Abstract
Historically, the ribosome has been viewed as a complex ribozyme with constitutive rather than regulatory capacity in mRNA translation. Here we identify mutations of the Ribosomal Protein L38 (Rpl38) gene in mice exhibiting surprising tissue specific patterning defects, including pronounced homeotic transformations of the axial skeleton. In Rpl38 mutant embryos, global protein synthesis is unchanged however the translation of a select subset of Homeobox mRNAs is perturbed. Our data reveal that RPL38 facilitates 80S complex formation on these mRNAs as a regulatory component of the ribosome to confer transcript-specific translational control. We further show that Rpl38 expression is markedly enriched in regions of the embryo where loss-of-function phenotypes occur. Unexpectedly, a ribosomal protein (RP) expression screen reveals dynamic regulation of individual RPs within the vertebrate embryo. Collectively, these findings suggest that RP activity may be highly regulated to impart a new layer of specificity in the control of gene expression and mammalian development.
Introduction
Control of gene expression in space and time plays an important role in enabling cells to “know” where they are in the developing embryo and what to become. Decades of research have demonstrated numerous layers of regulation, at both the transcriptional and post-transcriptional level, that coordinate this process. Our current understanding of translational regulation has been guided, to a large extent, by studies of early invertebrate development where decisive events occur prior to the initiation of zygotic transcription and are solely directed by translational control of pre-existing maternal mRNAs (Kuersten and Goodwin, 2003; Sonenberg and Hinnebusch, 2009; Thompson, 2007). In contrast, early mammalian development relies on almost immediate zygotic transcription and hence the requirement for translational control during development and pattern formation has been less certain.
The eukaryotic ribosome is a molecular machine composed of 4 RNA molecules and 79 distinct ribosomal proteins (RPs)(Laboratory, 2002). To date, the ribosome--although an immensely complex and amazing machine--has largely been thought to exert a constitutive rather than regulatory function in translating mRNAs. This is, however, surprising given the fact that almost every major molecular machinery involved in gene regulation, including the splicesome (Nilsen and Graveley) and chromatin associated histones (Campos and Reinberg, 2009) confer more specialized functions in gene expression. Despite the constitutive nature of the ribosome, several layers of specificity may exist. Ribosomal RNA (rRNA) and the 79 RPs are both extensively modified (Chow et al., 2007; Decatur and Fournier, 2003). Ribosome heterogeneity, including differences in RP composition and/or modifications, has been well documented in Dictyostelium discoideum, in which ribosomes of vegetative amoebae differ from those of spores and cells at different stages of development(Ramagopal, 1992). Unexpectedly, modifications in rRNA have been shown to confer more specialized functions in IRES-dependent translation control and defects in this process underlie the human X-linked Dyskeratosis Congenita syndrome(Bellodi et al.; Bellodi et al.; Yoon et al., 2006). In addition, Saccharomyces cerevisiae contain many duplicated genes encoding RPs, and these RP paralogues appear to be functionally distinct(Komili et al., 2007). Moreover, unexpected tissue specific phenotypes are associated with RP loss of function in many organisms. For example, in Arabidopsis thaliana mutations in RPs perturb various developmental processes (Byrne, 2009). In zebrafish, knockdown or mutation of multiple distinct RPs leads to a remarkable range of phenotypes associated with specific defects in brain, body trunk, eye and ear development and an increase in the incidence of peripheral nervous system tumors(Amsterdam et al., 2004; Uechi et al., 2006). In all of these cases, the mechanism(s) that would account for these specific phenotypes remains poorly understood or has been attributed to extra-ribosomal protein functions, such as activation of p53 (Warner and McIntosh, 2009). One important consideration is whether these phenotypes could be attributed to inherent differences in the rate of protein synthesis in one cell type or tissue versus another. For example, highly proliferating tissues may be more sensitive to changes in protein synthesis and when these thresholds are lowered, indirect effects on cell proliferation or programmed cell death may manifest as tissue specific morphological abnormalities. It has therefore remained ambiguous whether these morphological abnormalities reflect a direct role for RPs in control of embryonic development.
In the present study, we unexpectedly show that a component of the ribosome is required to establish the mammalian body plan. Our data reveal that the phenotypes associated with Rpl38 loss of function, in particular homeotic transformations i.e. the replacement of one skeletal structure with another, are consistent with a very specific and regulatory role for ribosomes during tissue patterning. Indeed, by optimizing genetic and molecular approaches to study translational control within the vertebrate embryo we uncover an important role for RPL38 in transcript-specific translational control. Moreover, our data reveal surprising heterogeneity in expression profiles between individual RPs in the vertebrate embryo. These findings highlight an unexpected role for RPs as regulators of key patterning events during embryonic development and strongly suggest that that RPs may be highly regulated to impart a new level of specificity in control of gene expression.
Results
The Rpl38 gene is mutated in Ts, Tss, and Rbt mice
Tail short (Ts) is a spontaneous, dominant mouse mutation discovered 60 years ago that exhibits pronounced skeletal patterning defects, including homeotic transformations, for which the causative mutation is unknown(Deol, 1961; Morgan, 1950). The name reflects the most obvious phenotype present in Ts/+ mice; a shorter and kinky tail that is associated with more pronounced skeletal patterning defects(Deol, 1961; Morgan, 1950). Two additional mouse mutants, Tail-short Shionogi (Tss) (Tsukahara et al., 2000) and Rabo torcido (Rbt) (Hustert et al., 1996), display virtually identical phenotypes to Ts and map to the distal part of chromosome 11 where the Ts locus is found. Homozygote Ts mutants are morphologically abnormal at the morula stage and die around the time of implantation (our unpublished observations). In the present study we backcrossed the Ts allele onto a C57BL/6J background. Ts/+ embryos display an abnormally short and kinky tail by E12.5 (Figure 1A), a midline facial cleft (Figure 1A) and/or cleft palate (22%) (data not shown), as well as exencephaly that can be accompanied by a wavy neural tube (30%) (Figure 4B and Figure S3 and data not shown). Ts/+ embryos also have various eye abnormalities, including failure in optic fissure closure as well as microphthalmy (small eye size) (Figure 1A and data not shown). The most pronounced phenotypes observed in Ts/+ embryos are axial skeletal patterning defects (Figure 1B and Figure 2). For example, Ts/+ mice show the presence of 14 instead of 13 ribs (Figure 1B) strongly suggesting that the first lumbar vertebra that normally does not possess a rib anlage is now transformed anteriorly into a thoracic vertebra. While this phenotype was noted in the original description of Ts/+ mutant mice in 1950, it was not interpreted as a homeotic transformation at that time (Deol, 1961; Morgan, 1950). Our complete analysis of skeletal patterning in Ts/+ mice reveals pronounced patterning defects as well as A-P directed homeotic transformations along the entire axial skeleton (Figure 2). Therefore, the gene mutated in Ts is an important regulator of A-P skeletal patterning.
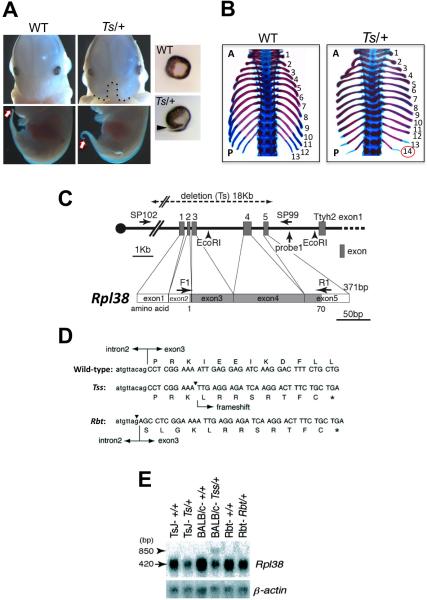
Figure 1. The Rpl38 gene is mutated in Ts, Tss, and Rbt mice.
(A) Ts/+ embryos show a midline facial cleft (black dotted line) and eye defects such as a failure in optic fissure closure (black arrow) at E13.5. Ts/+ embryos have an abnormally short and kinky tail (white arrow) by E12.5. (B) An example of the axial skeletal patterning defects in Ts/+ embryos at E18.5. Note the presence of 14 (red circle) instead of 13 ribs in Ts/+ embryos, indicating that the first lumbar vertebra that normally does not possess a rib anlage is now transformed anteriorly into a thoracic vertebra. The rib cage has been removed in these photographs. (C) The exon-intron organization of the Rpl38 gene. The region deleted in the Ts mutation is illustrated on the genome structure. The coding region of the Rpl38 gene is shadowed. The initiation codon ATG for the first methionine is located at the end of exon2. PCR and Southern Blot analysis of the genome structure of the Rpl38 locus in Ts mice with the indicated primers (Sp102 and Sp99) and probes (Probe 1 and Exon 4) is shown in Fig. S1B. (D) Rescue of Ts skeletal phenotypes in Ts/+; pCAGGS-Rpl38 mice. (E) Rpl38 mutations in Rbt and Tss. In exon3 of Tss, a deletion of a base A in exon3 (arrowhead). Rbt has a C to G transversion mutation at intron2 adjacent to the acceptor splice site near exon3 (arrowhead). (F) In Ts/+ Tss/+, Rbt/+ mice the expression of Rpl38 mRNA is reduced to almost half that of wild-type levels. See also Figure S1.
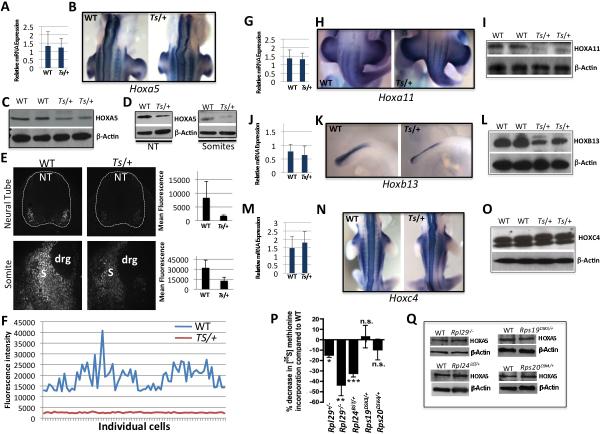
Figure 4. Analysis of Hox gene mRNA expression levels, boundaries, and protein levels in Wt and Ts/+ embryos compared to additional RP mouse deficiencies.
(A-O) Somite stage 40 (~E11.0) Wt and Ts/+ embryos were analyzed. Unless specified, all experiments have been performed utilizing the microdissected neural tube & somite tissue fragment, Western Blot (WB), whole mount in situ hybridization (WISH). (A) Hoxa5 qPCR. (B) Hoxa5 WISH showing the anterior boundary of expression. (C) HOXA5 WB. (D) HOXA5 WB in microdissected neural tube (NT) or Somites (S). (E) HOXA5 immunofluoresence (grey scale) of Wt and Ts/+ transverse tissue sections at the forelimb level. Neural tube (top panel) and somite (bottom panel) expression is shown from serial sections of the same embryo (10X). Dorsal root ganglion (drg). Quantification of mean fluorescence intensity is shown in the graphs to the right. (F) Quantification of florescent intensities of HOXA5 within individual cells (ticks on the X-axis) of the NT. (G) Hoxa11 qPCR. (H) Hoxa11 WISH showing the anterior boundary of expression. (I) HOXA11 WB. (J) Hoxb13 qPCR. (K) Hoxb13 WISH. (L) HOXB13 WB. (M) Hoxc4 qPCR. (N) Hoxc4 WISH showing the anterior boundary of expression. (O) HOXC4 WB. (P) Relative [S35] methionine incorporation to monitor the rates of de-novo protein synthesis in the neural tube and somites of stage 40 embryos with distinct RP deficiencies (see also Figure S4D). The graph shows quantification in n=3 embryos, *P=0.04, **P=0.007, ***P=0.03 (Student's t-test), not significant (n.s.). (Q) HOXA5 WB, a target mRNA translationally deregulated in Ts/+ embryos (see C-D), reveals no change in expression levels in other RP deficiencies. Data are presented as the average ± SEM. See also Figure S4.
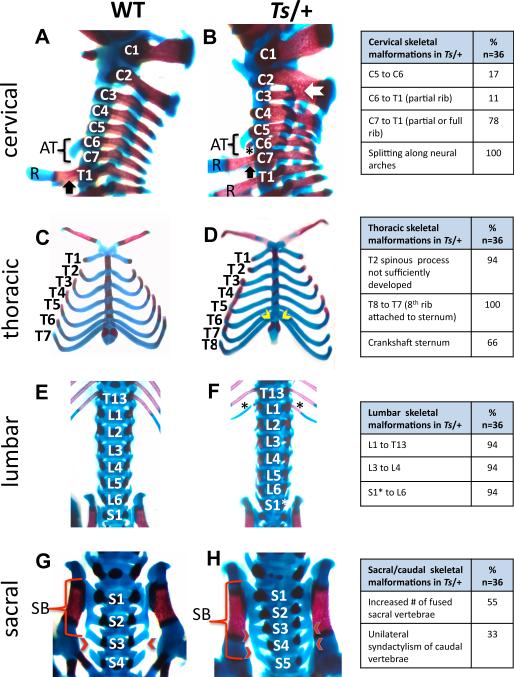
Figure 2. Homeotic transformations and skeletal patterning defects in Ts/+ embryos.
(A-H) Alizarin red and Alcian blue staining of WT and Ts/+ skeletons at E18.5. The frequency of patterning defects is presented in the table on the right (note that not all of these phenotypes are shown). Lateral view of the cervical (C) and upper thoracic (T) region. In Ts/+ embryos the anterior tuberculum (AT) normally a feature of C6 is instead present on C5. The star indicates an ectopic, partially formed rib on C6 in Ts/+ embryos. The black arrow indicates the presence of ribs (R) on the first thoracic vertebrae in Wt embryos that is a feature of C7 in Ts/+ embryos. Fusions between cervical vertebrae in Ts/+ embryos (white arrow). (C-D) Ventral view of the thorax reveals that in Ts/+ embryos there are eight pairs of vertebrosternal ribs instead of seven attached to the sternum (yellow arrow heads). The attachment site of ribs to the sternum is asymmetric in Ts/+ embryos resulting in a crankshaft sternum. (E-F) In the lumbar region, an extra pair of ribs (black asterisks) is present on the first lumbar vertebrae in Ts/+ embryos. A normal number of lumbar segments in Ts/+ embryos suggest that a partial transformation of S1 into a lumbar identity (S1*) has occurred. (G-H) Sacral vertebrae in Wt (G) and Ts/+ (H). In Wt embryos, the first three sacral vertebrae are fused to make the sacral bone (SB), while in Ts/+ embryos, the fourth and sometimes fifth sacral vertebrae are fused (red arrow heads). See also Figure S2.
To characterize the genetic defect responsible for the Ts phenotype, we positionally cloned the responsible gene (Figure S1A) and disclosed a 18kb deletion in Rpl38 encompassing all exons of the gene in Ts/+ mice (Figure 1C, Figure S1B, Figure S1C). Direct sequencing of Tss/+ and Rbt/+ mice also identified mutations in Rpl38. In Tss/+ mice, a deletion of a single base is present within exon 3. This results in a frameshift after codon four that is followed by a stop codon, which disrupts the production of RPL38 protein (Figure 1E and Figure S1D). In Rbt/+ mice, an insertion of the dinucleotide AG precedes the first codon in exon3. This insertion results in a change of the acceptor splice site, leading to a frameshift mutation with a very short theoretical translation product (12 amino acids) and a stop codon (Figure 1E). To further ensure that the Rpl38 gene indeed causes the Ts phenotype, we generated pCAGGS-Rpl38 transgenic mice and found that the majority of Ts phenotypes, including homeotic transformations, were no longer manifest in Ts/+; pCAGGS-Rpl38 mice (Figure 1D, Figure S1E). Importantly, Rpl38 expression is reduced by approximately 50% in Rbt/+, Tss/+, and Ts/+ mice (Figure 1F and Figure S1F) and is rescued to normal levels in Ts/+; pCAGGS-Rpl38 mice (Figure S1F). All of these results confirm that the phenotypes observed in Ts/+ mice are a direct consequence of the deletion in the Rpl38 gene.
RPL38 regulates axial skeletal patterning independently from control of Hox gene expression boundaries and transcript levels
The vertebrate skeleton is built on an intricate pattern of stereotyped anatomical elements, the vertebrae, which on the basis of their relative position along the anterior-posterior (A-P) axis in the developing embryo have distinct anatomical features(Iimura et al., 2009). The relative number and arrangement of these vertebrae types collectively give rise to the mammalian body plan(Burke et al., 1995). Vertebrae are grouped into five distinct types: cervical, thoracic, lumbar, sacral and caudal that extend from A-P along the body axis. Skeletal patterning defects, including homeotic transformations are present along the entire A-P axis of Ts/+ mice (Figure 2). Notable examples include transformations in the cervical region. In wild-type (WT) mice, the 6th cervical vertebra (C6) is readily distinguishable by the presence of a ventral process known as the anterior tuberculum, but in Ts/+ embryos it can be found on the 5th cervical vertebra (C5) (Figure 2A and 2B), illustrating a posterior transformation of the C5 vertebra towards the identity of C6. Furthermore, a rib anlage is not normally present on cervical vertebrae. However, in Ts/+ embryos, a minute or extensive rib anlage can develop on one or both processus transversus of C6 or C7, indicating a partial posterior transformation (Figure 2C and 2D). In the thoracic region, Ts/+ embryos display eight instead of seven vertebrosternal ribs (ribs attached to the sternum), and frequently their attachment is asymmetric resulting in a crankshaft sternum (Figure 2C and 2D). The fact that 14 instead of 13 ribs are associated with vertebrae in Ts/+ mice (Figure 1B) indicates that the first lumbar vertebra (L1) is transformed anteriorily and acquires a thoracic identity (Figure 2E and 2F). In the sacral region, normally three or four sacral vertebrae contribute to form the sacral bone, whereas in Ts/+ embryos at least four or even five transverse processes fuse and these alterations illustrate additional anterior transformations (Figure 2G and 2H). Additional abnormalities in the axial skeleton are observed to a variable extent and include fusions of vertebrae and/or laminae, splitting along neural arches (most predominately in the cervical region) as well as ectopic points of ossicification between cervical vertebrae (Figure 2 and data not shown). Therefore, the analysis of Ts/+ mice identifies RPL38 as an important regulator of axial skeletal patterning.
Mammals have 39 Hox genes that are clustered on four chromosomes(Kmita and Duboule, 2003). Targeted loss and gain of function analysis in the mouse have clearly demonstrated that Hox genes are the key regulators of morphology along the axial skeleton (2009; Deschamps and van Nes, 2005; Wellik, 2009). Surprisingly, a careful analysis of Hox gene expression boundaries and transcript levels (Figure S2A, Figure S2B and data not shown) revealed that they remain unchanged in Ts/+ embryos. A notable exception is the increase in Hoxb4 and Hoxd4 transcript levels in Ts/+ embryos (Figure S2B), and these results will be discussed in the context of post-transcriptional analysis of Hox gene expression (see below and Figure 3). Together, these findings show that Rpl38 is a critical regulator of skeletal patterning and the phenotypes observed in Ts/+ mice are unlikely to be the result of shifts in Hox gene expression boundaries and/or transcript levels.
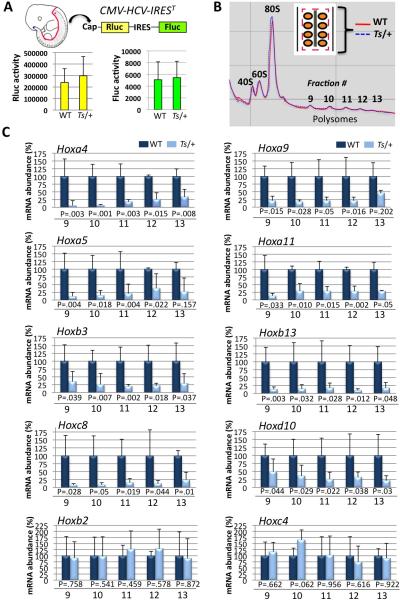
Figure 3. Analysis of global protein synthesis rates and polysome profiling within the neural tube and somites of Wt and Ts/+ embryos.
(A) Transgenic mice expressing a stable, genetically encoded translational reporter for cap- and IRES-dependent translation (CMV-HCV-IREST) were intercrossed with Ts/+ mice. No changes in CAP or IRES-dependent translation are evident within the neural tube and somites of Ts/+ embryos compared to WT. (B) Overlay of polysome profiles from Wt (red solid line) and Ts/+ (blue dotted line) somite stage 40 embryos (~E11.0). Schematic of micro-dissection strategy is shown (insert). Neural tube (grey) and somites (orange) were simultaneously microdissected as illustrated (red dashed line). (C) qPCR analysis of 8/39 Hox mRNAs that show differential association in polysome fractions (fraction numbers are depicted on the bottom of the graph) in Ts/+ tissue fragments, (n=5) for each genotype. The bottom two graphs are representative examples of 2/31 Hox mRNAs that do not show changes in polysome association in Ts/+ tissue fragments. P values (Student's t-test) for each polysome fraction are shown. Data are presented as the average ± SEM. See also Figure S3.
Polysome profiling reveals a subset of Hox mRNAs that are translationally regulated by RPL38, whereas global protein synthesis is unaffected in Ts /+ embryos
We next asked whether changes in translational control underlie the skeletal patterning defects observed in Ts/+ mice. Historically, it has been difficult to systematically assess translational control of gene expression directly within a developing vertebrate embryo. To overcome these limitations, we optimized unique approaches. To directly monitor global protein synthesis in-vivo, we employed a bioluminescent “translation reporter mouse (CMV-HCV-IREST)” that expresses a stable, genetically encoded translational reporter for cap (Rluc) and IRES-dependent translation (Fluc) (Bellodi et al.) (Figure 3A). We simultaneously microdissected the neural tube and somites (the precursors of vertebrae) from somite stage 40 CMVHCV-IREST and Ts/+; CMV-HCV-IREST embryos and measured Rluc and Fluc activity. This analysis revealed no major differences in general cap-dependent or IRES-dependent translational control in Ts/+ embryonic tissues (Figure 3A). We also confirmed that there is no change in global protein synthesis in Ts/+ embryos by [S35] methionine incorporation (Figure S3G and S3H) and direct immunostaining with the Y10b antibody, which labels rRNA (Figures S3A-S3F). Together, these findings demonstrate that changes in global protein synthesis are not observed in somites and neural tube of Ts/+ embryos, where patterning defects are manifest (Figure 2 and Figure 5).
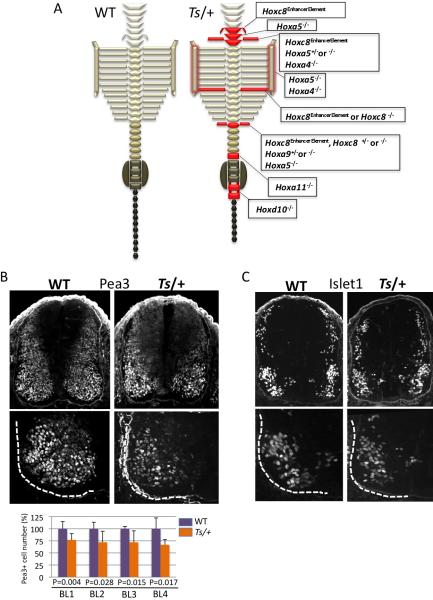
Figure 5. Comparison of the Ts/+ axial skeletal and neural tube patterning defects with Hox loss of function mutants.
(A) Schematic representation of the axial skeleton of WT and Ts/+ mice. Illustrated in red are transformations or patterning defects frequently observed in Ts/+ mice. The red shadow outlining the sternum indicates abnormal rib to sternum attachment in Ts/+ mice. Corresponding Hox mutants that show similar phenotypes are listed in boxes to the right of the Ts/+ axial skeleton schematic representation: Hoxc8 enhancer element knockout (Hoxc8EE)(Juan and Ruddle, 2003), Hoxa5 knockout (Hoxa5)(Jeannotte et al., 1993), Hoxa4 knockout (Hoxa4)(Horan et al., 1994), Hoxc8 knockout (Hoxc8)(Le Mouellic et al., 1992), Hoxa9 knockout (Hoxa9)(Fromental-Ramain et al., 1996), Hoxd10 knockout (Hoxd10)(Carpenter et al., 1997), Hoxa11 knockout (Hoxa11)(Small and Potter, 1993). See also Figure 2. (B) Analysis of Pea3 expression at the Branchial Level (BL) of the neural tube in serial transverse sections (BL1-BL4; caudal to rostral) at somite stage 40. The bottom sections show the ventral left quadrant of the neural tube that is outlined with a dashed white line. The graph shows quantification of Pea3+ LMC motor neurons (MNs) from n=7 WT and Ts/+ embryos and P values (Student's t-test) are indicated. (C) Islet 1 expression at the Branchial Level (BL). The bottom sections show the ventral left quadrant of the neural tube that is outlined with a dashed white line. No differences in Islet 1 expression are observed in Ts/+ embryos. See also Figure S5.
The relative translation rate of an mRNA can be inferred from the number of ribosomes (polysomes) it recruits and can be quantitatively analyzed by employing sucrose gradients to purify mRNAs associated with translationally active ribosomes (Rousseau et al., 1996; Sagliocco et al., 1996). Several limitations exist to performing polysome analysis from vertebrate embryonic tissues including the small size of mid-gestation embryos and the limiting number of cells that can be obtained from microdissected tissue fragments. To investigate whether qualitative changes in mRNA translation may underlie the skeletal patterning defects observed in Ts/+ embryos, we optimized microscale polysome analysis (see methods and Figure S3I-S3K). This approach enabled us to detect and collect polysome fractions employing small tissue fragments. We simultaneously microdissected the neural tube and somites from somite stage 40 (~E11.0) Wt and Ts/+ embryos for this analysis (Figure 3B). This microdissection strategy ensured that all A-P somites and the entire rostral-caudal extent of the neural tube was collected from each embryo in an identical and precise manner. We did not detect noticeable differences in the ratio of small to large ribosome subunits or polysome distribution comparing WT and Ts/+ ribosome profiles, in agreement with the fact that general protein synthesis is not impaired in Ts/+ samples (Figure 3B). We next optimized qPCR primers (Table S1) for all genes that belong to the Hox A, B, C, and D clusters and assayed their translational state in WT and Ts/+ embryos by detecting their mRNA distribution in polysome fractions (Figure 3C and data not shown). Consistent with the fact that general protein synthesis is not perturbed in Ts/+ tissue samples, the majority of Hox genes (a total of 31) did not show any difference in polysome association (Figure 3C and data not shown). Strikingly, however, we identified a specific subset of Hox genes [Hox a4; a5; a9; a11; b3; b13; c8; d10] that are markedly decreased in polysome association in Ts/+ embryos compared to WT (Figure 3C), and this defect is evident in both heavy and light polysomes. Interestingly, the Hox genes found translationally deregulated in Ts/+ embryos include members from each of the four Hox gene clusters (Figure 3C). Moreover, the decreases in Hoxa4 mRNA polysome association in Ts/+ embryos are consistent with the observed increases in Hoxd4 and Hoxb4 transcript levels (Figure S2B), as Hox4 family members have been shown to cross-regulate their expression levels (Zappavigna et al., 1991).
We next carried out additional experiments to validate our polysome analysis (Figure 4). Consistent with these findings (Figure 3C), we observed diminished HOXA5 protein levels in Ts/+ embryos compared to WT (Figure 4C) while no differences were observed in Hoxa5 mRNA levels or expression boundaries (Figure 4A-4B). Similar reductions in HOXA5 protein levels were found in both the neural tube and somites (Figure 4D). Moreover, quantitative HOXA5 immunofluoresence revealed markedly diminished expression in both the neural tube and somites along the A-P extent of Ts/+ embryos compared to WT (Figure 4E-4F). The systematic analysis of additional Hox family members, such as Hoxa11 and Hoxb13, similarly revealed marked decreases in protein levels, while no changes in mRNA expression levels or boundaries are evident in Ts/+ embryos (Figure 4G-4L). Moreover, mRNAs that showed no change in polysome association in Ts/+ embryos, such as Hoxc4, showed no change in expression (Figure 4M-O). Importantly, the polysome analysis and molecular characterization of Hox gene expression were carried out at an early stage (somite stage 40), much prior to when morphological defects are evident in Ts/+ embryos. Therefore the molecular defects in translational control of a subset of Hox genes precede the developmental defect in axial skeletal patterning. Moreover, the protein expression levels of Hox genes regulated by RPL38 are restored to normal levels in Ts/+; pCAGGS-Rpl38 embryos (Figure S4A), consistent with the rescue in skeletal patterning defects (Figure S1E). In agreement with the direct role of RPL38 in translational control of Hox mRNAs, we observed a similar decrease in HOX protein levels upon stable knockdown of RPL38 in C3H/10T1/2 cells, a multipotent mesenchymal stem cell line that is representative of the mesoderm/somite fate (Pinney and Emerson, 1989)(Figure S4B). Importantly, Hox mRNA levels (Figure S4B) and HOX protein stability (data not shown) are unperturbed upon RPL38 knockdown. In addition, we find that Hox mRNAs translationally deregulated in Ts/+ embryos can be efficiently translated in an in-vitro translation system (Figure S4C) revealing that the molecular defect cannot be at the level of the Hox transcripts themselves. Together, these findings show that RPL38 exerts a specialized function in translational control of a subset of Hox mRNAs.
A series of mouse RP deficiencies do not show Ts /+ phenotypes or changes in Hox mRNA translation, even when global protein synthesis is markedly reduced
We characterized five additional mouse mutants(Kirn-Safran et al., 2007; McGowan et al., 2008; Oliver et al., 2004) with specific RP deficiencies (Rps19DSK3/+, Rps20Dsk4/+, Rpl29+/−, Rpl29−/−, Rpl24BST/+) and found that they produce either no decrease, a small change ie ~10%, or up to a ~35% decrease in global protein synthesis in exactly the same cell/tissue types—neural tube and somites—that have been analyzed in Ts/+ embryos (Figure 4P and Figure S4D). In all of these RP deficiencies, including Rpl29−/− and Rpl24BST/+ embryos that possess a very significant (~30% or greater) decrease in global protein synthesis, no differences in the expression of Hox proteins are evident (Figure 4Q). This is consistent with the fact that none of these additional RP deficiencies(Kirn-Safran et al., 2007; McGowan et al., 2008; Oliver et al., 2004) produce the same tissue patterning phenotypes as in Ts/+ embryos (our unpublished observations). Therefore, these findings strongly support that RPL38 exerts a specialized function in translational control that is distinct from other components of the translational apparatus.
Tissue patterning defects in Ts /+ embryos are phenocopied by the loss of function of specific Hox mRNAs translationally regulated by RPL38
We next systematically compared the axial skeletal phenotypes of Ts/+ mice to those associated with loss of function of each of the Hox genes translationally regulated by RPL38. Strikingly, the majority of Ts/+ axial skeletal phenotypes can be fully accounted for by the combinatorial loss in expression of these specific Hox genes (Figure 5A). Moreover, many of the skeletal phenotypes manifest in Hox mutant mice occur in heterozygosity (Figure 5A), which indicates that maintenance of accurate Hox protein levels is critically required for accurate axial skeletal patterning. In addition, loss of some RPs triggers a p53-dependent stress response, which may account for certain morphological abnormalities(McGowan et al., 2008). Importantly, the axial skeletal patterning defects observed in Ts/+ mice (Figure 2) remained unchanged when the dose of p53 is reduced (Figure S5A).
We also further investigated whether the reductions in Hox protein expression within the neural tube of Ts/+ embryos (Figure 4) are associated with neural patterning defects. While Sonic hedgehog (Shh) specifically directs patterning of the ventral neural tube(Jacob and Briscoe, 2003), Hox genes control motor neuron fates along the rostrocaudal axis, in particular the specification of columnar and pool subtypes(Dasen and Jessell, 2009). A number of Hox genes, such as Hoxc8 specifically control the expression of motor pool markers such as Pea3, an ETS-class transcription factor (Dasen et al., 2005). In Ts/+ embryos, no changes are evident in Dorso-Ventral (D-V) patterning markers regulated by Shh (Figure S5B). Consistent with the fact that RPL38 controls Hoxc8 translation (Figure 3C), we observed a specific decrease in Pea3+ MNs within the brachial LMC of Ts/+ embryos (Figure 5B), while MN differentiation appears unaffected (Figure 5C). Altogether, these findings strongly suggest that the specific changes in MN markers (ie. Pea3 but not D-V patterning markers regulated by Shh) manifest in Ts/+ embryos phenocopies specific molecular changes associated with Hox loss of function in the neural tube.
Although RPL38 may regulate the translation of additional target mRNAs, this comparison strongly suggests that the axial skeletal and neural tube patterning phenotypes observed in Ts/+ mice are, to a large extent, the consequence of diminished protein levels of a specific subset of Hox genes. Moreover, these findings reveal new complexity in regulation of Hox gene expression at the level of translational control.
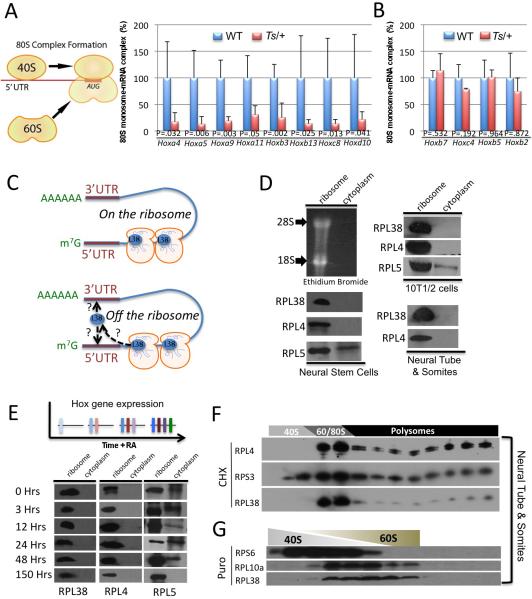
RPL38 exerts specialized control of translation through its association with the ribosome and regulates 80S-mRNA complex formation on specific Hox mRNAs
In order to delineate the molecular step at which RPL38 affects translation, we next performed sucrose gradient fractionation experiments in which we examined the abundance of Hox mRNAs in earlier, non-polysomal fractions comparing WT and Ts/+ samples. A specific and dramatic decrease in 80S-mRNA complex formation is observed on selective Hox mRNAs in Ts/+ samples (Figure 6A). Moreover, consistent with our polysome analysis (Figure 3C) only Hox mRNAs that show decreased polysome association in Ts/+ embryos are affected (Figure 6B). In certain cases, the decrease in 80S-mRNA complex formation was accompanied by a corresponding increase in mRNA accumulation in early non-ribosomal fractions that likely reflects an accumulation of mRNA not bound by ribosomal subunits (Figure S6A). Therefore, these findings show an important role for RPL38 in facilitating 80S complex formation on specific mRNAs at the earliest stages of translation initiation.
Figure 6. RPL38 controls 80S complex formation on selective Hox mRNAs from the ribosome.
(A-B) 80S monosome–mRNA complex formation assessed in-vivo in microdissected neural tube & somites from somite stage 40 WT and Ts/+ embryos, n=5. qPCR analysis of Hox mRNAs from fractions 7-8 of sucrose gradients, corresponding to the 80S monosome, shows diminished complex formation on specific Hox mRNAs found to be decreased at the translation level in Ts/+ embryos (A) but not others (B). P values (Student's t-test) are shown. (C) Two representative models for control of 80S complex formation by RPL38, on or off the ribosome. (D-E) Neural stem cells (NSC), the murine embryonic mesenchymal cell line C3H/10T1/2 (10T1/2), microdissected neural tube & somites from E11.5 embryos, and embryonic stem cell lines treated with Retinoic Acid for the indicated amount of time (hours) were fractioned on a sucrose cushion to separate the ribosomal fraction from ribosome- free cytosol and both fractions were subject to immunoblot analysis. (F-G) Sucrose gradient fractionation of neural tube & somites from E11.0 embryos and subsequent Western Blot analysis with representative small and large RPs. Note that Rpl38 is only present in fractions containing the 60S, 80S, and polysomes. (G) Upon puromycin (Puro) treatment that specifically releases nascent polypeptides from ribosomes and dissociates ribosomal subunits, RPL38 is only found associated with the 60S and is localized to the same fractionations as another RP belonging to the large subunit. Data are presented as the average ± SEM. See also Figure S6.
An important question raised from these studies is whether RPL38 regulates 80S complex formation on Hox mRNAs as part of the ribosome or whether it may have an extra-ribosomal function (Figure 6C). For example, the regulated release of RPL13a from the large ribosomal subunit and its subsequent binding to the 3’UTR of a subset of mRNAs has been shown to mediate translational control(Mazumder et al., 2003). To investigate whether RPL38 may exist in a cytosolic, nonribosomal form we performed ribosome sucrose cushion experiments in which ribosomal complexes are separated from the ribosome-free cytosol. This analysis revealed that RPL38 is exclusively found in the ribosome fraction in-vivo within the neural tube/somites of embryos, in stem cells reflective of either neural tissue or mesoderm alone (Figure 6D), as well as in embryonic stem cells where Hox gene expression is temporally activated by Retinoic Acid treatment (Figure 6E and Figure S6B)(Simeone et al., 1990; Simeone et al., 1991). This localization is similar to other RPs, such as RPL4. On the contrary, RPL5, which has previously been shown to have extra-ribosomal functions(Zhang et al., 2006), is localized to both ribosomal and ribosome-free cytosolic fractions (Figure 6D). Linear sucrose gradient fractionation that allows for better separation of ribosomal subunits from monosomes and polysomes also revealed that RPL38 is exclusively localized to fractions containing the 60S, 80S, and polysomes in vivo in the neural tube and somites of developing embryos (Figure 6F).
Treatment of cells with Puromycin specifically releases nascent polypeptides from ribosomes and dissociates ribosomes into individual subunits(Blobel and Sabatini, 1971). If RPL38 existed in a non-ribosomal complex we might expect to see localization in either different or additional fractions under these conditions. As shown in Figure 6G, RPL38 is exclusively found in the 60S fractions and it is localized in an identical fashion to another representative large RP. Taken together, these findings strongly suggest that RPL38 selectively controls translation of specific Hox mRNAs as a component of the ribosome by facilitating 80S complex formation. This is also consistent with recent eukaryotic 80S Cryo-EM studies that have localized RPL38 to a highly dynamic region of the ribosome (see discussion).
A quantitative gene expression screen for the majority of RPs reveals distinct profiles of expression in the vertebrate embryo
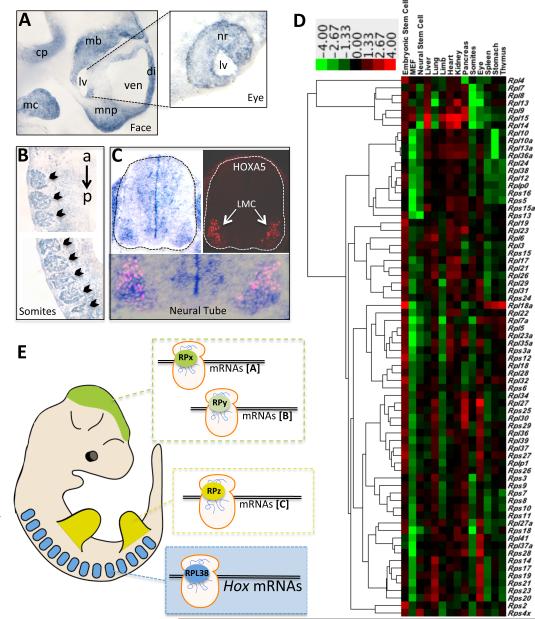
RPs are considered to be abundantly expressed, ubiquitous proteins. However, the tissue specific patterning defects observed in Ts/+ mice prompted us to examine Rpl38 expression during embryonic development. Unexpectedly, we observed a very marked enrichment of Rpl38 transcripts in specific tissues (Figures 7A-C) where phenotypes are observed in Ts/+ embryos (Figures 1 and 2). For example, Rpl38 expression is enriched in the maxillary arch, mandibular component of the first branchial arch and the frontonasal process (Figure 7A and not shown). The maxillary prominences grow and fuse with the frontonasal processes, giving rise to the upper lip and primary palate where cleft palate defects is observed in Ts/+ embryos (Figure 1A). Rpl38 is also highly expressed in the neural retina of the eye (Figure 7A), where eye defects are observed in Ts/+ embryos (Figure 1A). Most importantly, consistent with the axial skeletal patterning defects observed in Ts/+ embryos, Rpl38 expression is substantially enriched within developing somites, the vertebrae precursors (Figures 7B). Rpl38 expression is coincident with somite formation (not shown) and is evident along the entire A-P axis during all stages of somitogenesis (Figure 7B). During somite differentiation Rpl38 expression is evident in the sclerotome (mesenchymal cells that will become vertebral cartilage), dermamyotome (mesenchymal cells that will become dermis) and myotome (muscle cell precursors) although Rpl38 expression is substantially reduced in the dorsal most region of the dermamyotome (not shown). In addition, Rpl38 expression is also very robust within the neural tube (Figure 7C) and enriched expression is evident in the ventricular zone and most notably within the lateral motor column (LMC) where many Hox genes are expressed (Figure 7C). By performing Rpl38 in-situ hybridization on serial sections that were also stained for HOXA5, a target Hox mRNA translationally regulated by RPL38, we observed striking overlap in expression within the LMC (Figure 7C). Therefore, Rpl38 expression is enriched in specific tissues such as the developing eye, face, neural tube, and somites where tissue patterning defects are observed in Ts/+ embryos. However, the expression of Rpl38 does not always fully dictate where loss of function phenotypes will occur. For example, Rpl38 is also enriched in the kidney where no apparent phenotypes are observed in Ts/+ embryos (data not shown).
Figure 7. Ribosomal protein expression during organogenesis.
(A-C) RPL38 in situ hybridization on tissue sections at E11.5. (A) Rpl38 expression in the developing mouse face. Enriched RPL38 expression is observed in the medial nasal process (mns), mandibular component of the first branchial arch (mc), cerebellar plate (cp), diencephalon (di), midbrain (mb). Insert is from a more lateral serial section of the eye from the head region showing that RPL38 expression is enriched in the neural retina (nr). Third ventricle (ven), Lens vesicle (lv). (B) Sagittal section showing markedly enriched RPL38 expression in all A-P somite derivates (black arrowhead). The upper and lower panels are representative serial sections of the same embryo at more anterior (a) and posterior (p) positions. (C) Transverse tissue section of the neural tube (outlined in a dotted line) at the brachial level. To the right is a serial section stained with a HOXA5 antibody. The overlay of RPL38 and HOXA5 expression is shown in the bottom panel, where strong overlap is observed in motor neurons within the lateral motor column (LMC). (D) Heatmap diagram displaying the hierarchical clustering of the expression of 72 ribosomal proteins. The columns in the diagram are separated by tissue and primary cell type; each row is a ribosomal protein, large (l) or small (s) subunit. The color bar at the top indicates the color-coding of gene expression from +4 to -4 in log2 space. (E) Proposed model of RP specificity in control of gene expression during embryogenesis. The enriched expression of specific ribosomal proteins (RP) in different tissues may confer translational specificity to distinct classes of mRNAs (a,b,c). Brain (green), Limbs (red), Somites (blue). (See also Table S3)
We next addressed whether other RPs belonging to the large and small ribosome subunit may display heterogeneous expression during organogenesis. To this end, we carried out a quantitative gene expression-profiling screen of 72 RPs (Table S2) that were hierarchically clustered based on their expression levels in 14 tissue and cell types within the vertebrate embryo. Strikingly, we observed tremendous heterogeneity in RP expression among different RPs and within different tissues over a 250 fold range, expressed in log2 space (Figure 7D and Table S3). This heterogeneity was also observed in expression levels of RP paralogues that share high sequence homology. A notable exception is Rpl10 and Rpl10a that form a primary cluster based on expression. In addition, more general patterns emerged such as the enriched expression of the majority of RPs in embryonic stems cells compared to more differentiated cell types, such as murine embryonic fibroblasts. The heterogeneity in expression profiles of RPs, including RPL38, suggests that RPs in different regions of the embryo are highly regulated and may exert more specialized functions. Collectively these results provide a comprehensive gene expression map of RPs during embryonic development and reveal unexpected differences in the expression levels of ribosome components.
Discussion
Here we show a surprising role of the ribosome in vertebrate tissue patterning and control of gene expression. At the molecular level, we uncover an important function for RPL38 in translational control of a subset of Hox genes that underlies Ts/+ tissue-patterning defects. An elaborate set of regulatory mechanisms are responsible for the spatial and temporal expression of all 39 Hox A, B, C and D genes imposed by their tandem localization on four chromosomal clusters (Deschamps and van Nes, 2005; Kmita and Duboule, 2003). As these Hox expression boundaries are established very early in embryonic development, as a consequence of colinear transcriptional activation, translational control may provide an important means to maintain and refine these patterns of expression overtime. Indeed, post-transcriptional control may be an important mechanism to regulate Hox expression as the distribution of certain Hox transcripts and proteins appears to be distinct(Brend et al., 2003; Dasen et al., 2003) and accumulating evidence exists for posttranscriptional silencing by microRNAs (Yekta et al., 2008). Our present study demonstrates a critical role for translational control of Hox gene expression that is required to establish the mammalian body plan and is regulated by RPL38 activity.
RPs are generally considered to be abundant, ubiquitously expressed proteins. Unexpectedly, our data reveal that Rpl38 transcripts are markedly enriched in a tissue specific manner; with a striking overlap between Rpl38 expression and regions of the embryo where Ts/+ phenotypes are observed (Figure 7A-C). Moreover, a quantitative expression-profiling screen reveals that RP expression is dynamically regulated during embryonic development (Figure 7D). The complexity and magnitude of the overall heterogeneity in RP expression profiles (Figure 7D) suggest specialized functions for individual RPs. For some RPs, dramatic differences in expression levels may reflect extra-ribosomal functions (Warner and McIntosh, 2009). However, for other RPs such as Rpl38, these expression patterns may indicate a specific role in translational control as constituents of the ribosome. It is also tempting to speculate that heterogeneity in RP expression profiles may indicate heterogeneity in ribosome composition. The RP mRNA expression profiling described here (Figure 7D) will help in directing further characterization of ribosome composition at the protein level in distinct cell/tissue types. These studies will be important in clarifying whether “specialized ribosomes” exist at the level of heterogeneity in ribosome composition. It is also possible that specialized ribosome activity, irrespective of ribosome make-up, adds an important layer of specificity in translational control. This is illustrated by our current studies in which we identify a specialized role of RPL38, as a constituent of the ribosome, in translational control of a subset of Hox mRNAs. Interestingly, RPs have also been shown to undergo numerous post-translational modifications including phosphorylation, acetylation, neddylation, and methylation(Chow et al., 2007). While the function(s) of these modifications remain poorly understood, they could act to further refine specialized ribosome functions.
Heterogeneity in RP expression levels and translational specificity may also explain, at least in part, the unexpectedly tissue-specific phenotypes associated with RP loss of function observed in Arabidopsis (Byrne, 2009), zebrafish(Uechi et al., 2006), mouse(Anderson et al., 2007; Kirn-Safran et al., 2007; Oristian et al., 2009; Panic et al., 2006) and humans(Lipton and Ellis). For example, accumulating evidence links mutations in a number of RPs to congenital birth defects, including Diamond–Blackfan anemia (DBA) (Lipton and Ellis). Interestingly, in addition to anemia, DBA patients display specific birth defects including limb defects, cleft palate, growth failure, and a predisposition to cancer.
In the context of the present study, we have mechanistically traced the tissue patterning defects observed in Ts/+ embryos to a specific role of RPL38 in translational control of a subset of Hox genes. Global protein synthesis is not perturbed by loss of RPL38 function within the neural tube and somites of Ts/+ embryos, where defects in translation control of a subset of Hox genes are observed (Figure 3A and Figure S3G-H). The specialized role for RPL38 in translational control is further supported by the direct comparison of Ts/+ phenotypes with additional RP deficiencies in the mouse that do not result in the same phenotypes or affect Hox mRNA translation, even when general rates of protein synthesis are minimally or profoundly decreased (Figure 4P and Figure S4D). At the molecular level we have shown that RPL38 acts to facilitate 80S complex formation during the earliest steps in translation initiation on selective Hox mRNAs while part of the ribosome (Figure 6). In the future it will be important to determine whether unique elements in target 5’- or 3’- UTRs may be recognized by RPL38 either directly or through RNA-binding protein intermediaries and serve as regulatory modules to assemble initiation complexes. Interestingly, both the Antennapedia and Ultrabithorax homeotic genes in Drosophila are translationally controlled through unique regulatory elements positioned in the 5’UTR of these mRNAs, that promote spatially and temporally restricted patterns of Hox protein expression during development (Oh et al., 1992; Ye et al., 1997). Many RPs are positioned on the surface of eukaryotic ribosomal subunits, and it is tempting to speculate that they may make contacts with important regulatory regions of specific mRNAs to exert more specialized functions in translational control (Mauro and Edelman, 2007). For example, RPS25 may make specific contacts with IRES-elements present in viral mRNAs to facilitate translation initiation(Landry et al., 2009; Nishiyama et al., 2007).
Interestingly, RPL38 is a eukaryotic specific RP. Very recent eukaryotic 80S Cryo-EM studies have localized RPL38 on the surface of the ribosome near a region of rRNA, known as expansion segment 27 (ES27) that is a dramatically extended, highly mobile rRNA element that has undergone tremendous evolutionary change(Armache et al.; Armache et al.). ES27 is not present in prokaryotic ribosomes, it is 100 nucleotides in yeast, and extended by ~700 nucleotides in mammals with a hitherto unknown function(Hassouna et al., 1984). Moreover, this region of the ribosome is highly dynamic and undergoes multiple conformational changes(Armache et al.; Armache et al.; Beckmann et al., 2001) that appear to be regulated by RPL38(Armache et al.). These findings suggest that RPL38 activity may, at least in part, impinge on translational specificity of subsets of mRNAs by controlling specific conformational changes in the ribosome.
The “RNA World model” has stipulated that ancestral proto-ribosomes may have contained only RNA in their structures(Agmon, 2009). In keeping with this model, our results suggest that RPs may have conferred greater specificity to the RNA-based translational machinery in control of gene expression (Figure 7E). Therefore, ribosome mediated translational specificity may add an important layer of regulation to the control of gene expression and mammalian development.
Experimental Procedures
Mice
The TSJ/Le-Ts/+ and Rpl24BST/+ strain was purchased from the Jackson Laboratory (Bar Harbor Maine, USA). The TSJ/Le-Ts/+ strain was backcrossed onto a pure C57BL6 background to generate the C57BL6-Ts/+ strain used in this study. BALB/c-Tss/+ mice originated and were maintained in the Aburabi Laboratories, Shionogi & Co., Ltd. in Koka Shiga, Japan. The Rbt mutation was found at the Pasteur Institute in Paris in the laboratory of Dr. Jean-Louis Guenet. Mice used in this study were obtained by in vitro fertilization using frozen sperm of the Rbt mutant stock provided by GSF Research Center in Neuherberg, Germany and C57BL/6J oocytes. Rpl29+/− mice were obtained from Dr. Catherine Kirn-Safran (University of Delaware) and Rps19DSK3/+ and Rps20DSK4/+ mice from Dr. Greg Barsh (Stanford).
Construction of a physical map and generation of pCAGGS-Rpl38 mice
Mouse YAC and BAC libraries and clones were purchased from Research Genetics (Alabama, USA). YACs containing markers linked to the Ts locus were identified from the database at the Whitehead Institute/MIT Center for Genome Research. YAC-end DNA fragments were subcloned using inverse-PCR methods and the end- sequences were used to synthesize oligonucleotide primers for further screening of the 129/SvJ BAC library. End sequences of BAC inserts were determined by direct sequencing. To generate pCAGGS-Rpl38 mice, cDNA of the Rpl38 gene without 5’ and 3’UTR sequences was inserted into the pCAGGS expression vector that consists of CMV-IE enhancer, Chicken b-actin promoter and Rabbit b-globin polyA signal. Additional methods are described in Extended Experimental Procedures.
Dissection of Somites and Neural Tube
WT and Ts/+ embryos were staged precisely by counting the number of somites. Embryos at somite stage 40 (~E 11.0) were employed for the majority of studies. All dissections were performed in media (DMEM F12 1:1, 10% FBS, and 1% penn-strep) in a Sylgard dissection dish (Sylgard 184 Silicone Elastomer Kit; Dow Corning). Additional methods are described in Extended Experimental Procedures.
Polysome fractionation and RNA/Protein isolation
For each polysome set, somites and neural tube from three WT and three Ts/+ 40-somite stage embryos were employed (see Dissection of Somites and Neural Tube). A minimum of four polysome sets (n=4) per genotype was used for polysome profiling analysis (See Figure 4). In brief, cellular extracts were centrifuged at 10,000 rpm for 5 min at 4°C and the supernatant was carefully isolated and loaded onto 10-50% sucrose gradients containing 0.1 mg/ml heparin and 2mM DTT and centrifuged at 37,000 rpm for 2.5 h at 4°C (SW40 rotor). The sucrose gradient was subsequently fractionated with a gradient fractionation system (ISCO) connected to a UV detector to monitoring absorbance at 252nm. As small embryonic tissue fragments were employed, a limiting factor was the analog output of absorbance that fell below the range that could be accurately recorded with a chart recorder. We therefore employed a digital voltmeter that was attached to the spectrometer to digitally record the full range of data as that enabled better resolution of the polysome profile. RNA was isolated from polysomal fractions using the PureLink RNA Mini Kit (Invitrogen). Additional methods are described in Extended Experimental Procedures.
Ribosomal protein expression profiling
The following mouse tissues – lung, limb, liver, heart, kidney, pancreas, somites, eye, spleen, stomach, and thymus – were micro-dissected from WT embryos at E11.5. The pancreas was dissected from WT embryos at E13.5. All dissections were performed in media (DMEM F12 1:1, 10% FBS, and 1% penn-strep) in a Sylgard dissection dish (Sylgard 184 Silicone Elastomer Kit; Dow Corning). The clustering of ribosomal protein expression in different embryonic tissues was generated and visualized using Cluster 3.0 and Java TreeView software. Additional methods are described in Extended Experimental Procedures.
Ribosome Fractionation
Ribosome and nonribosomal fractions were collected essentially as described (Mazumder et al., 2003). Additional methods are described in Extended Experimental Procedures.
Supplementary Material
Acknowledgements
We are very grateful to Dr. Davide Ruggero for helpful discussions and CMV-HCV-IRES transgenic mice; we thank Katrina Cabaltera for technical assistance; Dr. Gail Martin and members of her lab, Drs. Davide Ruggero, Tom Kornberg, and Ross Metzger for critical reading of the manuscript; Drs. Rudi Balling and Toshio Uchiumi for discussion and comments; Dr. Lee Niswander for helpful discussions on neural tube patterning; Drs. Jens Schuster and Cristina Al-Khalili Szigyarto for the RPL38 antibody and Dr. Lucie Jeannotte for the Hoxa5 antibody. We are grateful to Dr. Jean-Louis Guenet for providing Rbt and Dr. Tsutomu Hirasawa for providing Tss; Dr. Catherine Kirn-Safran for Rpl29+/− mice; Dr. Greg Barsh for Rps19DSK3/+ and Rps20DSK4/+ mice. The monoclonal antibodies against Shh, Nkx2.2, Isl1, Pax6 and Pax7 were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, USA. This work was supported by the Program for Breakthrough Biomedical Research, UCSF (M.B), the March of Dimes Basil O'Connor Scholar Research Award (M.B.), and Special coordination funds for promoting Science and Technology from the MEXT, Japan (T.S.).
REFERENCES
- (2009). Hox genes (San Diego, Academic Press).
- Agmon I. The dimeric proto-ribosome: structural details and possible implications on the origin of life. Int J Mol Sci. 2009;10:2921–2934. doi: 10.3390/ijms10072921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci U S A. 107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci U S A. 107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci U S A. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brend T, Gilthorpe J, Summerbell D, Rigby PW. Multiple levels of transcriptional and post-transcriptional regulation are required to define the domain of Hoxb4 expression. Development. 2003;130:2717–2728. doi: 10.1242/dev.00471. [DOI] [PubMed] [Google Scholar]
- Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- Byrne ME. A role for the ribosome in development. Trends Plant Sci. 2009;14:512–519. doi: 10.1016/j.tplants.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Carpenter EM, Goddard JM, Davis AP, Nguyen TP, Capecchi MR. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development. 1997;124:4505–4514. doi: 10.1242/dev.124.22.4505. [DOI] [PubMed] [Google Scholar]
- Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Deol MS. Genetical studies on the skeleton of the mouse XXVIII. Tail-short. ProcRSocLondon(Biol) 1961;155:78–95. [Google Scholar]
- Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Doll e P, Chambon P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 1996;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- Hassouna N, Michot B, Bachellerie JP. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan GS, Wu K, Wolgemuth DJ, Behringer RR. Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc Natl Acad Sci U S A. 1994;91:12644–12648. doi: 10.1073/pnas.91.26.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustert E, Scherer G, Olowson M, Guenet JL, Balling R. Rbt (Rabo torcido), a new mouse skeletal mutation involved in anteroposterior patterning of the axial skeleton, maps close to the Ts (tail-short) locus and distal to the Sox9 locus on chromosome 11. Mamm Genome. 1996;7:881–885. doi: 10.1007/s003359900261. [DOI] [PubMed] [Google Scholar]
- Iimura T, Denans N, Pourquie O. Establishment of Hox vertebral identities in the embryonic spine precursors. Curr Top Dev Biol. 2009;88:201–234. doi: 10.1016/S0070-2153(09)88007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannotte L, Lemieux M, Charron J, Poirier F, Robertson EJ. Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Dev. 1993;7:2085–2096. doi: 10.1101/gad.7.11.2085. [DOI] [PubMed] [Google Scholar]
- Juan AH, Ruddle FH. Enhancer timing of Hox gene expression: deletion of the endogenous Hoxc8 early enhancer. Development. 2003;130:4823–4834. doi: 10.1242/dev.00672. [DOI] [PubMed] [Google Scholar]
- Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, Carson DD. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn. 2007;236:447–460. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Laboratory CSH. The Ribosome, Vol LXVI 1edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2002. [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mouellic H, Lallemand Y, Brulet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992;69:251–264. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- Lipton JM, Ellis SR. Diamond Blackfan anemia 2008-2009: broadening the scope of ribosome biogenesis disorders. Curr Opin Pediatr. 22:12–19. doi: 10.1097/MOP.0b013e328334573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan WC. A new tail-short mutation in the mouse whose lethal effects are conditioned by the residual genotypes. J Hered. 1950;41:208–215. doi: 10.1093/oxfordjournals.jhered.a106131. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Scott MP, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oristian DS, Sloofman LG, Zhou X, Wang L, Farach-Carson MC, Kirn-Safran CB. Ribosomal protein L29/HIP deficiency delays osteogenesis and increases fragility of adult bone in mice. J Orthop Res. 2009;27:28–35. doi: 10.1002/jor.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, Grabusic K, Volarevic S. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney DF, Emerson CP., Jr. 10T1/2 cells: an in vitro model for molecular genetic analysis of mesodermal determination and differentiation. Environ Health Perspect. 1989;80:221–227. doi: 10.1289/ehp.8980221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopal S. The Dictyostelium ribosome: biochemistry, molecular biology, and developmental regulation. Biochem Cell Biol. 1992;70:738–750. doi: 10.1139/o92-113. [DOI] [PubMed] [Google Scholar]
- Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagliocco FA, Moore PA, Brown AJ. Polysome analysis. Methods Mol Biol. 1996;53:297–311. doi: 10.1385/0-89603-319-8:297. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763–766. doi: 10.1038/346763a0. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Nigro V, Faiella A, D'Esposito M, Stornaiuolo A, Mavilio F, Boncinelli E. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech Dev. 1991;33:215–227. doi: 10.1016/0925-4773(91)90029-6. [DOI] [PubMed] [Google Scholar]
- Small KM, Potter SS. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev. 1993;7:2318–2328. doi: 10.1101/gad.7.12a.2318. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B, Wickens M, Kimble J. Translational control in development. In: Mathews NSMB, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 507–544. [Google Scholar]
- Tsukahara K, Hirasawa T, Makino S. Tss (Tail-short Shionogi), a new short tail mutation found in the BALB/cMs strain, maps quite closely to the Tail-short (Ts) locus on mouse chromosome 11. Exp Anim. 2000;49:131–135. doi: 10.1538/expanim.49.131. [DOI] [PubMed] [Google Scholar]
- Uechi T, Nakajima Y, Nakao A, Torihara H, Chakraborty A, Inoue K, Kenmochi N. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS One. 2006;1:e37. doi: 10.1371/journal.pone.0000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM. Hox genes and vertebrate axial pattern. Curr Top Dev Biol. 2009;88:257–278. doi: 10.1016/S0070-2153(09)88009-5. [DOI] [PubMed] [Google Scholar]
- Ye X, Fong P, Iizuka N, Choate D, Cavener DR. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Renucci A, Izpisua-Belmonte JC, Urier G, Peschle C, Duboule D. HOX4 genes encode transcription factors with potential auto-and cross-regulatory capacities. EMBO J. 1991;10:4177–4187. doi: 10.1002/j.1460-2075.1991.tb04996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.