Abstract
Adenosine diphosphate ribosyltransferases (ARTDs; ARTD1–17 in humans) are emerging as critical regulators of cell function in both normal physiology and disease. These enzymes transfer the ADP-ribose moiety from its substrate, nicotinamide adenine dinucleotide (NAD+), to amino acids of target proteins. The functional redundancy and overlapping target specificities among the 17 ARTDs in humans make the identification of direct targets of individual ARTD family members in a cellular context a formidable challenge. Here we describe the rational design of orthogonal NAD+ analogue-engineered ARTD pairs for the identification of direct protein targets of individual ARTDs. Guided by initial inhibitor studies with nicotinamide analogues containing substituents at the C-5 position, we synthesized an orthogonal NAD+ variant and found that it is used as a substrate for several engineered ARTDs (ARTD1, −2, and −6) but not their wild-type counterparts. Comparing the target profiles of ARTD1 (PARP1) and ARTD2 (PARP2) in nuclear extracts highlighted the semi-complementary, yet distinct, protein targeting. Using affinity purification followed by tandem mass spectrometry, we identified 42 direct ARTD1 targets and 301 direct ARTD2 targets. This represents a powerful new technique for identifying direct protein targets of individual ARTD family members, which will facilitate studies delineating the pathway from ARTD activation to a given cellular response.
Adenosine diphosphate ribosylation (ADPr) is a post-translational modification that plays a major role in a wide array of cellular processes (e.g., energy metabolism, transcription, and genomic maintenance).1 In humans, ADP-ribose transfer is catalyzed by a family of 17 diphtheria toxin-like ADP-ribosyl-transferases (ARTDs) that share a conserved catalytic domain.2 ARTDs catalyze the transfer of the ADP-ribose moiety from nicotinamide adenine dinucleotide (NAD+) to their target proteins.3 The ARTD family has been recently subclassified on the basis of the ability of the individual ARTD enzymes to catalyze the transfer of a single ADP-ribose unit (mono-ARTDs: ARTD7, 8, 10–12, 14–17) or multiple ADP-ribose units (poly-ARTDs: ARTD1–3, 5–6) onto target proteins.2
The best-understood ARTD is ARTD1 (also known as PARP1), which was previously thought to be the sole enzyme responsible for poly-ADPr in cells.4 ARTD1 is involved in stress signaling, such as the DNA damage repair pathway. While protein targets of ADP-ribosylation in stress-signaling pathways have been identified,5,6 in many cases it is not clear if these are direct targets of ARTD1. This is because of the functional redundancy and overlapping target specificities among ARTDs. Non-radioactive NAD+ derivatives, such as 6-biotin-NAD+ 7 and 6-alkyne-NAD+ (6-a-NAD+),8 for use in copper-catalyzed conjugation to an azidoalkyl reporter (click chemistry), have been useful for globally identifying targets of ADPr. But these reagents are insufficient to identify the direct protein targets of ARTDs since they are substrates for all ARTDs. Given that up to 17 unique ARTDs are expressed within the cell at any given time,9 identifying direct targets for each ARTD family member remains an essential challenge toward parsing the functional role for each of these individual enzymes.
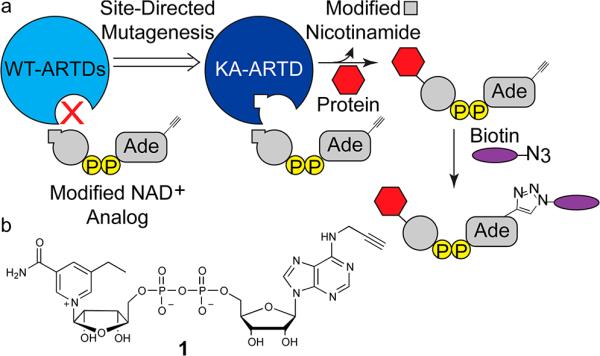
To address this challenge, we implemented a “bump-hole” strategy for identifying the direct protein targets of ARTDs. This strategy has been successfully used for the identification of the direct targets of kinases,10 acetyltransferases,11 and methyltransferases,12 as well as for the incorporation of non-natural amino acids via modified aminoacyl-tRNA synthetases.13 We envisioned that a unique hydrophobic pocket could be engineered in the nicotinamide-binding site of ARTDs such that they could bind an orthogonal NAD+ variant containing a substituent at the C-5 position on the nicotinamide moiety of NAD+. This orthogonal NAD+ variant would also contain an alkyne tag at the N-6 position on the adenosine moiety for copper-catalyzed conjugation to a biotin–azide probe (Figure 1a). We hypothesized that a substituent (e.g., alkyl or benzyl) at the C-5 position on the nicotinamide moiety would be sufficient to exclude the orthogonal NAD+ variant from the active site of wild-type ARTDs.
Figure 1.
(a) Schematic for the design of modified NAD+ analogues that are preferentially utilized by engineered ARTDs (KA-ARTD). P = phosphate. (b) 5-Et-6-a-NAD+ (1) used in this study.
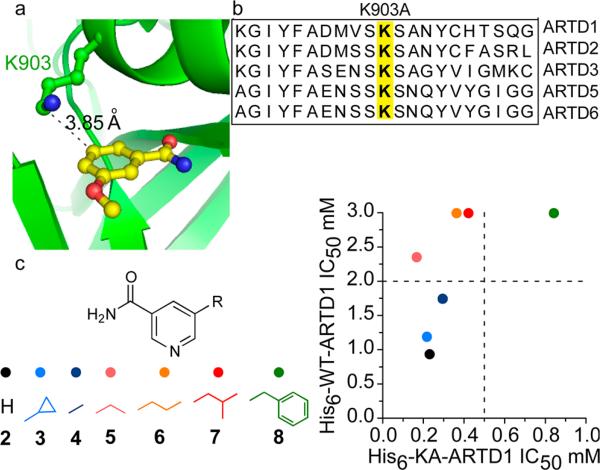
We first identified a position within the poly-ARTD nicotinamide-binding site that we could mutate to a smaller amino acid to create a hydrophobic pocket that would accommodate orthogonal NAD+ variants containing a substituent at the C-5 position. Multiple crystal structures are available for the poly-ARTD sub-class bound to nicotinamide analogues, providing salient atomic details for the nicotinamide-binding site.14–16 The crystal structure of ARTD1 bound to the nicotinamide analogue 3-methoxybenzamide (PDB ID: 3PAX)14 reveals a lysine residue (K903) that forms van der Waals contacts with the C-5 position of the benzamide ring (Figure 2a). While we considered other positions within the active site, we focused on K903 since it is conserved throughout the poly-ARTD sub-class, thus providing a potentially general strategy for identifying the direct targets of poly-ARTDs (Figure 2b). Due to its established, well-characterized enzymatic activity, we began by engineering ARTD1.17–19 To accommodate substituents at the C-5 position of the nicotinamide ring, we mutated K903 to an alanine (K903A) (Figure S1). Rather than starting with a synthetically challenging orthogonal NAD+ variant, we reasoned that we could first test simple nicotinamide analogues as inhibitors, since nicotinamide and the nicotinamide nucleotide of NAD+ bind in the same orientation in the active site of ARTDs. We therefore synthesized a small panel of nicotinamide analogues containing various substituents at the C-5 position (Figure 2c and Schemes S1 and S2).
Figure 2.
(a) The key residue, K903, in ARTD1 (green), near the C-5 position of 3-methoxybenzamide (yellow) (PDB ID: 3PAX).14 (b) Sequence alignment of the nicotinamide binding site of the poly-ARTDs. (c) Comparative inhibition of WT-ARTD1 and KA-ARTD1 by select nicotinamide analogues. IC50 values (mM) for each analogue are plotted comparing the WT-ARTD1 (y-axis) and KA-ARTD1 (x-axis) variants.
To determine if nicotinamide analogues with substituents at the C-5 position are more selective for K903A ARTD1 (KA-ARTD1) than for wild-type ARTD1 (WT-ARTD1), we monitored ADP-ribose transfer to the ARTD1 target,20 Histone H1, using the previously described clickable NAD+ derivative, 6-a-NAD+ 8 (Scheme S3). 6-a-NAD+ contains an alkyne at the N-6 position of the adenosine ring that can be conjugated to an azide reporter via click chemistry for monitoring ADPr.8 We sought a C-5-substituted nicotinamide analogue that was at least 10-fold more selective for KA-ARTD1 compared to WT-ARTD1. We found that all C-5-substituted nicotinamide analogues were more selective for KA-ARTD1 than for WT-ARTD1 (Figures 2c and S2). The most potent and selective analogue of KA-ARTD1 was 5-ethyl-nicotinamide (IC50 = 165 ± 11 μM), with 14-fold selectivity over WT-ARTD1 (IC50 = 2359 ± 368 μM) (Figures 2c and S2). Together, these results demonstrate that ARTD1 can be engineered to create a unique, inhibitor-sensitizing mutation in the nicotinamide-binding site.
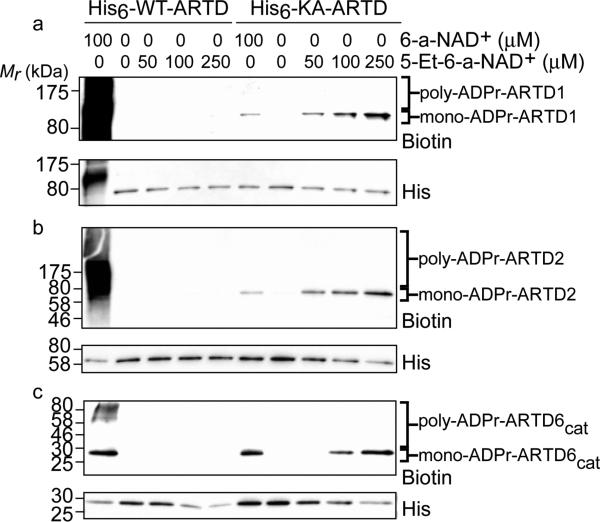
We next sought to prepare an orthogonal NAD+ variant that would be a substrate for KA-ARTD1 but not WT-ARTD1. Guided by the results of our inhibitor studies, we synthesized the orthogonal NAD+ variant 5-Et-6-a-NAD+ (1), which contains an ethyl substituent at the C-5 position of the nicotinamide ring and an alkyne at the N-6 position of the adenosine ring (Figure 1b and Scheme S4). To investigate the substrate selectivity of 5-Et-6-a-NAD+, we monitored auto-ADP-ribosylation of WT- and KA-ARTD1 by click conjugation to a biotin–azide reporter. Incubation of KA-ARTD1 with increasing concentrations of 5-Et-6-a-NAD+ resulted in auto-ADPr of KA-ARTD1 (Figure 3a). By contrast, no modification of WT-ARTD1 was detected with up to 250 μM 5-Et-6-a-NAD+ (Figure 3a).
Figure 3.
Orthogonal auto-ADPr of sensitized ARTD(s) using modified NAD+ variants. The concentration of the NAD+ analogue is indicated. Samples were subjected to immunoblot detection with streptavidin (biotin) to detect modified protein. His-tag (His) detection served as a loading control. Poly-ADPr and mono-ADPrmodified fractions are indiciated: (a) ARTD1, (b) ARTD2, and (c) ARTD6cat.
Whereas extensive poly-ADPr of WT-ARTD1 was observed using 6-a-NAD+, as evidenced by the biotinylated smear above the molecular weight of WT-ARTD1, only mono-ADPr of KA-ARTD1 was observed using both 6-a-NAD+ and 5-Et-6-a-NAD+, as evidenced by the single biotinylated band at the molecular weight of KA-ARTD1 (Figure 3a). Using a poly-ADP-ribose specific antibody (10H) and the native NAD+ substrate, we confirmed that KA-ARTD1 functions as a mono-ARTD (Figure S3). We cannot rule out the possibility that the altered activity of KA-ARTD1 may complicate efforts to identify all of the ARTD1 substrates. However, the apparent activity of the mono-ARTD is sufficient to transfer modified ADP-ribose onto target substrates for initial identification. In fact, the exclusive mono-ADPr activity of KA-ARTD1 provides an unexpected benefit: lack of poly-ADPr actually decreases the complexity of samples submitted for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, decreasing noise in the fragmentation pattern. Indeed, a previous study demonstrated that converting ARTD1 to a mono-ARTD by mutating the catalytic glutamate to a glutamine greatly facilitated the identification of ADP-ribosylation sites by LC-MS/MS.21
To further establish the generalizability of our approach, we next sought to determine if our engineered ARTD enzyme–5-Et-6-a-NAD+ pair could work for other poly-ARTD family members. We focused on ARTD2 and ARTD6 (also known as Tankyrase 2) (Figure S1) because of their well-characterized enzymatic activity. Similar to WT-ARTD1, no modification of WT-ARTD2 and WT-ARTD6 was detected by up to 250 μM 5-Et-6-a-NAD+ (Figure 3b,c). Mutation of the conserved lysine in the nicotinamide binding pocket of ARTD2 and ARTD6 to alanine (KA-ARTD2 and KA-ARTD6, respectively) conferred sensitivity to 5-Et-6-a-NAD+, as evidenced by the single biotinylated band at the molecular weight of the KA-ARTDs (Figure 3b,c). Taken together, these experiments demonstrate that we have successfully created a poly-ARTD wide-engineered enzyme–modified substrate pair.
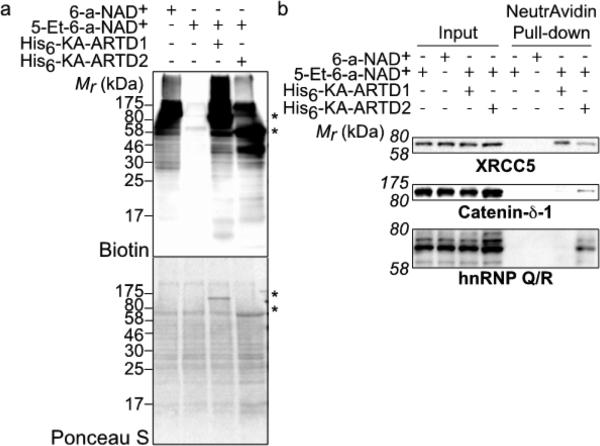
Having demonstrated that we can engineer poly-ARTDs to accept an orthogonal NAD+ variant that is not used by their WT counterpart, we next determined if we could label direct targets of ARTD1 in a cellular context. We treated nuclear extracts from human embryonic kidney (HEK) 293T cells with 5-Et-6-a-NAD+ (250 μM) alone or in the presence of KA-ARTD1. As a positive control for ADP-ribose detection, we treated nuclear extracts with 6-a-NAD+ alone. Protein targets of ADP-ribosylation were detected by click chemistry with biotin–azide. In contrast to 6-a-NAD+ (100 μM) treatment, which resulted in labeling of several proteins due to the endogenous activity of various ARTDs (predominately ARTD1),8 no labeling was detected with 5-Et-6-a-NAD+ treatment (Figure 4a). Treatment of nuclear extracts with both 5-Et-6-a-NAD+ (250 μM) and KA-ARTD1 resulted in extensive labeling (Figure 4a). The band pattern produced is similar to that seen with endogenous enzymes and 6-a-NAD+, though there are notable differences between total ADP-ribosylation and direct ADPr attributable to KA-ARTD1.
Figure 4.
(a) Lysate labeling by ARTDs and modified NAD+ analogues. The faint bands observed in the 5-Et-NAD+-only lane correspond to endogenous biotinylated proteins. The membrane stained with Ponceau S serves as a loading control. The asterisks mark the KA-ARTD1 (upper) and KA-ARTD2 (lower) bands. (b) Immunoblot detection of the LC-MS/MS-identified targets (XRCC5, Catenin-δ-1, hnRNP Q/R) following NeutrAvidin enrichment.
As previous studies suggest that ARTD2 is capable of compensating for the loss of ARTD1,22 we applied our method to compare the direct protein targets of ARTD1 and ARTD2. Nuclear extracts treated with 5-Et-6-a-NAD+ (250 μM) and KA-ARTD2 resulted in a band pattern that was similar—with notable differences in band presence and intensity—to the pattern produced from KA-ARTD1 (Figure 4a). These results suggests that, while ARTD1 and ARTD2 share redundant functions in the cell,22 they do have distinct protein targets.
We next sought to use our strategy to identify the direct protein targets of ARTD1 and ARTD2 in nuclear lysates using LC-MS/MS. HEK 293T nuclear extracts were treated with either 6-a-NAD+ alone (total ADP-ribosylated proteins), 5-Et-6-a-NAD+ alone (background control), or 5-Et-6-a-NAD+ in the presence of either KA-ARTD1 (ARTD1-specific protein targets) or KA-ARTD2 (ARTD2-specific protein targets). Following conjugation with biotin–azide, biotinylated proteins were enriched using NeutrAvidin agarose and proteolyzed, and eluted peptides were subjected to LC-MS/MS (Figure S4). We identified 339 proteins across the multiple conditions (Tables S1–S5). Among the 339 proteins, 42 proteins were uniquely identified as direct ARTD1 targets, and 301 were direct ARTD2 targets (thresholds discussed in Supporting Information). There is a 52% and 7% overlap between direct ARTD1 or ARTD2 targets, respectively, and total ADP-ribosylated targets (Figure S5), with an additional 25 and 17 targets identified as ADP-ribosylated but not directly by ARTD1 or ARTD2. Twenty-two of the ARTD1 targets and 133 of the ARTD2 targets were previously identified as targets of ADP-ribosylation under cell stress paradigms,5,6,8,23–25 suggesting that we have identified bona fide ARTD1 and ARTD2 targets.
To confirm our LC-MS/MS results, we selected the ARTD1 and ARTD2 direct target, XRCC5 (also known as Ku80),23 and the ARTD2-specific targets, Catenin-δ-1 and hnRNP Q/R, for identification using NeutrAvidin enrichment (Figure S6) followed by immunoblot detection with target specific antibodies. Nuclear lysate treated with 5-Et-6-a-NAD+ in the absence of KA-ARTD1 or KA-ARTD2 resulted in a complete lack of enrichment for the selected targets (Figure 4b). Enrichment of XRCC5 was observed with both KA-ARTD1 and KA-ARTD2 (Figure 4b). By contrast, enrichment of Catenin-δ-1 and hnRNP Q/R was observed only with KA-ARTD2 (Figure 4b). Taken together, these results highlight our ability to identify unique, direct targets of a given ARTD family member in a complex mixture.
In this study we developed new orthogonal NAD+ analogue-engineered ARTD pairs to efficiently label and identify family-member-specific ARTD protein targets. By initially focusing on simple C-5-substituted nicotinamide analogues as selective inhibitors of the engineered ARTD1 mutant, KA-ARTD1, we were able to quickly identify a complementary interaction, which guided the design of the orthogonal NAD+ variant, 5-Et-6-a-NAD+. The results from the nicotinamide inhibitor assay mirrored the results achieved when the modified 6-a-NAD+ analogue was tested for specificity in auto-ADP-ribosylation reactions of ARTD1. By enriching for tagged proteins in experiments with either KA-ARTD1 or KA-ARTD2 and the modified NAD+ analogue, we identified sets of direct, bona fide ARTD1 and ARTD2 targets via LC-MS/MS.
Our strategy proved applicable to multiple members of the poly-ARTD sub-class and revealed how two closely related poly-ARTD family members, namely ARTD1 and ARTD2, exhibited distinct targeting patterns in nuclear lysate. Although we focused primarily on the well-characterized poly-ARTDs, we envision that the strategy described here could be applicable for the identification of the direct targets of mono-ARTDs, as the nicotinamide binding site is conserved throughout the family. Given the difficulties in delineating the targeting specificities for this highly homologous enzyme class, our method provides a powerful new tool toward advancing our understanding of ARTD function in the cell.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeffrey Huang for assistance in cloning the ARTD2 gene and John Kilmek for the MS/MS analysis. We thank Samie Jaffrey, Tom Scanlan, and members of the Cohen laboratory for many helpful discussions and for advice on the manuscript.
Footnotes
Supporting Information
Experimental procedures, synthesis of compounds, and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Krishnakumar R, Kraus WL. Mol. Cell. 2010;39:8. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Trends Biochem. Sci. 2010;35:208. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Burkle A. FEBS J. 2005;272:4576. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 4.Chambon P, Weill JD, Mandel P. Biochem. Biophys. Res. Commun. 1963;11:39. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Wang J, Ding M, Yu Y. Nature Methods. 2013;10:981. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 6.Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Mol. Cell. 2013;52:272. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J. Methods Enzymol. 1997;280:255. doi: 10.1016/s0076-6879(97)80117-6. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Kim JH, Frizzell KM, Kraus WL, Lin H. J. Am. Chem. Soc. 2010;132:9363. doi: 10.1021/ja101588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czapski GA, Adamczyk A, Strosznajder RP, Strosznajder JB. Neurochem. Int. 2013;62:664. doi: 10.1016/j.neuint.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Hertz NT, Wang BT, Allen JJ, Zhang C, Dar AC, Burlingame AL, Shokat KM. Curr. Protoc. Chem. Biol. 2010;2:15. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Mi J, Feng Y, Ngo L, Gao T, Yan L, Zheng YG. J. Am. Chem. Soc. 2013;135:7791. doi: 10.1021/ja311636b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam K, Bothwell I, Chen Y, Sengelaub C, Wang R, Deng H, Luo M. J. Am. Chem. Soc. 2012;134:5909. doi: 10.1021/ja2118333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 14.Ruf A, de Murcia G, Schulz GE. Biochemistry. 1998;37:3893. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 15.Karlberg T, Hammarstrom M, Schutz P, Svensson L, Schuler H. Biochemistry. 2010;49:1056. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- 16.Narwal M, Venkannagari H, Lehtio L. J. Med. Chem. 2012;55:1360. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
- 17.Kawaichi M, Ueda K, Hayaishi O. J. Biol. Chem. 1981;256:9483. [PubMed] [Google Scholar]
- 18.Beneke S, Alvarez-Gonzalez R, Burkle A. Exp. Gerontol. 2000;35:989. doi: 10.1016/s0531-5565(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 19.Nottbohm AC, Dothager RS, Putt KS, Hoyt MT, Hergenrother PJ. Angew. Chem. 2007;46:2066. doi: 10.1002/anie.200603988. [DOI] [PubMed] [Google Scholar]
- 20.Lindgren AE, Karlberg T, Thorsell AG, Hesse M, Spjut S, Ekblad T, Andersson CD, Pinto AF, Weigelt J, Hottiger MO, Linusson A, Elofsson M, Schuler H. ACS Chem. Biol. 2013;8:1698. doi: 10.1021/cb4002014. [DOI] [PubMed] [Google Scholar]
- 21.Tao Z, Gao P, Liu HW. J. Am. Chem. Soc. 2009;131:14258. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 22.Boehler C, Gauthier L, Yelamos J, Noll A, Schreiber V, Dantzer F. Methods Mol. Biol. 2011;780:313. doi: 10.1007/978-1-61779-270-0_19. [DOI] [PubMed] [Google Scholar]
- 23.Henrie MS, Kurimasa A, Burma S, Menissier-de Murcia J, de Murcia G, Li GC, Chen DJ. DNA Repair. 2003;2:151. doi: 10.1016/s1568-7864(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 24.Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, Corda D, Di Girolamo M. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4243. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, Kelly I, Boutin M, Moon KM, Foster LJ, Poirier GG. Nucleic Acids Res. 2012;40:7788. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.