Abstract
Despite intense efforts by the medical and pharmaceutical communities, Staphylococcus aureus continues to be a pervasive pathogen that causes a myriad of diseases and a high level of morbidity and mortality among infected patients. Thus, discovering or designing novel therapeutics able to kill both drug-resistant and drug-sensitive S. aureus remains a top priority. Bacteriolytic enzymes, mostly from phage, have shown great promise in preclinical studies, but little consideration has been given to cis-acting autolytic enzymes derived from the pathogen itself. Here, we use the S. aureus autolysin LytM as a proof of principal to demonstrate the antibacterial potential of endogenous peptidoglycan-degrading enzymes. While native LytM is only marginally bactericidal, fusion of LytM to the lysostaphin cell wall binding domain enhances its anti-staphylococcal activity approximately 540-fold, placing it on par with many phage lysins currently in preclinical development. The potential to therapeutically co-opt a pathogen's endogenous peptidoglycan recycling machinery opens the door to a previously untapped reservoir of antibacterial drug candidates.
Keywords: MRSA, antimicrobial enzyme, peptidoglycan hydrolysis, pentaglycine, M23 peptidase, lysin
In this study, we develop a novel anti-S. aureus therapeutic agent by engineering S. aureus' own enzyme to make it deadly against this pervasive pathogen.
In this study, we develop a novel anti-S. aureus therapeutic agent by engineering S. aureus' own enzyme to make it deadly against this pervasive pathogen.
INTRODUCTION
The spread of drug-resistant bacterial pathogens, compounded by a deficit in antibiotic development pipelines, is fueling a growing public health crisis (Taubes 2008; Centers for Disease Control and Prevention 2013). Staphylococcus aureus has been of particular concern, given its prevalence (30% of the healthy human population are carriers), infectious potential, high mortality rates and observed resistance towards many first-line antibiotics (World Health Organization 2014). The spread of both community- and hospital-acquired methicillin-resistant S. aureus (MRSA) has created a sense of urgency in the search for improved monitoring and treatment strategies, and the emergence of S. aureus resistance to second-line therapies such as vancomycin (Howden et al., 2009) and daptomycin (Patel et al., 2006) has served to underscore the mounting severity of the situation. Long-term solutions to this challenge will require improved stewardship of antibiotics combined with the discovery and development of novel antibacterial agents able to kill contemporary strains while minimizing the emergence of new resistance phenotypes.
Bacteriolytic enzymes that catalytically degrade cell wall peptidoglycan represent an intriguing alternative to conventional small molecule antibiotics (Szweda et al., 2012). As a whole, these bactericidal agents benefit from numerous advantageous properties including low effective molar dosages, rapid action, synergy with conventional antibiotics, exquisite selectivity for specific pathogens and inherently low susceptibility to new resistance phenotypes (Pastagia et al., 2013). Currently, four major sources of lytic enzymes have been studied in some depth. First and most notably are the bacteriophage endolysins (Nelson et al., 2011), which represent a diverse reservoir of biotherapeutic candidates that are now advancing to human trials. Second are the bacteriophage virion-associated peptidoglycan hydrolases, which have a distinct role in the natural phage lifecycle, are functionally homologous to phage endolysins, and are also interesting drug candidates (Rodríguez-Rubio et al., 2013). Third, secreted ‘bacterial warfare’ enzymes belonging to the class III, group A bacteriocins are highly active against their target organisms, presumably due to their key role in bacterial ecology (Yang et al., 2014). Staphylococcus simulans lysostaphin (ssLys) is the most extensively studied member of the bacteriocin drug candidates (Kokai-Kun 2012). Fourth are the more promiscuous lysozymes derived from the innate immune systems of higher organisms. For example, human lysozyme has proven to be efficacious in both native and genetically engineered formats (Bhavsar et al., 2010, 2011; Teneback et al., 2013; Griswold et al., 2014). As a whole, these various trans-acting lytic enzymes have yielded promising results and represent a growing field of research.
The majority of highly active lysins have a multi-domain architecture composed of separate catalytic and cell wall binding domains (CWBDs). The CWBDs serve to target the enzymes to the bacterial outer surface, thereby facilitating direct interaction between the enzymes’ catalytic domains and their peptidoglycan substrate. This targeting function is of paramount importance in the activity of multi-domain lysins, as deletion of CWBDs greatly reduces the lytic activity of otherwise potent enzymes (Gargis et al., 2010; Frankel and Schneewind 2012). Conversely, the selectively and activity of numerous lysins have been manipulated and enhanced via genetic fusion of different catalytic and CWBD components (Nelson et al., 2011; Pastagia et al., 2013).
In contrast to mounting interest in such trans-acting bacteriolytic enzymes, there has been little analysis of cis-acting autolysins as a possible source of next generation antibacterial agents. Autolysins are a diverse family of peptidoglycan-degrading enzymes utilized by bacteria in processes such as autolysis, peptidoglycan recycling, cell division and biofilm formation. While there exists ample study of their physiological significance (Takahashi et al., 2001; Kajimura et al., 2005; Bose et al., 2011; Frankel et al., 2011; Egan and Vollmer 2012), to date there has been little systematic analysis of their therapeutic potential. One barrier to therapeutic applications is the tightly regulated activity of endogenous autolysins. Bacterial evolution has precisely tuned autolysin activity via control of expression, localization and inherent catalytic proficiency (Schlag et al., 2010; Frankel and Schneewind 2012). As a whole, autolysins appear to have lower intrinsic activity than bactericidal lysins, and we speculate that suboptimal (from a therapeutic perspective) association with the cell wall is a key contributor (see, for example, Sabala et al., 2012). Experience with phage endolysin engineering suggests that chimeragenesis with appropriate CWBDs could address this limitation.
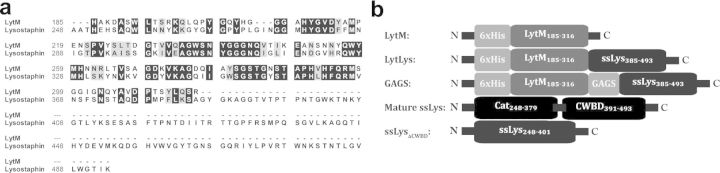
As a proof of concept, we have examined the antibacterial potential of recombinantly produced LytM, a well-studied S. aureus autolysin (Ramadurai and Jayaswal 1997; Odintsov et al., 2004; Firczuk, Mucha and Bochtler 2005; Singh, Carlos and Singh 2010). LytM has a high degree of homology to the catalytic domain of ssLys (Fig. 1a). Both proteins are produced as inactive pre-proenzymes that are proteolytically processed into mature active forms in vivo. Likewise, both enzymes are zinc-dependent glycyl-glycine endopeptidases that cleave the pentaglycine crosslink found in the peptidoglycan of some Staphylococcus species, particularly S. aureus (Browder et al., 1965; Schleifer and Kandler 1972; Bardelang et al., 2009). Importantly, previous studies have demonstrated that exogenously added LytM displays little activity towards live S. aureus cells, and that the protein has negligible therapeutic efficacy in a mouse eczema infection model (Sabala et al., 2012). However, because of its considerable homology to ssLys, we believed LytM possessed untapped anti-Staphylococcal potential that might be unveiled via fusion to an appropriate CWBD. Here, we revisited the enzyme's antibacterial activity in the context of engineered fusions with the ssLys CWBD.
Figure 1.
Sequences and modular structures of LytM chimeras. (A) Sequence alignment of mature, catalytically active LytM (Firczuk et al., 2005) and lysostaphin (Thumm and Gotz 1997) generated with BioEdit v7.1.3.0 using a Blosum62 matrix. Identical residues are highlighted in black, similar residues are in gray and dissimilar residues are black text on white. (B) Schematics of the engineered constructs. ssLys represents the abbreviation ‘S. simulans lysostsaphin’. Numbers represent the amino acid positions corresponding to the full-length sequences (LytM UniProt O33599; lysostaphin UniProt P10547). LytM constructs consist of an N-terminal hexahistidine tag followed by a ser-met-ala linker. The ‘GAGS’ construct bears a synthetic gly-ala-gly-ser linker between the catalytic and CWBDs.
MATERIALS AND METHODS
Materials and reagents
Oligonucleotides (25 nmol scale, standard desalting) were from Integrated DNA Technology (San Diego, CA, USA). Commercial lysostaphin (ssLys) and SYPRO Orange 5000× Protein Stain were from Sigma (St Louis, MO, USA). MicroAmp® Fast Optical 0.1 mL 96-Well Plates and MicroAmp® Optical Adhesive Film were from Applied Biosystems (Bedford, MA, USA). Restriction enzymes and PCR reagents were purchased from New England BioLabs (Ipswich, MA, USA). Plasmid purification kits and Ni-NTA resin were from Qiagen (Valencia, CA, USA). Growth medium was from Becton Dickinson (Franklin Lakes, NJ, USA). PCR cleanup and gel extraction kits were from Zymo Research (Irvine, CA, USA). SP Fast Flow was from GE Healthcare Life Sciences (Little Chalfont, Buckinghamshire, UK). SYTOX® Green nucleic acid stain was purchased from Life Technologies (Carlsbad, CA). MBECTM Biofilm Inoculator 96-well plates were purchased from Innovotech (Edmonton, AB, Canada). Unless noted, all other chemicals and reagents were from Fisher Scientific (Pittsburgh, PA, USA).
Bacterial strains
During cloning, plasmids were maintained in Escherichia coli strain DH5α [F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK− mK+), λ–] and proteins were expressed in E. coli strain BL21(DE3) [F− ompT hsdSB (rB− mB−) gal dcm (DE3)]. Staphylococcus aureus strains SA113 and 49521 were purchased from ATCC. All other S. aureus clinical isolate strains were the kind gift of Dr Ambrose Cheung.
Molecular cloning
Whole-cell lysate of S. aureus SA113 was used as a template for LytM185–316 cloning. Briefly, the gene was amplified by PCR using 5′-primer CACCATTCCATGGCGCATGCGAAAGACGCAAGCTGG first and then with 5′-primer TACTACATATGCATCACCACCATCACCATTCCATGGC second, which appended a 5′ NdeI site, a 6xHis tag and a ser-met-ala linker sequence, and 3′-primer GCTCAATACTGGATCCTTATCTACTTTGCAAGTATGA was used in both reactions, which appended a 3′ BamHI site. Splice overlap extension PCR was used to silently mutate an internal NcoI site within the LytM gene at amino acid 200 (proline, CCA to CCT) to facilitate future cloning (5′-primer CAACTACAACCTTATGGACAATATCACGGTGG and 3′-primer GTCCATAAGGTTGTAGTTGTTTACGACTTGTTAAC). The resulting PCR product was double digested with NdeI and BamHI, ligated into similarly digested pET26b vector, transformed by electroporation into BL21(DE3), plated on LB agar supplemented with 30 μg mL−1 kanamycin (LB-Kan30), and clones were sequence verified. For fusion constructs, the coding sequence for the ssLys CWBD (amino acids 385–493) was amplified by PCR from pRG5 (ATCC: 67076). The ssLys catalytic domain (ssLysΔCWBD) consisting of amino acids 248–400 (UniProt P10547) was amplified from a plasmid encoding a codon-optimized, aglycosylated, S126P point mutant (Zhao et al., 2014) using primers ATCGCTCGAGAAAAGAGCTGCTACCCACGAGCACTCCGCT and CGATGAATTCTTAGGTGTTTGGGGTTGGGGTGACGGT. The amplicon was cloned into the XhoI and EcoRI sites of vector pPIC9, transformed into DH5α and sequence verified. Sequence-verified plasmid was linearized with SacI, transformed into Pichia pastoris strain GS115, and expressed and purified as described elsewhere (Zhao et al., 2014). Gene fusions were generated using splice overlap extension PCR of LytM185–316 (5′-primer ACCATTCCATGGCGCATGCGAAAGAC and 3′-primer CTGCTTTTCCATATCCTGCTCTACTTTGCAAGTATGAC) and ssLys385–493 (5′-primer GTCATACTTGCAAAGTAGAGCAGGATATGGAAAAGCAG and 3′ primer CTCGAATTCGGATCCTTACTTTATAGTTCCCCAAAGAACACC), either with or without the encoded GAGS linker. The same cloning procedures described above were performed.
Purification
Escherichia coli BL21(DE3) expression hosts were grown overnight with shaking at 37°C in LB-Kan30. Cells were subcultured 1:100 into fresh medium, grown 2.5 h at 37°C, equilibrated 20 minutes at 25°C, induced with 1 mM IPTG and left to shake overnight. Harvested cells were suspended in lysis buffer (20 mM Tris, 20 mM imidazole, 500 mM NaCl, pH 7.5) and lysed by sonication. Soluble material was harvested by centrifugation at 30 000× g for 20 min, the supernatant was 0.22 micron filtered and the filtrate was incubated with Ni-NTA resin for 1 h at 4°C. The resin was then washed with 6 mL of lysis buffer and eluted with 20 mM Tris, 200 mM imidazole and 500 mM NaCl, pH 7.5. The eluent was diluted 1:10 with 10 mM KH2PO4 pH 7.3 (cation buffer) and flowed over a gravity column packed with 500 μL SP-Sepharose Fast Flow resin. The column was washed with 6 mL of 50 mM NaCl cation buffer followed by 3 mL of 100 mM NaCl cation buffer, and protein was eluted with 500 μL aliquots of 200 mM NaCl cation buffer. Quantification was performed using A280 from a NanoDrop spectrophotometer. The following molar extinction coefficients at A280 were obtained from the ExPASy ProtParam Tool: 32890 M−1 cm−1 for LytM, 63830 M−1 cm−1 for LytLys and GAGS, and 67840 M−1 cm−1 for commercial ssLys. Protein purity was assessed by SDS-PAGE analysis on 12% acrylamide gels.
Lytic assays
Staphylococcus aureus was grown either to mid-log or to saturation in tryptic soy broth (TSB) at 37°C with shaking. Cells were harvested, washed once with PBS pH 7.3 (Green and Sambrook 2012) and then suspended in PBS pH 7.3 to a final OD600 of 3. The fluorometric kinetic microplate assay was performed essentially as described (Scanlon et al., 2010). Briefly, reactions were assembled in black 96-well plates, and the wells consisted of 250 μL PBS containing cells at a final OD600 of 1.5, SYTOX® Green at 5 μM and enzyme at the specified concentrations. Mean fluorescence intensity readings were taken every 20–30 s on a SpectroMax Gemini plate reader (Molecular Devices, Sunnyvale, CA) using an excitation of 504 nm and emission of 523 nm. Kinetic data were analyzed by determining the slope of the steepest linear region in the lysis curve. Turbidity assays were performed and analyzed essentially as described (Lee and Yang 2002) and otherwise employing reaction conditions analogous to the fluorometric assay described above (with the exception that the SYTOX® dye was omitted and the plate was read with a Spectramax 190 at A450).
Melting temperatures
Differential scanning fluorimetry was performed essentially as reported (Niesen, Berglund and Vedadi 2007) with modifications as described previously (Osipovitch et al., 2012).
Minimal inhibitory concentration (MIC) determination
MICs were determined by 1:2 dilutions of enzyme into wells of a polypropylene 96-well plate (CoStarTM 3879) containing approximately 40 000 SA113 cells per 100 μL of MHIIB media supplemented with 2% NaCl and 0.1% BSA. Plates were grown overnight at 37°C with shaking at 900 rpm on an orbital shaker. MICs are defined as the lowest concentration of enzyme in which there was no visible growth of bacteria at 20 h.
Minimum biofilm eradication concentration (MBEC)
Saturated overnight cultures of SA113 in TSB were subcultured 1:100 and grown with shaking for 2.5 h at 37°C. Subcultures were then diluted 1:10 and 100 μL of culture was added per well into Innovotech MBEC 96-well plates, which include lids bearing pendant polymer pegs on which biofilms are grown (Olson et al., 2002; Harrison et al., 2010). Biofilms were grown on the pendant pegs of MBEC lids by incubating the plates without shaking at 37°C overnight. Lids on which peg-adherent biofilms had been grown were washed 1x by suspension in PBS for 60 s before being transferred to a fresh 96-well plate containing 150 μL of enzyme dilutions in MHIIB supplemented with 2% NaCl and 0.1% BSA. Treatments were incubated for 2 h at 37°C before being washed once with PBS. Poorly adherent cells and biofilm are removed from the MBEC lid substrate during this washing process. Biofilms were then transferred to a 96-well plate containing 150 μL of 1x alamarBlue® cell viability reporter (Nakayama et al., 1997; Pettit, Weber and Pettit 2009) in MHIIB. After 1 h incubation at 37°C, plates were read with 560 nm excitation and 590 nm emission. Data were normalized to highest cell-viability signal and a four-parameter curve fit was performed in Prism v.5 software (La Jolla, CA) to determine the EC50.
RESULTS AND DISCUSSION
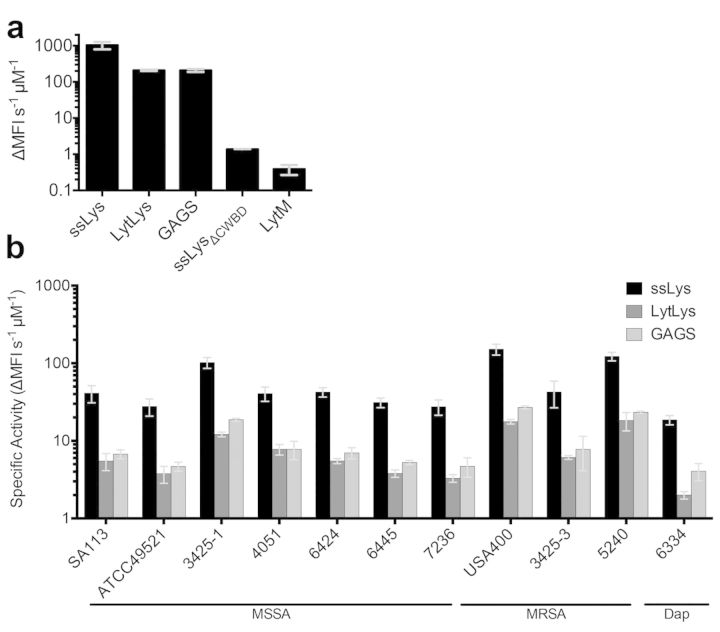
To benchmark our molecular engineering efforts, we first characterized the inherent activity of native LytM. The mature, active form of LytM (Firczuk et al., 2005) (Fig. 1b) was recombinantly expressed in E. coli, purified to near homogeneity (Fig. S1, Supporting Information) and analyzed for antibacterial activity against live S. aureus. LytM expressed reasonably well with purified yields of 1–2 mg L–1of shake flask culture. The kinetics of LytM-mediated bacterial lysis were orders of magnitude slower than those of the potent bacteriocin ssLys (Fig. 2a). In antibacterial assays, the slow LytM kinetics manifested as an inability to achieve an MIC and a failure to clear established S. aureus biofilms (Table 1). Indeed, these data correspond well with prior analyses of LytM (Sabala et al., 2012). Thus, despite its physiologic importance as an autolysin, LytM's exogenous activity versus live S. aureus is minimal.
Figure 2.
Bacterial lysis kinetics as determined by a SYTOX® fluorescence assay. (A) Specific activities of the various enzymes against SA113 cells in exponential growth phase. ssLys is commercial lysostaphin and ssLysΔCWBD is the catalytic domain of lysostaphin. (B) Specific activity of the enzymes against various strains of S. aureus taken from stationary phase cultures. Error bars represent one standard deviation from biological duplicates measured in triplicate. SA113 is a laboratory strain; ATCC 49521, 3425–1, 4051, 6424, 6445, 7236 are methicillin-sensitive clinical isolates (MSSA); USA400, 3425–3, and 5240 are methicillin-resistant clinical isolates (MRSA); and 6334 is a daptomycin (Dap) resistant clinical isolate.
Table 1.
Measurements of enzyme stability and activity.
| Enzyme | Melting temperature (°C)a | MIC | |||||
|---|---|---|---|---|---|---|---|
| SA113b | ATCC49521c | MBECd | |||||
| μg mL−1 | nM | μg mL−1 | nM | μg mL−1 | nM | ||
| ssLys | 47.1 ± 0.6 | 0.04 ± 0.01 | 1.5 ± 4 | 0.016 | 0.6 | 0.010–0.018 | 0.37–0.67 |
| LytLys | 54.5 ± 0.4 | 0.42 ± 0.1 | 15 ± 4 | 1.25 | 46 | 0.21–0.44 | 7.6–16 |
| GAGS | 54.9 ± 0.3 | 0.9 ± 0.3 | 33 ± 11 | 1.25 | 46 | 0.28–0.49 | 10–18 |
| LytM | 67 ± 2 | ≫100 | ≫6400 | ND | ND | NDA | NDA |
aValues represent mean ± standard deviation.
bValues represent mean ± standard deviation of triplicate measures performed in two biologic replicates.
cNo variance was detected. Values represent triplicate measures performed in two biologic replicates.
dValues represent 95% confidence interval of triplicate measures performed in four biologic replicates.
MIC: Minimal inhibitory concentration.
MBEC: Minimum biofilm eradication concentration.
ND: Not determined.
NDA: No detectable activity.
Many peptidoglycan hydrolases contain CWBDs that increase the enzymes’ affinity for the bacterial cell wall (Schlag et al., 2010; Frankel and Schneewind 2012), and we speculated LytM's poor anti-staphylococcal performance stemmed from its lack of a CWBD. For example, ssLys's activity is compromised by removal of its CWBD (Gargis et al., 2010), and we ourselves have observed a 750-fold decrease in the bacterial lysis kinetics of truncated ssLys (ssLysΔCWBD; amino acids 248–400; Fig. 2a). Recent studies with chimeric phage lysins have shown that lytic activity can be enhanced by fusion with appropriate CWBDs (Donovan et al., 2006; Becker et al., 2009b; Fernandes et al., 2012; Schmelcher et al., 2012; Mao et al., 2013), and we therefore hypothesized that fusion of LytM to the ssLys CWBD would increase the autolysin's bacteriolytic activity. Using the alignment shown in Fig. 1a, we designed two fusions whereby the ssLys catalytic domain was replaced with the mature form of LytM. The first, LytLys, is a direct fusion of ssLys amino acids 384–493 to the C-terminal arginine of LytM (Fig. 1b). The second fusion, GAGS, includes the four amino acid linker gly-ala-gly-ser for added flexibility between the domains (Fig. 1b).
The genetically engineered LytM-CWBD fusions were recombinantly expressed in E. coli and purified in a manner similar to LytM. Greater than 95% purity was easily obtained in a two-step purification with average final yields of 1–2 mg L–1 culture (Fig. S1, Supporting Information). Differential scanning fluorimetry demonstrated that the chimeric proteins exhibited thermostability intermediate to that of ssLys and LytM (Table 1). Fluorogenic microplate assays using exponential growth phase S. aureus strain SA113 revealed that the fusions exhibited tremendously enhanced cell wall degradation kinetics, with an increase in specific activity of ∼540-fold versus native LytM (Fig. 2a). Bacterial lysis was similarly assessed for a small panel of stationary phase S. aureus strains, including many clinical isolates (Fig. 2b). Compared to exponential growth phase, bacteria from overnight saturated cultures can have thicker cell walls (Zhou and Cegelski 2012), which may explain their slower rates of enzyme-mediated lysis (compare, for example, Fig. 2a to strain SA113 in Fig. 2b). Regardless, all 11 strains, including several drug-resistant strains, were sensitive to each of the lytic enyzmes, and the fusions were, on average, only 6-fold less active than ssLys. There was a consistent trend of higher activity for the linker-containing GAGS construct compared to LytLys, although the difference was not statistically significant for any one strain, suggesting that this subtle modification to linker length does not strongly influence reaction kinetics.
The improved cell wall degradation kinetics of the LytM fusion proteins were found to translate into radically enhanced antibacterial activity relative to the non-targeted wild-type enzyme (Table 1). Unlike LytM, the fusions were able to inhibit the growth of S. aureus in an MIC assay and were likewise able to clear established biofilms. To provide additional context, small molecule chemotherapeutics typically have anti-S. aureus MIC values in the range of single digit μg ml−1 to sub-μg ml−1 (Table 2). Highly active antimicrobial peptides, a clinically relevant pool of antibacterial biomolecules, tend to have similarly low μg ml−1 MIC values, although on a molar basis the larger antimicrobial peptides trend towards higher potency. The LytM fusion proteins of this study exhibit sub-μg ml−1 MIC values that, on a molar basis, translate roughly to an order of magnitude greater potency than chemotherapeutics and antimicrobial peptides. To compare LytM fusion kinetics to other lysin therapeutic candidates, a conventional turbidity reduction assay was performed with ssLys and GAGS, yielding specific activities of 1.3 ± 0.1 ΔA450 min−1 μM−1 (0.16 ± 0.01 ΔA450 min−1 μg−1) and 0.28 ± 0.03 ΔA450 min−1 μM−1 (0.033 ± 0.003 ΔA450 min−1 μg−1), respectively. We note that, in the absence of standardized assay conditions, comparison of lytic rates from the literature should be approached with caution. Variables such as the density of the cell suspension, bacterial strain, growth phase, buffer formulation, assay temperature and enzyme concentration can profoundly influence results. Nonetheless, the kinetics of GAGS and LytLys are seemingly on par with or exceed that of many top-performing lysins, including λSa2 (Becker et al., 2009b), 2638A (Abaev et al., 2013) and LysK (Becker et al., 2009a).
Table 2.
Literature MIC values of antibacterial agents.
| Antimicrobial agent | MIC | Strain | Reference | |
|---|---|---|---|---|
| μg mL−1 | nM | |||
| Small Molecules | ||||
| Rifampin | 0.02 | 24 | SA113 | Lechner, Lewis and Bertram (2012) |
| Clindamycin | 0.1 | 240 | SA113 | Yamakawa, Mitsuyama and Hayashi (2002) |
| Methicillin | 0.72 | 1900 | SA113 | Peschel et al. (2000) |
| Vancomycin | 1.4 | 970 | SA113 | Peschel et al. (2000) |
| Daptomycin | 25 | 15000 | SA113 | Lechner et al. (2012) |
| Antimicrobial Peptidesa | ||||
| Bacteriocin E50–52 | 0.2 | 50 | Clinical isolate | Svetoch et al. (2008) |
| Scolopin 2 | 0.5 | 200 | ATCC2576 | Peng et al. (2010) |
| Polypemusin I | 0.5 | 200 | Not given | Zhang, Rozek and Hancock (2001) |
| Buforin-2 (5–13) [RLLR]3 | 1 | 400 | Not given | Park et al. (2000) |
| Dermcidin, DCD-1L | 1 | 200 | Not given | Schittek et al. (2001) |
| Melittin (12–26) [G1L] | 1.1 | 590 | Not given | Yan et al. (2003) |
| Gaegurin 5 (1–13)[F1W] | 1.6 | 1100 | Not given | Won et al. (2004) |
| Enterocin E-760 | 1.6 | 260 | Not given | Line et al. (2008) |
aRepresentative top-ranking antimicrobial peptides from the Database of Antimicrobial Activity and Structure of Peptides (Gogoladze 2014).
CONCLUSION
Autolysins currently represent an untapped resource in the search for novel antibacterial therapeutics, and ongoing research continues to expand the list of autolysin candidates that might be harvested from the proteomes of various pathogens. We speculate, however, that suboptimal cell wall localization poses a general barrier to the use of autolysins as antibacterial therapies. As shown here, enhanced molecular targeting of the S. aureus autolysin LytM using the ssLys CWBD vastly improved its bacteriolytic and bactericidal properties. Thus, appropriately engineered chimeric fusion proteins may allow the biopharma community to better capitalize on this promising reservoir of antibacterial agents.
SUPPLEMENTARY DATA
Acknowledgments
We would like to thank Professor Ambrose Cheung for the kind gift of S. aureus clinical isolates and Dr Emily K. Schaeffer for critically reviewing the manuscript. We would like to also thank Kristina Blazanovic and Dr Hongliang Zhao for providing the ssLysΔCWBD protein, and Nathan Hidook for providing the graphical abstract.
FUNDING
This work was supported in part by R21 grant 1R21AI098122 from the National Institutes of Health NIAID to KEG.
Conflict of interest statement. None declared.
REFERENCES
- Abaev I, Foster-Frey J, Korobova O, et al. Staphylococcal phage 2638A endolysin is lytic for Staphylococcus aureus and harbors an inter-lytic-domain secondary translational start site. Appl Microbiol Biot. 2013;97:3449–56. doi: 10.1007/s00253-012-4252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelang P, Vankemmelbeke M, Zhang Y, et al. Design of a polypeptide FRET substrate that facilitates study of the antimicrobial protease lysostaphin. Biochem J. 2009;418:615–24. doi: 10.1042/BJ20081765. [DOI] [PubMed] [Google Scholar]
- Becker SC, Dong S, Baker JR, et al. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol Lett. 2009a;294:52–60. doi: 10.1111/j.1574-6968.2009.01541.x. [DOI] [PubMed] [Google Scholar]
- Becker SC, Foster-Frey J, Stodola AJ, et al. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene. 2009b;443:32–41. doi: 10.1016/j.gene.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Bhavsar T, Liu M, Hardej D, et al. Aerosolized recombinant human lysozyme ameliorates Pseudomonas aeruginosa-induced pneumonia in hamsters. Exp Lung Res. 2010;36:94–100. doi: 10.3109/01902140903154608. [DOI] [PubMed] [Google Scholar]
- Bhavsar T, Liu M, Liu X, et al. Aerosolized recombinant human lysozyme enhances the bactericidal effect of tobramycin in a hamster model of Pseudomonas aeruginosa-induced pneumonia. Exp Lung Res. 2011;37:536–41. doi: 10.3109/01902148.2011.609578. [DOI] [PubMed] [Google Scholar]
- Bose JL, Lehman MK, Fey PD, et al. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PloS one. 2011;7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browder HP, Zygmunt WA, Young JR, et al. Lysostaphin: enzymatic mode of action. Biochem Bioph Res Co. 1965;19:383–9. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Antibiotic Resistance Threats in the United States. 2013 http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf (28 October 2013, date last accessed) [Google Scholar]
- Donovan DM, Dong S, Garrett W, et al. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl Environ Microb. 2006;72:2988–96. doi: 10.1128/AEM.72.4.2988-2996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann NY Acad Sci. 2012;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Fernandes S, Proença D, Cantante C, et al. Novel chimerical endolysins with broad antimicrobial activity against methicillin-resistant Staphylococcus aureus. Microb Drug Resist. 2012;18:333–43. doi: 10.1089/mdr.2012.0025. [DOI] [PubMed] [Google Scholar]
- Firczuk M, Mucha A, Bochtler M. Crystal structures of active LytM. J Mol Biol. 2005;354:578–90. doi: 10.1016/j.jmb.2005.09.082. [DOI] [PubMed] [Google Scholar]
- Frankel MB, Hendrickx AP, Missiakas DM, et al. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem. 2011;286:32593–605. doi: 10.1074/jbc.M111.258863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel MB, Schneewind O. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J Biol Chem. 2012;287:10460–71. doi: 10.1074/jbc.M111.336404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargis SR, Heath HE, LeBlanc PA, et al. Inhibition of the activity of both domains of lysostaphin through peptidoglycan modification by the lysostaphin immunity protein. Appl Environ Microb. 2010;76:6944–6. doi: 10.1128/AEM.01066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogoladze G, Grigolava M, Vishnepolsky B, et al. DBAASP: database of antimicrobial activity and structure of peptides. FEMS Microbiol Lett. 2014;357:63–8. doi: 10.1111/1574-6968.12489. [DOI] [PubMed] [Google Scholar]
- Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; Cold Spring Harbor; 2012. [Google Scholar]
- Griswold KE, Bement JL, Teneback CC, et al. Bioengineered lysozyme in combination therapies for Pseudomonas aeruginosa lung infections. Bioengineered. 2014;5:143–7. doi: 10.4161/bioe.28335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JJ, Stremick CA, Turner RJ, et al. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat Protoc. 2010;5:1236–54. doi: 10.1038/nprot.2010.71. [DOI] [PubMed] [Google Scholar]
- Howden BP, Davies JK, Johnson PD, et al. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2009;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura J, Fujiwara T, Yamada S, et al. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol. 2005;58:1087–101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- Kokai-Kun JF. Antimicrobial drug discovery : emerging strategies/edited by George Tegos and Eleftherios Mylonakis. In: Tegos George, Mylonakis Eleftherios., editors. Series: Advances in molecular and cellular microbiology. Vol. 22. Wallingford, Oxfordshire: CABI; 2012c. pp. 147–165. [Google Scholar]
- Lechner S, Lewis K, Bertram R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J Mol Microb Biotech. 2012;22:235–44. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Yang D. Determination of lysozyme activities in a microplate format. Anal Biochem. 2002;310:223–4. doi: 10.1016/s0003-2697(02)00320-2. [DOI] [PubMed] [Google Scholar]
- Line JE, Svetoch EA, Eruslanov BV, et al. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. AntimicrobAgents Ch. 2008;52:1094–100. doi: 10.1128/AAC.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Schmelcher M, Harty WJ, et al. Chimeric Ply187 endolysin kills Staphylococcus aureus more effectively than the parental enzyme. FEMS Microbiol Lett. 2013;342:30–6. doi: 10.1111/1574-6968.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama GR, Caton MC, Nova MP, et al. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods. 1997;204:205–8. doi: 10.1016/s0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Schmelcher M, Rodriguez-Rubio L, et al. Endolysins as antimicrobials. Adv Virus Res. 2011;83:299–365. doi: 10.1016/B978-0-12-394438-2.00007-4. [DOI] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, et al. Latent LytM at 1.3A resolution. J Mol Biol. 2004;335:775–85. doi: 10.1016/j.jmb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Olson ME, Ceri H, Morck DW, et al. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66:86–92. [PMC free article] [PubMed] [Google Scholar]
- Osipovitch DC, Parker AS, Makokha CD, et al. Design and analysis of immune-evading enzymes for ADEPT therapy. Protein Eng Des Sel. 2012;25:613–23. doi: 10.1093/protein/gzs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CB, Yi KS, Matsuzaki K, et al. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. P Natl Acad Sci USA. 2000;97:8245–50. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastagia M, Schuch R, Fischetti VA, et al. Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol. 2013;62:1506–16. doi: 10.1099/jmm.0.061028-0. [DOI] [PubMed] [Google Scholar]
- Patel JB, Jevitt LA, Hageman J, et al. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin Infect Dis. 2006;42:1652–3. doi: 10.1086/504084. [DOI] [PubMed] [Google Scholar]
- Peng K, Kong Y, Zhai L, et al. Two novel antimicrobial peptides from centipede venoms. Toxicon. 2010;55:274–9. doi: 10.1016/j.toxicon.2009.07.040. [DOI] [PubMed] [Google Scholar]
- Peschel A, Vuong C, Otto M, et al. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Ch. 2000;44:2845–7. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit RK, Weber CA, Pettit GR. Application of a high throughput Alamar blue biofilm susceptibility assay to Staphylococcus aureus biofilms. Ann Clin Microbiol Antimicrob. 2009;8:28. doi: 10.1186/1476-0711-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadurai L, Jayaswal RK. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J Bacteriol. 1997;179:3625–31. doi: 10.1128/jb.179.11.3625-3631.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rubio L, Martínez B, Donovan DM, et al. Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. Crit Rev Microbiol. 2013;39:427–34. doi: 10.3109/1040841X.2012.723675. [DOI] [PubMed] [Google Scholar]
- Sabala I, Jonsson I-MM, Tarkowski A, et al. Anti-staphylococcal activities of lysostaphin and LytM catalytic domain. BMC Microbiol. 2012;12:97. doi: 10.1186/1471-2180-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon TC, Teneback CC, Gill A, et al. Enhanced antimicrobial activity of engineered human lysozyme. ACS Chem Biol. 2010;5:809–18. doi: 10.1021/cb1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittek B, Hipfel R, Sauer B, et al. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat Immunol. 2001;2:1133–7. doi: 10.1038/ni732. [DOI] [PubMed] [Google Scholar]
- Schlag M, Biswas R, Krismer B, et al. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol. 2010;75:864–73. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–77. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher M, Powell AM, Becker SC, et al. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl Environ Microb. 2012;78:2297–305. doi: 10.1128/AEM.07050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Carlos MR, Singh K. Physiological significance of the peptidoglycan hydrolase, LytM, in Staphylococcus aureus. FEMS Microbiol Lett. 2010;311:167–75. doi: 10.1111/j.1574-6968.2010.02087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetoch EA, Eruslanov BV, Perelygin VV, et al. Diverse antimicrobial killing by Enterococcus faecium E 50–52 bacteriocin. J Agr Food Chem. 2008;56:1942–8. doi: 10.1021/jf073284g. [DOI] [PubMed] [Google Scholar]
- Szweda P, Schielmann M, Kotlowski R, et al. Peptidoglycan hydrolases-potential weapons against Staphylococcus aureus. Appl Microbiol Biot. 2012;96:1157–74. doi: 10.1007/s00253-012-4484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J, Komatsuzawa H, Yamada S, et al. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol Immunol. 2001;46:601–12. doi: 10.1111/j.1348-0421.2002.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Taubes G. The bacteria fight back. Science. 2008;321:356–61. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Teneback CC, Scanlon TC, Wargo MJ, et al. Bioengineered lysozyme reduces bacterial burden and inflammation in a murine model of mucoid Pseudomonas aeruginosa lung infection. Antimicrob Agents Ch. 2013;57:5559–64. doi: 10.1128/AAC.00500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm G, Gotz F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol. 1997;23:1251–65. doi: 10.1046/j.1365-2958.1997.2911657.x. [DOI] [PubMed] [Google Scholar]
- Won HS, Jung SJ, Kim HE, et al. Systematic peptide engineering and structural characterization to search for the shortest antimicrobial peptide analogue of gaegurin 5. J Biol Chem. 2004;279:14784–91. doi: 10.1074/jbc.M309822200. [DOI] [PubMed] [Google Scholar]
- World Health Organization Antimicrobial resistance: global report on surveillance. 2014 [Google Scholar]
- Yamakawa T, Mitsuyama J, Hayashi K. In vitro and in vivo antibacterial activity of T-3912, a novel non-fluorinated topical quinolone. J Antimicrob Chemoth. 2002;49:455–65. doi: 10.1093/jac/49.3.455. [DOI] [PubMed] [Google Scholar]
- Yan H, Li S, Sun X, et al. Individual substitution analogs of Mel (12–26), melittin's C-terminal 15-residue peptide: their antimicrobial and hemolytic actions. FEBS Lett. 2003;554:100–4. doi: 10.1016/s0014-5793(03)01113-x. [DOI] [PubMed] [Google Scholar]
- Yang S-C, Lin C-H, Sung CT, et al. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol. 2014;5:241. doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Rozek A, Hancock REW. Interaction of cationic antimicrobial peptides with model membranes. J Biol Chem. 2001;276:35714–22. doi: 10.1074/jbc.M104925200. [DOI] [PubMed] [Google Scholar]
- Zhao H, Blazanovic K, Choi Y, et al. Gene and protein sequence optimization for high-level production of fully active and aglycosylated lysostaphin in Pichia pastoris. Appl Environ Microb. 2014;80:2746–53. doi: 10.1128/AEM.03914-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Cegelski L. Nutrient-dependent structural changes in S. aureus peptidoglycan revealed by solid-state NMR spectroscopy. Biochemistry. 2012;51:8143–53. doi: 10.1021/bi3012115. [DOI] [PMC free article] [PubMed] [Google Scholar]