Abstract
Sumoylation is essential for progression through mitosis, but the specific protein targets and functions remain poorly understood. In this study, we used chromosome spreads to more precisely define the localization of SUMO-2/3 to the inner-centromere and protein scaffold of mitotic chromosomes. We also developed methods to immunopurify proteins modified by endogenous, untagged SUMO-2/3 from mitotic chromosomes. Using these methods we identified 149 chromosome-associated SUMO-2/3 substrates by nLC-ESI-MS/MS. Approximately one-third of the identified proteins have reported functions in mitosis. Consistent with SUMO-2/3 immunolocalization, we identified known centromere and kinetochore associated proteins, as well as chromosome scaffold associated proteins. Notably, >30 proteins involved in chromatin modification or remodeling were identified. Our results provide insights into the roles of sumoylation as a regulator of chromatin structure and other diverse processes in mitosis. Furthermore, our purification and fractionation methodologies represent an important compliment to existing approaches to identify sumoylated proteins using exogenously expressed and tagged SUMOs.
Keywords: Chromosome, mitosis, proteomics, SUMO
INTRODUCTION
Small ubiquitin-related modifiers (SUMOs) are covalently conjugated to other proteins and regulate essential cellular processes including transcription, DNA repair and mitosis [1]. Like phosphorylation and ubiquitylation, sumoylation is now recognized as an important regulator of multiple events in mitosis. Studies from yeast to humans have demonstrated that sumoylation is critical for centromere and kinetochore function, chromosome condensation and sister chromatid segregation [2, 3]. The best understood functions have come from targeted analyses of a limited number of SUMO-modified proteins. For example, sumoylation of topoisomerase IIα at centromeres has been shown to be critical for proper decatenation of sister chromatids at the metaphase to anaphase transition [4, 5]. Sumoylation of kinetochore-associated proteins has also been shown to be critical for kinetochore assembly and function [6-9]. Mitotic functions for sumoylation outside of kinetochores and centromeres, however, remain largely unexplored.
Vertebrates express three predominant SUMO paralogs (SUMO-1, SUMO-2, SUMO-3) [1]. While SUMO-2 and SUMO-3 share 97% identity and are referred to as SUMO-2/3, SUMO-1 shares ~50% identity with SUMO-2/3. In mammalian cells, SUMO-1 and SUMO-2/3 are uniquely regulated and conjugated to distinct proteins during mitosis [9]. SUMO-1 modified proteins, including RanGAP1, localize to the mitotic spindle in early mitosis and to the spindle midzone in late mitosis. In contrast, SUMO-2/3 modified proteins localize to centromeres and kinetochores in early mitosis and appear to coat chromosome arms as cells progress from metaphase to telophase. Although the substrates and functions of SUMO-2/3 modification on chromosome arms are unknown, sumoylation is tightly linked to chromatin structure and gene expression in other cell cycle stages [10]. Thus, sumoylation may help regulate the dramatic changes in chromosome required for progression through mitosis [11].
To better understand the functions of sumoylation in mitosis, we have developed a two-step approach for purifying and identifying proteins modified by endogenous SUMO-2/3 and associated with mitotic chromosomes. Combined with mass spectrometry, we identified 149 mitotic chromosome-associated SUMO-2/3 substrates. Identified proteins included kinetochore, centromere and chromatin scaffold-associated proteins, and proteins involved in chromatin remodeling and modification. Our findings are consistent with sumoylation affecting progression through mitosis by acting on a large number of factors to control kinetochore function and chromatin structure.
MATERIALS AND METHODS
Cell culture and synchronization
For immunofluorescence microscopy, HeLa cells were cultured using standard conditions. For immunopurifications, HeLa cells were grown in suspension at 37°C and 5% CO2 in Minimum Essential Medium (Sigma) supplemented with 5% fetal bovine serum, 1% penicillin-streptomycin, and 2 mg/ml sodium bicarbonate. Cells were synchronized overnight using 100 ng/ml nocodazole (Sigma), followed by a two-hour release. For double thymidine synchronizations, cells were treated in Dulbecco’s Modified Eagle Medium (Gibco/Invitrogen) supplemented with 5% fetal bovine serum and 1% HEPES with 2 mM thymidine (Sigma, T9250-5G) for 18 hours, released in thymidine-free media for 5 hours, followed by an additional 2 mM thymidine treatment for 18 hours and a final release.
Antibodies
SUMO-2/3 monoclonal antibody (8A2) [9] was purified from mouse ascites fluid as described [12] and immobilized on Affigel-10 beads (BioRad) according to the manufacturer’s protocol. 6.5 mg of purified 8A2 antibody (experimental) or 6.5 mg of mouse control IgG (Protein Mods LLC, Wisconsin) was used for each purification from 4 L of HeLa cell culture. Other antibodies used: CREST human auto-antibodies, Dr. Ted Salmon (University of North Carolina, NC); anti-TIF1β (ADI-KAM-TF200), Enzo Life Sciences; anti-topoisomerase IIα (sc-13058), Santa Cruz Biotechnology; anti-SMC4, Dr. Tatsuya Hirano (Riken, Japan); anti-phospho-histone H3-Ser10 (06-570), Upstate-Millipore; anti-histone 3 (39163), Active Motif; anti-Hsp90 (610418), BD Transduction Laboratories; anti-KIF4A (GTX115579), Genetex; anti-Smc3 (PA5-29131), Thermo Scientific; anti-APC4 (A301-176A), Bethyl Laboratories; anti-GAPDH (GTX100118), Genetex.
Immunofluorescence microscopy
HeLa cell chromosome spreads were prepared as previously described [13]. Permeabilization buffers were supplemented with 20 mM N-ethylmaleimide (NEM) to inhibit isopeptidases. Immunostaining was performed as described previously [14]. Images were collected using a Zeiss Observer.Z1 fluorescence microscope with an Apotome VH optical sectioning grid and were processed using AxioVision Software.
Chromosome fractionation
Synchronized HeLa cells were harvested by centrifugation and mitotic chromosomes were purified essentially as described [15]. The lysis buffer was supplemented with 10 mM NEM to inhibit isopeptidases. After dounce lysis, lysates were spun at 200 × g for 5 minutes to remove intact cells and nuclei. Lysates were layered onto a 15% sucrose cushion and spun for 30 min at 2,000 × g. Pellets were resuspended in RIPA Buffer (20 mM Tris-HCl pH=7.5, 150 mM sodium chloride, 2 mM EDTA, 1% sodium deoxycholate, 1% triton X-100) supplemented with 1% SDS, 10 mM NEM and protease inhibitors (5 μg/ml leupeptin, 5 μg/ml pepstatin A, and 1 mM PMSF) and used as the soluble chromosome fraction.
Immunopurifications
The chromosome fraction was sonicated (3 × 15 sec) and diluted 1:10 in RIPA buffer supplemented with 10 mM NEM and protease inhibitors. Samples were spun at 50,000 × g, 2 hrs and supernatants were passed through a 0.22 μm filter. Samples were split equally between SUMO-2/3 or mouse IgG antibody beads and rocked at 4°C. Beads were washed 4 × with RIPA buffer supplemented with 0.1% SDS and 4 × with elution buffer (20 mM Tris-HCl pH = 7.5, 500 mM sodium chloride, 2 mM EDTA, 0.1% sodium deoxycholate, 0.1% triton X-100 and 0.1% SDS). Samples were eluted in elution buffer containing 0.5 mg/ml of an 8A2 epitope-specific peptide (IRFRFDGQPINE) and TCA precipitated.
For APC4 immunopurifications, synchronized cells were lysed in RIPA buffer [10 mM Tris pH 8.0, 1 mM EDTA, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS, and 150 mM NaCl, supplemented with 10 mM NEM, and a protease inhibitor cocktail (Complete Ultra Tablets, Mini EDTA-free, Roche) for 15 min on ice and cleared by centrifugation (15 min, 18,000 × g)]. Lysates were divided equally and immunopurifications were performed using APC4 or control rabbit IgG antibodies. Lysates were incubated with antibodies immobilized on Protein A beads (Thermo Scientific) at 4°C for 1 hr and washed 6x in cold RIPA Buffer. Bound proteins were eluted using SDS-sample buffer.
Mass spectrometry
Proteins were separated by SDS-PAGE electrophoresis and subjected to in-gel trypsin digestion. Peptides were extracted and analyzed on an Orbitrap mass spectrometer (Thermo Fisher Scientific). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD000381 [16]. Detailed experimental procedures are provided in the Supplemental Information.
RESULTS
SUMO-2/3 localization
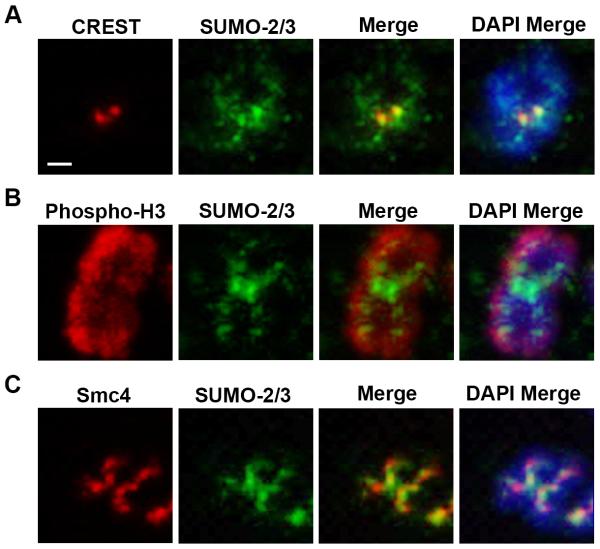
We previously demonstrated that SUMO-2/3 localize to chromosomes throughout mitosis using fixed, intact mitotic cells [9]. To investigate the localization of SUMO-2/3 on mitotic chromosomes more precisely, we analyzed HeLa cell chromosome spreads by immunofluorescence microscopy (Figure 1 and Supplemental Figure 1). Co-staining with human CREST auto-antibodies that recognize CENP-A, CENP-B, and CENP-C reveals that SUMO-2/3 localizes to the paired sister chromatid centromeres (Figure 1A). Because SUMO-2/3 was also detected on the chromosome arms, we conducted co-localization studies with antibodies to a histone marker, phospho-H3 (Figure 1B), and a chromosome scaffold marker, the condensin complex subunit Smc4 (Figure 1C). The SUMO-2/3 signal did not co-localize with the phospho-H3, demonstrating that it is not globally present throughout mitotic chromosomes. However, SUMO-2/3 partially overlaps with Smc4. This finding is consistent with a report showing SUMO-2/3 on the chromosome protein scaffold in cells treated with topoisomerase inhibitors [17].
Figure 1.
SUMO-2/3 localizes to the mitotic chromosome protein scaffold and centromeres. Mitotic chromosome spreads were labeled using DAPI and stained with SUMO-2/3 antibodies and either (A) CREST, (B) phospho-histone 3 or (C) Smc4 antibodies. Chromosomes were analyzed by immunofluorescence microscopy. Bar = 1 μm.
Purification and identification of mitotic chromosome-associated SUMO-2/3 substrates
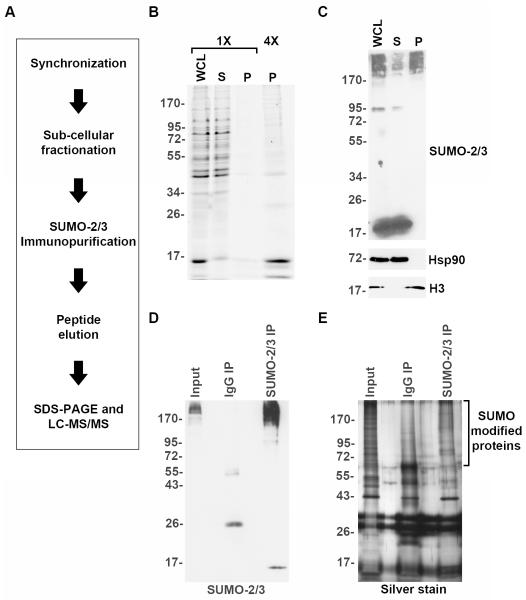
To define chromosome-associated functions of SUMO in mitosis, we developed a protocol for chromosome isolation and immunopurification of proteins modified by endogenous, untagged SUMO-2/3 (Figure 2A). HeLa cells were synchronized in mitosis through nocodazole treatment and released to obtain a predominantly prometaphase population of mitotic cells (Supplemental Figure 2A and 2B). Although mitotic synchronization did not reach 100%, further enrichment was achieved during cellular fractionation when mitotic chromosomes were separated from intact interphase nuclei (see Methods). Mitotic chromosomes were subsequently isolated from cell lysates by differential centrifugation and analyzed by SDS-PAGE followed by Coomassie Blue staining (Figure 2B) or immunoblotting (Figure 2C). The fractionation successfully separated cytosolic Hsp90 from nuclear histone H3 (Figure 2C). Immunoblotting also revealed that unconjugated SUMO-2/3 remained in the soluble protein fraction, while high-molecular mass SUMO-2/3 modified proteins were found in both the soluble and pellet fractions (Figure 2C). Thus, by performing cellular synchronization and fractionation, we enriched for high molecular mass SUMO-2/3 modified proteins associated with mitotic chromosomes and removed unconjugated SUMO-2/3.
Figure 2.
Purification of SUMO-2/3 modified proteins associated with mitotic chromosomes. (A) Schematic diagram illustrating the experimental strategy for purifying and identifying mitotic SUMO-2/3 modified proteins. (B) Cells were synchronized in mitosis and mitotic chromosomes were purified. Equivalent amounts of the resulting cell fractions (WCL=whole cell lysate; S=soluble fraction; P=Mitotic chromosome pellet) were separated by SDS-PAGE and analyzed by Coomassie Blue staining. For clarity, four times (4X) the chromosome pellet was also analyzed. (C) Immunoblot analysis of cell fractions with antibodies specific for heat shock protein 90 (Hsp90), histone 3 (H3) and SUMO-2/3. (D) Mitotic chromosome-associated proteins were solubilized and immunopurified using SUMO-2/3 specific antibodies or control IgG beads. Proteins were analyzed by immunoblotting with SUMO-2/3 specific antibodies. (E) SDS-PAGE and silver stain analysis of control and SUMO-2/3 immunopurified proteins.
To identify sumoylated proteins, chromosome pellets were solubilized and proteins were immunopurified using the SUMO-2/3 specific monoclonal antibody 8A2 or purified mouse IgG as a control. Purified proteins were eluted using an 8A2 epitope-specific peptide and analyzed by SDS-PAGE. Immunoblotting demonstrated that high molecular weight SUMO-2/3 modified proteins were specifically purified with the 8A2 antibody and not IgG (Figure 2D). Silver stain analysis also revealed that the 8A2 antibody uniquely enriched for high molecular mass proteins relative to the IgG control (Figure 2E).
Immunopurifications were conducted from two independently prepared fractions of mitotic chromosomes. Purified proteins were separated by SDS-PAGE and eluted peptides were analyzed by nLC-ESI-MS/MS (Supplemental File 1). The significance analysis of interactome (SAINT) approach was used to identify proteins unique to the SUMO-2/3 immunopurifications [18]. SAINT utilizes statistical analysis of the total number of spectra obtained for individual proteins in experimental and control immunopurifications to determine the probability that a protein is unique to the experimental sample. Using this method, we identified 149 proteins specific to SUMO-2/3 immunopurifications (Supplemental File 2).
Bioinformatic analysis of identified proteins
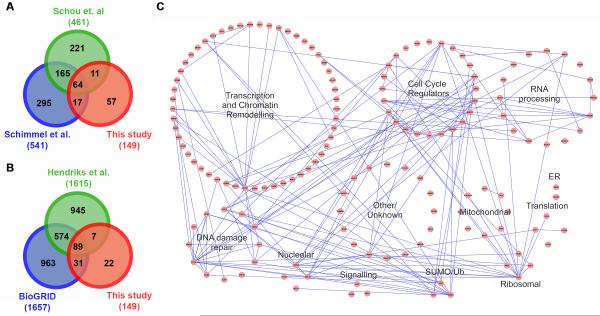
We compared the proteins identified in our study to those of two recent proteomic studies reporting the identification of mitotic SUMO-2/3 substrates (Figure 3A) [19, 20]. In the first study, proteins modified by an exogenously expressed and tagged variant of SUMO-2 were purified from whole cell lysates prepared from HeLa cells synchronized in mitosis using CDK1 inhibition and release [19]. Of the 149 proteins we identified, 54% were also found in this study. Similarly, ~50% of the proteins identified in our study overlapped with proteins identified in a second study also using whole cell lysates prepared from a HeLa cell line expressing an exogenous tagged SUMO-2 variant. In this second study, taxol and an Aurora B kinase inhibitor were used to enrich for mitotic cells [20]. We also compared proteins identified in our study to those identified in a more comprehensive proteome-wide SUMO substrate identification study [21] and to SUMO conjugates reported in the online interaction repository BioGRID [22]. This analysis revealed that >85% of the proteins identified in our study have previously been reported as SUMO conjugates (Figure 3B).
Figure 3.
Bioinformatic analysis of identified SUMO-2/3 modified proteins. (A) A Venn diagram of comparisons between SUMO-2/3 modified proteins identified in this study and two previous proteomic studies [19, 20]. Comparison with proteins identified by Schimmel et al. includes only proteins identified in early and late mitosis. (B) A Venn diagram of comparisons between SUMO-2/3 modified proteins identified in this study, reported in the online repository BioGRID [22] and identified by Hendriks et al. [21]. (C) Functional pathways and protein-protein interactions were analyzed using Cytoscape [48]. Proteins are represented as individual circles forming larger circles that represent the indicated functional pathways. Connecting lines indicate protein-protein interactions.
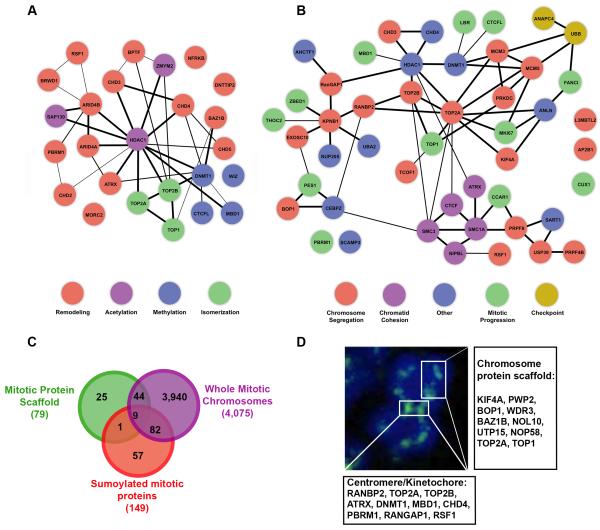
Lastly, we compared the proteins identified in our mitotic chromosome fraction to proteins identified in proteomic studies of the non-histone protein chromosome scaffold or whole mitotic chromosomes [23, 24]. Approximately 62% of the proteins that we identified were observed in these two studies (Figure 4C). Notably, multiple proteins identified in our study correspond to verified constituents of the chromosome protein scaffold (SAFB, KIF4A, BOP1 and the topoisomerases) or residents of centromeres or kinetochores (ATRX, RANBP2 and PBRM1) (Figure 4D).
Figure 4.
Chromatin remodeling and mitotic functions of identified chromosome-associated, SUMO-2/3 modified proteins. (A) Identified proteins with chromatin modifying or remodeling activity were analyzed using STRING [49]. Individual proteins are represented as circles with connecting lines indicating known or predicted interactions. (B) Identified proteins with documented connections to mitotic functions were analyzed using STRING. Individual proteins are represented as circles with connecting lines indicating known or predicted interactions. (C) A Venn diagram comparing the proteins identified in this study to proteins identified in proteomic studies of the mitotic chromosome scaffold and of whole mitotic chromosomes [23, 24]. (D) A schematic illustrating the proteins identified in this study and their known localizations to the chromosome protein scaffold, centromeres and kinetochores.
To gain further functional insights, a global analysis of the interaction networks and functional pathways of the proteins we identified was performed (Figure 3C, Supplemental File 2). Consistent with affecting diverse processes in mitosis, proteins identified in our study regulate transcription, DNA repair, RNA processing and cell cycle progression. Of interest, >40% of the identified proteins are involved in chromatin modification or ATP-dependent remodeling (including ARID4A, ATRX, CHD2, 3, 4 and 5, RSF1, PBRM1 and MORC2), suggesting a role for sumoylation in regulating the dynamic changes in chromatin structure and function that occur during mitosis (Figure 4A). In addition, bioinformatics analysis and literature based searches revealed that more than one-third of the identified proteins have ties to mitotic processes, including chromosome segregation, sister chromatid cohesion and checkpoint signaling (Figure 4B). It is also important to note that a subset of proteins may represent contaminants, including ribosomal and mitochondrial proteins. Further validation of individual proteins will be necessary, however, as some ribosomal proteins, for example, have been reported to have chromatin-associated functions [25].
Validation of identified proteins
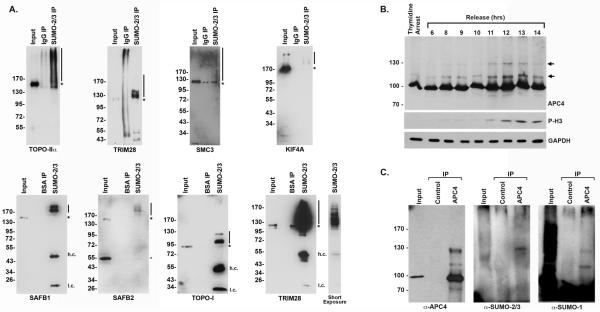
To validate that proteins identified in our study are bona-fide SUMO-2/3 substrates, we performed immunoblot analysis using antibodies specific for seven different proteins: topoisomerase IIα, TRIM28 (KAP-1), SMC3, SAFB1, SAFB2, Topoisomerase 1, and KIF4A (Figure 5A). Topoisomerase IIα was included as a positive control because it is known to be sumoylated in mitosis [4]. TRIM28, SAFB1, SAFB2, Topoisomerase 1 and SMC3, in contrast have only been shown to be sumoylated in asynchronous cell populations, while KIF4A was not previously identified as a SUMO substrate. All seven proteins were detected at their predicted molecular masses in the starting mitotic chromosome fraction. In addition, all seven proteins were also detected in the SUMO-2/3 immunopurification migrating at higher than expected molecular masses consistent with sumoylation. Proteins were not detected in control immunopurifications.
Figure 5.
Validation of identified SUMO substrates. (A) SUMO-2/3 modified proteins were immunopurified from isolated chromosome fractions (SUMO-2/3 IP) and analyzed by immunoblotting with antibodies to topoisomerase IIα, Trim28, KIF4A, SAFB1, SAFB2, Topoisomerase 1, and Smc3 as indicated. The starting chromosome fraction and equivalent fractions of proteins purified on control beads (containing either IgG or BSA, as indicated) are included. The asterisks mark the molecular weight of the unmodified proteins and lines indicate the sumoylated forms of the protein. h.c. denotes heavy chain, while l.c. denotes light chain. (B) HeLa cells were synchronized in S phase using a double-thymidine arrest and released for varying lengths of time. Whole cell lysates were analyzed by immunoblotting for APC4, mitotic phosphorylated histone H3 (P-H3) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control. Arrows indicate modified forms of APC4 peaking during mitosis. (C) Immunopurifications were performed from HeLa whole cell lysates using APC4 and control IgG antibodies. Lysates (Input) and immunopurified proteins were analyzed by immunoblotting with antibodies as indicated.
In addition to validating these SUMO substrates, we also analyzed the sumoylation of the APC4 subunit of the anaphase promoting complex/cyclosome (APC/C). Although APC4 has been suggested through quantitative mass spectrometry studies to be sumoylated in mitosis, its modification has not been more formally analyzed [19, 20]. To evaluate APC4 sumoylation throughout the cell cycle, we synchronized HeLa cells using a double-thymidine block and analyzed lysates at varying time points following release by immunoblotting for APC4 and the mitosis-specific marker, phosphorylated histone H3 (P-H3) (Figure 5B). This analysis revealed the presence of a major, unmodified form of APC4 at all time points (migrating at ~90 kDa) and the presence of two higher molecular mass forms of modified APC4 that peaked during early mitosis as indicated by P-H3 levels. Immunopurification and immunoblot analysis revealed that these higher molecular mass forms of APC4 react with SUMO-1 and SUMO-2/3 specific antibodies.
DISCUSSION
Development of methodologies for purifying proteins modified by SUMO-2/3
The identification of sumoylated proteins is challenging, as most SUMO substrates are modified at relatively low levels. To overcome this challenge, many groups have developed proteomic approaches for identifying sumoylated proteins that involve expression of exogenous, tagged SUMO variants. Although enabling efficient purification schemes, unnatural SUMO expression levels and protein tags introduce unavoidable caveats concerning specificity and function [26, 27]. To avoid these caveats, we have developed an effective strategy for the immunopurification and identification of proteins modified by endogenous, untagged SUMO-2/3. In addition to using SUMO-2/3 specific antibodies, we also introduced a combination of cell synchronization and fractionation procedures into our purification scheme to specifically enrich for chromosome-associated proteins modified in mitosis. In this regard, our approach is distinct from a related SUMO-2/3 immunopurification and substrate identification scheme recently reported [28]. Overall, our approach is fast and effective, although the identification of proteins modified by endogenous untagged SUMOs may require greater amounts of starting material compared to other approaches (~109 cells for endogenous SUMO vs ~107-108 for tagged exogenous SUMO). Thus, this approach may not be ideal for all studies, but represents an important complimentary approach for substrate identification and validation. Notably the overlap between proteins identified in our study and mitotic SUMO substrates identified in recent quantitative mass spectrometry studies was high, with >50% overlap between the most abundant proteins identified. This overlap validates not only our findings, but also serves as an important validation of proteins identified in studies using tagged, over-expressed SUMO variants.
Identification of mitotic SUMO-2/3 modified proteins: chromatin structure and function
We have found that SUMO-2/3 are associated with centromeres and the chromosome protein scaffold under normal cellular conditions. While the association of SUMO-2/3 with chromosomes could reflect sumoylation of a relatively limited number of related proteins, such as histones, we identified 149 sumoylated proteins with a broad range of functions. Thus our findings indicate that sumoylation regulates diverse chromosome-associated activities in mitosis. Among the proteins that we identified were 10 proteins previously shown to be present in the protein chromosome scaffold, including topoisomerase I and IIα [23]. Topoisomerases are likely to be major targets of scaffold sumoylation, as suggested by findings that treating cells with the topoisomerase inhibitors increases topoisomerase I and IIα sumoylation and the intensity of SUMO-2/3 detected on the protein scaffold [17]. Other scaffold proteins that we identified include three nucleolar proteins (NOL10, UTP15, NOP58), the transcription factor BAZ1B, and two proteins involved in proper chromosome segregation, KIF4A and BOP1 [29, 30]. Understanding how sumoylation of these factors affects their chromosome association and activities in mitosis will be an important question for future studies.
In addition to scaffold proteins, >40% of the proteins that we identified are involved in chromatin modification or remodeling. Of interest, a subset of these proteins are associated with pericentric heterochromatin, including ATRX [31, 32], CHD4 [33], RSF1 [34], DNMT1 [32], HDAC1 [32], and MBD1 [32]. In addition to playing roles at sites for kinetochore assembly, the centromere and pericentromere regions of chromosomes also encode for RNA transcripts involved in a variety of functions, including maintenance of heterochromatin structures [35]. Intriguingly, active expression of these transcripts occurs during mitosis (when transcription is otherwise globally suppressed) and is critical for optimal kinetochore assembly and function [36]. SUMO-2/3 are readily detected at centromeres in mitosis (Figure 1), and thus our findings highlight a potentially important function in regulating chromatin structure, and potentially gene expression, within these domains.
With few exceptions, transcription is largely suppressed in mitosis as a consequence of chromosome condensation and eviction of RNA polymerases and transcription factors. SUMO-1 and SUMO-2/3 are likely to be important, generic modulators of gene expression in interphase, as they are chromatin associated throughout the genome and enriched at the promoters of transcribed genes [37]. The fate of sumoylated, chromatin-associated factors upon entry into mitosis has not been carefully evaluated, although sumoylation in general is suppressed during early mitotic stages [9]. Thus, desumoylation may accompany or promote chromatin condensation and transcription inhibition. Notably, a subset of chromatin-associated factors are retained and act as mitotic bookmarks to reestablish interphase chromatin and transcription after mitotic exit [38]. It is therefore intriguing to speculate that chromatin remodeling and transcription factors that we identified in our study may function in bookmarking, and that sumoylation may play a role in their retention at promoters during mitosis. Chromatin modifying and remodeling factors that we identified may also represent proteins that are sumoylated during late stages of mitosis, as part of the process of reestablishing interphase chromatin structure and function.
Identification of mitotic SUMO-2/3 modified proteins: regulation of mitotic progression
SUMO-2/3 are enriched at centromeres and kinetochores, where sumoylation also functions to regulate sister chromatid decatenation, cohesion, and kinetochore assembly and function [2, 3]. We identified 7 kinetochore and centromere associated proteins, including RanGAP1, RanBP2, topoisomerase IIα, MBD1 and DNMT1 whose functions are SUMO-regulated [4, 39-41]. A number of previously described kinetochore-associated SUMO-2/3 substrates were not identified due to two possible explanations. First, many centromere and kinetochore proteins are present in low copy numbers, so the sumoylated forms of these proteins may have been below our level of detection. Proteins may also have been missed because our synchronization method produced a predominantly prometaphase population of cells. Evidence indicates that modification of a number of proteins may be transient and limited to unique phases of mitosis, so proteins modified in phases of mitosis under-represented in our study may have been missed [4, 7, 9].
In total, we identified ~50 putative SUMO-2/3 modified proteins with previously characterized regulatory roles in mitosis ranging from control of chromosome alignment to regulation of anaphase initiation. Of interest, we identified proteins associated with the anaphase-promoting complex (APC), including CCAR1 and a core subunit of the APC itself, APC4 [42]. Our further analysis indicated that APC4 sumoylation peaks during mitosis, suggesting that sumoylation might regulate mitotic APC/C activity. Intriguingly, sumoylation regulates APC/C activation during mitosis in yeast, suggesting that this may be an important and conserved function [43].
In addition, several factors involved in sister chromatid cohesion were identified, including the cohesin subunits, SMC1A and SMC3, the cohesin loader NIPBL and the cohesin-interacting protein CTCF. Studies in yeast have demonstrated that transient sumoylation of cohesin subunits is critical for establishment of sister chromatid cohesion during S phase [44]. In addition to affecting cohesin loading, sumoylation may also promote dissolution of cohesion through SUMO-targeted degradation of cohesion subunits at the metaphase to anaphase transition [45]. Although sumoylation has been implicated in regulating effects of CTCF on transcription during interphase, functions in mitosis remain unexplored [46, 47]. Of interest, CTCF and cohesins interact to control chromatin looping and long-range chromatin interactions relevant to the process of chromosome condensation in mitosis [31]. Understanding how sumoylation may affect interactions between CTCF and cohesins in mitosis will be another important question for future studies.
Concluding remarks
We have identified 149 putative SUMO-2/3 substrates associated with mitotic chromosomes. Our findings expand the repertoire of proteins regulated through sumoylation in mitosis and provide a foundation for more detailed studies aimed at deciphering the effects of sumoylation on chromosome structure, function and segregation. Our findings and methodologies also provide valuable validation and complementation to proteomic studies involving tagged, exogenously expressed SUMO variants.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Matunis lab. We would also like to acknowledge the PRIDE team for managing the mass spectrometry proteomics data. B.R. holds the Canada Research Chair in Proteomics and Molecular Medicine. Work in the Raught lab was supported by a grant from the Canadian Institutes of Health Research (MOP-130340). This work was also supported by grants from the National Institutes of Health (GM060980 to M.J.M.), (U54-RR-020839 to R.J.C. and M.J.M.) and the Johns Hopkins Bloomberg School of Public Health Sommer Scholars program (C.C.-P.)
ABBREVIATIONS
- SUMO
Small ubiquitin-related modifier
- Hsp90
Heat shock protein 90
- SENP
Sentrin-specific protease
- NEM
N- ethylmaleimide
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have declared no conflict of interest.
REFERENCES
- [1].Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annual review of biochemistry. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- [2].Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell division. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wan J, Subramonian D, Zhang XD. SUMOylation in control of accurate chromosome segregation during mitosis. Current protein & peptide science. 2012;13:467–481. doi: 10.2174/138920312802430563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. The Journal of cell biology. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Molecular cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- [6].Fernandez-Miranda G, Perez de Castro I, Carmena M, Aguirre-Portoles C, et al. SUMOylation modulates the function of Aurora-B kinase. Journal of cell science. 2010;123:2823–2833. doi: 10.1242/jcs.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Montpetit B, Hazbun TR, Fields S, Hieter P. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. The Journal of cell biology. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. The Journal of cell biology. 2010;188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang XD, Goeres J, Zhang H, Yen TJ, et al. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Molecular cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cubenas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Developmental cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ohta S, Wood L, Bukowski-Wills JC, Rappsilber J, Earnshaw WC. Building mitotic chromosomes. Current opinion in cell biology. 2011;23:114–121. doi: 10.1016/j.ceb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Harlow E, Lane D. Antibodies : a laboratory manual, Cold Spring Harbor Laboratory. Cold Spring Harbor; NY: 1988. [Google Scholar]
- [13].Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- [14].Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. The Journal of cell biology. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- [16].Vizcaino JA, Cote RG, Csordas A, Dianes JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Agostinho M, Santos V, Ferreira F, Costa R, et al. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer research. 2008;68:2409–2418. doi: 10.1158/0008-5472.CAN-07-2092. [DOI] [PubMed] [Google Scholar]
- [18].Choi H, Larsen B, Lin ZY, Breitkreutz A, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schimmel J, Eifler K, Sigurethsson JO, Cuijpers SA, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Molecular cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- [20].Schou J, Kelstrup CD, Hayward DG, Olsen JV, Nilsson J. Comprehensive Identification of SUMO2/3 Targets and Their Dynamics during Mitosis. PloS one. 2014;9:e100692. doi: 10.1371/journal.pone.0100692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nature structural & molecular biology. 2014 doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stark C, Breitkreutz BJ, Reguly T, Boucher L, et al. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gassmann R, Henzing AJ, Earnshaw WC. Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma. 2005;113:385–397. doi: 10.1007/s00412-004-0326-0. [DOI] [PubMed] [Google Scholar]
- [24].Ohta S, Bukowski-Wills JC, Sanchez-Pulido L, Alves Fde L, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sudakin V. Purification of the mitotic checkpoint complex (MCC) and the anaphase promoting complex/cyclosome (APC/C) from HeLa cells. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5449. pdb prot5449. [DOI] [PubMed] [Google Scholar]
- [26].Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nature protocols. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- [27].Andersen JS, Matic I, Vertegaal AC. Identification of SUMO target proteins by quantitative proteomics. Methods in molecular biology. 2009;497:19–31. doi: 10.1007/978-1-59745-566-4_2. [DOI] [PubMed] [Google Scholar]
- [28].Becker J, Barysch SV, Karaca S, Dittner C, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nature structural & molecular biology. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- [29].Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Killian A, Le Meur N, Sesboue R, Bourguignon J, et al. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene. 2004;23:8597–8602. doi: 10.1038/sj.onc.1207845. [DOI] [PubMed] [Google Scholar]
- [31].McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Craig JM, Earle E, Canham P, Wong LH, et al. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Human molecular genetics. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- [33].Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–457. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. The Journal of cell biology. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hall LE, Mitchell SE, O’Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2012;20:535–546. doi: 10.1007/s10577-012-9297-9. [DOI] [PubMed] [Google Scholar]
- [36].Chan FL, Marshall OJ, Saffery R, Kim BW, et al. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome research. 2013 doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kadauke S, Blobel GA. Mitotic bookmarking by transcription factors. Epigenetics & chromatin. 2013;6:6. doi: 10.1186/1756-8935-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lyst MJ, Nan X, Stancheva I. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. The EMBO journal. 2006;25:5317–5328. doi: 10.1038/sj.emboj.7601404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [40].Lee B, Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. The Biochemical journal. 2009;421:449–461. doi: 10.1042/BJ20090142. [DOI] [PubMed] [Google Scholar]
- [41].Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. The Journal of cell biology. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. The Journal of cell biology. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dieckhoff P, Bolte M, Sancak Y, Braus GH, Irniger S. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Molecular microbiology. 2004;51:1375–1387. doi: 10.1046/j.1365-2958.2003.03910.x. [DOI] [PubMed] [Google Scholar]
- [44].Almedawar S, Colomina N, Bermudez-Lopez M, Pocino-Merino I, Torres-Rosell J. A SUMO-dependent step during establishment of sister chromatid cohesion. Current biology : CB. 2012;22:1576–1581. doi: 10.1016/j.cub.2012.06.046. [DOI] [PubMed] [Google Scholar]
- [45].D’Ambrosio LM, Lavoie BD. Pds5 Prevents the PolySUMO-Dependent Separation of Sister Chromatids. Current biology : CB. 2014 doi: 10.1016/j.cub.2013.12.038. [DOI] [PubMed] [Google Scholar]
- [46].Kitchen NS, Schoenherr CJ. Sumoylation modulates a domain in CTCF that activates transcription and decondenses chromatin. Journal of cellular biochemistry. 2010;111:665–675. doi: 10.1002/jcb.22751. [DOI] [PubMed] [Google Scholar]
- [47].MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Molecular and cellular biology. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lopes CT, Franz M, Kazi F, Donaldson SL, et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Franceschini A, Szklarczyk D, Frankild S, Kuhn M, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.