by the loss of microRNA processing. This finding is of great importance and encouraging to plan and perform follow-up studies now addressing the role of specific microRNAs in development and maintenance of renal architecture.
Interestingly, a small number of microRNAs is either independent from Dicer or from the Drosha/Dgcr8 complex [24,25]. As an example the so-called mirtrons are processed by the spliceosome in the nucleus instead of Drosha/Dgcr8 [26]. As for Dicer miR-451 is not processed by this enzyme but depends on Ago2 in its maturation [27-29]. Consequently, our study does not only confirm the crucial role of microRNAs in renal development but also narrows down the list by excluding any small RNAs processed by only one of the two enzymes.
In contrast to our previous study on Dicer knockout animals with a penetrance of the phenotype of about 66% [3] the Dgcr8 knockout described in this study has a complete penetrance with no Dgcr8 knockout animal surviving longer than 8 weeks. Whether this may be due to a partial rescue of the Dicer knockout animals by another enzyme – e.g. for microRNAs that are generally processed by Dicer but may be processed by Ago2 as well – remains elusive and will be subject to future studies.
In summary, our results underline the relevance of microRNAs during kidney development and will encourage further functional studies examining single microRNAs and their target mRNA interactions – such as miR-20 and its targets PKD1 and PKD2 [3,6,30,31] - as regulators of renal organogenesis. This will be fundamental for gaining a better understanding of developmental defects in human kidney formation as observed in CAKUT – the predominant cause of end-stage renal disease in children.
Conclusions
MiRNAs are key regulators of intracellular signaling and development. In this study we show that loss of Dgcr8 dependent miRNAs in the kidney epithelium leads to severe hydronephrosis, kidney cysts and rapid kidney failure. This confirms an essential role for miRNAs in renal development and disease.
Acknowledgments
This work was funded by the Deutsche Nierenstiftung and the Deutsche Forschungsgemeinschaft MU3629/2-1 to R.-U.M., SFB829 to T.B. and SCHE1562-2 to B.S. We thank Elaine Fuchs and Peter Igarashi for providing the Dgcr8 fl/fl and KspCre mouse lines, respectively and members of the laboratory for helpful discussion. We would like to thank Sonja Kunath and Nadine Urban for their support with the animal experiments and Martyna Brütting for excellent technical help. We thank the CECAD Imaging Facility for their technical support with the Slidescanner.
Abbreviations
- CAKUT
Congenital anomalies of the kidney and urinary tract
- Dgcr8
DiGeorge syndrome critical region 8
- PKD
Polycystic kidney disease
Additional file
A Genotyping after weaning at 3–4 weeks of age reveals that the knockout mice did not reach weaning at a Mendelian ratio suggesting death before the timepoint of weaning and genotyping. In line with this finding several mice of unknown genotype had died before weaning. B Glomerular density is reduced in Dgcr8 knockout kidneys (n = 3 per group; error bars represent SEM; ** = p <0.01 using an unpaired Student’s t-test; 5 high power fields of the kidney cortex were counted per animal) C Table showing the number of mice revealing either kidney cysts or hydronephrosis at weaning.
Footnotes
Competing interests
The authors declare that they have no competing interests. The results presented in this paper have not been published previously in whole or part.
Authors’ contributions
MPB and RUM were involved in all experimental steps and drafting of the manuscript. CD and SH were involved in histological analysis and image aquisition. TB and BS coordinated the study and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Malte P Bartram, Email: malte.bartram@uk-koeln.de.
Claudia Dafinger, Email: claudia.dafinger@uk-koeln.de.
Sandra Habbig, Email: sandra.habbig@uk-koeln.de.
Thomas Benzing, Email: thomas.benzing@uk-koeln.de.
Bernhard Schermer, Email: bernhard.schermer@uk-koeln.de.
Roman-Ulrich Müller, Email: roman-ulrich.mueller@uk-koeln.de.
References
- 1.Tüfekci KU, Meuwissen RLJ, Genç S. The role of microRNAs in biological processes. Methods Mol Biol Clifton NJ. 2014;1107:15–31. doi: 10.1007/978-1-62703-748-8_2. [DOI] [PubMed] [Google Scholar]
- 2.Pastorelli L, Wells S, Fray M, Smith A, Hough T, Harfe B, et al. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome. 2009;20:140–51. doi: 10.1007/s00335-008-9169-y. [DOI] [PubMed] [Google Scholar]
- 3.Bartram MP, Höhne M, Dafinger C, Völker LA, Albersmeyer M, Heiss J, et al. Conditional loss of kidney microRNAs results in congenital anomalies of the kidney and urinary tract (CAKUT) J Mol Med Berl Ger. 2013;91:739–48. doi: 10.1007/s00109-013-1000-x. [DOI] [PubMed] [Google Scholar]
- 4.Nagalakshmi VK, Ren Q, Pugh MM, Valerius MT, McMahon AP, Yu J. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79:317–30. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu JYS, Sims-Lucas S, Bushnell DS, Bodnar AJ, Kreidberg JA, Ho J. Dicer function is required in the metanephric mesenchyme for early kidney development. Am J Physiol Renal Physiol. 2014;306:F764–72. doi: 10.1152/ajprenal.00426.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel V, Hajarnis S, Williams D, Hunter R, Huynh D, Igarashi P. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J Am Soc Nephrol JASN. 2012;23:1941–8. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol JASN. 2008;19:2150–8. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional micrornas leads to rapid glomerular and tubular injury. J Am Soc Nephrol JASN. 2008;19:2069–75. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol JASN. 2008;19:2159–69. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johanson TM, Lew AM, Chong MMW. MicroRNA-independent roles of the RNase III enzymes drosha and dicer. Open Biol. 2013;3:130144. doi: 10.1098/rsob.130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teta M, Choi YS, Okegbe T, Wong G, Tam OH, Chong MMW, et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Dev Camb Engl. 2012;139:1405–16. doi: 10.1242/dev.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011;80:719–30. doi: 10.1038/ki.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol CB. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao X, Somlo S, Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol JASN. 2002;13:1837–46. doi: 10.1097/01.ASN.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 20.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins RH, Martin J, Phillips AO, Bowen T, Fraser DJ. Pleiotropy of microRNA-192 in the kidney. Biochem Soc Trans. 2012;40:762–7. doi: 10.1042/BST20120085. [DOI] [PubMed] [Google Scholar]
- 22.Luhur A, Chawla G, Wu Y-C, Li J, Sokol NS. Drosha-independent DGCR8/Pasha pathway regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2014;111:1421–6. doi: 10.1073/pnas.1318445111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Cáceres JF. DGCR8 HITS-CLIP reveals novel functions for the microprocessor. Nat Struct Mol Biol. 2012;19:760–6. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie M, Steitz JA. Versatile microRNA biogenesis in animals and their viruses. RNA Biol. 2014;11(6):673–81. doi: 10.4161/rna.28985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong MMW, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–60. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dueck A, Meister G. MicroRNA processing without Dicer. Genome Biol. 2010;11:123. doi: 10.1186/gb-2010-11-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel V, Williams D, Hajarnis S, Hunter R, Pontoglio M, Somlo S, Igarashi P. miR-17∼92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci. 2013;110(26):10765–70. doi: 10.1073/pnas.1301693110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marrone AK, Stolz DB, Bastacky SI, Kostka D, Bodnar AJ, Ho J. MicroRNA-17 ~ 92 is required for nephrogenesis and renal function. J Am Soc Nephrol JASN. 2014;25:1440–52. doi: 10.1681/ASN.2013040390. [DOI] [PMC free article] [PubMed] [Google Scholar]


