SUMMARY
RING1B, a Polycomb Group (PcG) protein, binds methylated chromatin through its association with another PcG protein called Polycomb (Pc). However, RING1B can associate with nonmethylated chromatin suggesting an alternate mechanism for RING1B interaction with chromatin. Here, we demonstrate that two proteins with little sequence identity between them, the Pc cbox domain and RYBP, bind the same surface on the C-terminal domain of RING1B (C-RING1B). Pc cbox and RYBP each fold into a nearly identical, intermolecular beta sheet with C-RING1B and a loop structure which are completely different in the two proteins. Both the beta sheet and loop are required for stable binding and transcription repression. Further, a mutation engineered to disrupt binding on the Drosophila dRING1 protein prevents chromatin association and PcG function in vivo. These results suggest that PcG targeting to different chromatin locations relies, in part, on binding partners of C-RING1B that are diverse in sequence and structure.
INTRODUCTION
The Polycomb Group (PcG) of gene silencers are chromatin-associated multiprotein complexes that maintain the genomic program of cells (Schwartz and Pirrotta, 2008). In stem cells, PcG complexes bind to hundreds of genomic loci, where they repress genes that promote differentiation (Boyer et al., 2006; Lee et al., 2006). A fundamental question regarding PcG function is how these complexes bind specifically to so many sites. Such targeting would appear to require highly specific and elaborate regulatory mechanisms since it is ultimately responsible for maintaining the intricate balance between pluripotency and differentiation.
Several of the multiprotein PcG complexes have been isolated and shown to have distinct repressive functions. Polycomb repression complex 1 (PRC1) is composed of four core components: Polycomb (Pc), RING1A or RING1B (dRING1 or Sex combs extra in Drosophila), Bmi-1 or Mel-18 (Posterior sex combs, Psc, in Drosophila), and Polyhomeotic (Ph) (Francis et al., 2001; Shao et al., 1999). In vitro experiments suggest that PRC1 may mediate repression through chromatin compaction and the inhibition of chromatin remodeling enzymes (Francis et al., 2001, 2004; Shao et al., 1999). Alternatively, the ligation of ubiquitin to lysine 119 of histone H2A (Wang et al., 2004a), which is catalyzed by a heterodimer formed by the RING finger domains of RING1B and Bmi-1 (Buchwald et al., 2006; Li et al., 2006), may maintain RNA polymerase II in an inactive, but poised position at repressed genes (Stock et al., 2007). A potential way for these two distinct repressive mechanisms of PRC1 could exist was recently suggested by the identification of a PRC1-like complex in Drosophila called dRAF (Lagarou et al., 2008). The dRAF complex is distinct from PRC1 but still contains two of the PRC1 core components (Psc and dRING1) and is a competent Ub ligase. Thus, it may be possible for the PRC1-like dRAF complex to function as a ubiquitin E3 ligase while PRC1 may function to create higher order repressive chromatin structures.
What is largely unresolved with regards to PcG function are the precise mechanisms of targeting PcG complexes to hundreds of locations within genomes. PcG complexes bind to cis-regulatory DNA elements called Polycomb response elements (PREs). In Drosophila, a PcG protein called Pleiohomeotic (Pho), the only PcG protein capable of binding a specific DNA sequence, plays a key role in directing PRC1 to the PREs (Wang et al., 2004b). Pho can cooperate with PRC1 to bind PREs that are depleted of histones forming a repressive structure whereby the DNA is wrapped around the Pho/PRC1 complex (Mohd-Sarip et al., 2005, 2006). Pho is also a component of a distinct PcG complex called PhoRC which includes the PcG protein dSfmbt (Klymenko et al., 2006). The methylated histone binding ability of the dSfmbt MBT domain (Grimm et al., 2009; Klymenko et al., 2006) may allow PhoRC to utilize the combination of the DNA binding ability of Pho and methylated histone binding of dSfmbt to bind specifically to hundreds of sites within the Drosophila genome (Oktaba et al., 2008). While Pho is clearly important for PRE PcG targeting, a PRE prediction algorithm using the DNA binding sequence of Pho and other PcG-associated proteins (Ringrose et al., 2003) failed to detect most PcG binding sites identified from several Drosophila genome-wide studies (Negre et al., 2006; Schwartz et al., 2006; Tolhuis et al., 2006). Predicting PREs in vertebrate genomes have proved to be a challenging task and the identification of two mammalian PREs have only been recently reported (Sing et al., 2009; Woo et al., 2010). Like the PREs in Drosophila, the mammalian PREs contain DNA sequences that are recognized by the homolog of Pho, Yin Yang 1 (YY1).
The vertebrate PRC1 component that likely plays a key role in directing PRC1 to the YY1 binding sites is RING1A or RING1B. The C-terminal domain of RING1A or RING1B (C-RING1A or C-RING1B) binds to the C-terminal region of an adaptor protein called RYBP (RING1 YY1 binding protein) while a nonoverlapping N-terminal portion of RYBP associates with YY1 (Garcia et al., 1999). This type of protein-protein interaction would allow targeting of PRC1 to specific DNA sequences in the mammalian genome. The important role played by RYBP in PcG function is reflected in its requirement for repression mediated through the mammalian PRE (Woo et al., 2010).
RING1B also plays a role in an alternative mechanism of targeting PRC1 to chromatin. The posttranslational trimethylation of histone H3 at lysine 27 (H3K27Me3) is important in PRC1 targeting. The N-terminal chromo domain of Pc binds H3K27Me3 (Cao et al., 2002; Fischle et al., 2003; Min et al., 2003), providing one avenue by which PRC1 can be targeted to chromatin. The C-terminal cbox domain of Pc directly associates with C-RING1B (Schoorlemmer et al., 1997), facilitating assembly of PRC1 at the histone posttranslational modification. It should be noted that the chromo domain from other vertebrate Pc orthologs are capable of binding different histone methylations besides H3K27Me3 (Bernstein et al., 2006). For example, cbx7 can bind both H3K9Me3 as well as H3K27Me3. Because of this and perhaps of alternative targeting mechanisms for PRC1, there are many instances where RING1B is bound to chromatin at locations other than H3K27Me3. There are several examples of this phenomenon. (1) In preimplantation embryos, paternal heterochromatin that lacks H3K27Me3 can still bind maternal RING1B and other PRC1 components (Puschendorf et al., 2008). (2) In embryonic stem cells that are deficient in the enzyme that methylates H3K27 and thus lack H3K27Me3, RING1B can still bind to the X chromosome (Schoeftner et al., 2006). (3) In mouse embryonic stem cells, RING1B was found to bind to the promoter region of 244 genes (of the 1219 total genes that exhibit RING1B binding) where the H3K27Me3 was not detected (Boyer et al., 2006). While it is possible that RING1B is present at these sites but not detected, alternative recruitment mechanisms may also be in play. Moreover, the Drosophila ortholog dRING1 has been shown to localize to sites on polytene chromosomes that are independent of other PRC1 components (Gorfinkiel et al., 2004).
C-RING1B was predicted to have (Sanchez-Pulido et al., 2008) and confirmed to possess a ubiquitin fold (Bezsonova et al., 2009) but how it is capable of having multiple binding partners remains unknown. Here, we provide evidence suggesting how RING1B, and by extension, PRC1, can be specifically targeted to different sites in the genome. We show that two different binding partners to C-RING1B, the Pc cbox domain and RYBP, can be very different in their sequence and bind with different conformations to RING1B. Yet, despite these differences, these proteins bind to the same binding site on RING1B. This finding has important implications for the types of multiprotein PRC1-like complexes that can form in vivo and how they might be targeted to chromatin.
RESULTS
C-RING1B cbx7 cbox Structure
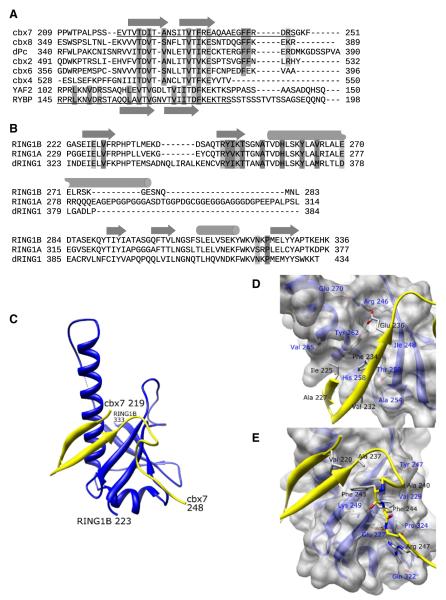
In order to better understand the protein-protein interactions of C-RING1B, we first determined the structure of C-RING1B bound to a Pc cbox domain. Of the five vertebrate Pc orthologs, we chose the cbox domain from cbx7 for crystallization because of its strong affinity for C-RING1B (Kd = 9.2 nM) (Wang et al., 2008). To facilitate crystallization, we used C-RING1B and cbx7 cbox residues (223–333 and 219–248, respectively) which showed the least amount of conformational flexibility when in complex with each other based on NMR relaxation measurements (Figures 1A and 1B; see Figure S1 available online). We determined the crystal structure of the C-RING1B/cbx7 219–248 complex and refined it to 1.7 Å resolution (Figures 1C–1E; Table S1). In the crystal, there are two C-RING1B/cbx7 cbox complexes in the asymmetric unit associated via the interaction between each of their long central helices. Our mutagenesis data (Demeler et al., 2010) as well as the structure of C-RING1B alone (Bezsonova et al., 2009) indicate that this interaction is the homodimerization interface of C-RING1B when C-RING1B is devoid of either the Pc cbox or RYBP. Because this interaction occurs only in the absence of a binding partner (Czypionka et al., 2007; Wang et al., 2008), the two C-RING1B molecules in the asymmetric unit are likely the result of the high concentration of protein required for crystallization and not reflective of what occurs inside cells. Our analysis of the structure is thus focused on a single heterodimer unit. The C-RING1B structure is composed of an extensive beta sheet region and a long central helix that extends from residue Val 256 to Glu 277. Residues 219–238 of the cbx7 cbox domain form an antiparallel beta sheet which makes an extended, intermolecular beta sheet with C-RING1B. The combined beta sheet structure packs against the C-RING1B central helix. In the cbox beta sheet (Figure 1D), the side-chain aromatic ring of cbx7 Phe 234 is most notable as it stacks against the side-chain aromatic ring of RING1B Tyr 262. Additionally, cbx7 beta sheet residues Thr 223, Ile 225, Ala 227, and Val 232 are clustered around Phe 234, packing against a hydrophobic pocket formed by RING1B residues Ile 248, Thr 250, Ala 254, His 258, Leu 259, Tyr 262, and Val 265. In addition to the backbone hydrogen bonds between C-RING1B and cbx7 that comprise the intermolecular beta sheet structure, there is a side-chain mediated polar interaction between cbx7 Glu 236 and RING1B residues Arg 246 and Tyr 262. Furthermore, cbx7 residues 239–248 form a loop structure that contacts C-RING1B (Figure 1E). In the cbox loop region, two consecutive cbx7 Phe residues, 243 and 244, pack into a hydrophobic pocket formed by RING1B residues Val 229, Tyr 247, and Pro 324. The side chains of RING1B residues Lys 249 and Glu 227 are in an extended conformation and are hydrogen bonded to the backbone atoms of the cbox loop residues. These extended conformations allow the methylene groups on these residues to help form the binding pocket for Phe 243 and 244. The guanidinium group of cbx7 Arg 247 forms a salt bridge interaction with the RING1B Glu 227 side chain while also hydrogen bonding to the carboxamide group of RING1B Gln 322.
Figure 1. C-RING1B/cbx7 cbox Structure.
(A) ClustalW sequence alignment of the cbox domains from all five human and Drosophila Pc proteins and the region of the human RYBP sequence known to associate with C-RING1B (Garcia et al., 1999). The underlined sequences represent the residues used in the crystallization.
(B) The C-terminal domains from human RING1A, RING1B, and Drosophila RING1 proteins. Secondary structures are indicated as arrows (beta sheet) and cylinders (helix). Residues that mediate hydrophobic and polar interactions are dark and light shaded, respectively.
(C) Structure of the C-RING1B cbx7 cbox complex. The RING1B is blue, cbx7 is yellow. Termini are labeled. The dotted line indicates RING1B residues 285 and 288 which could not be modeled because of weak density.
(D and E) A closeup view of the (D) beta sheet and (E) the loop interactions. Cbx7 residues 219–224 are deleted in (D) for clarity. C-RING1B residues are labeled in blue, cbx7 residues are labeled in black.
Both the cbox beta Sheet and Loop Are Required for Binding C-RING1B and for Repression
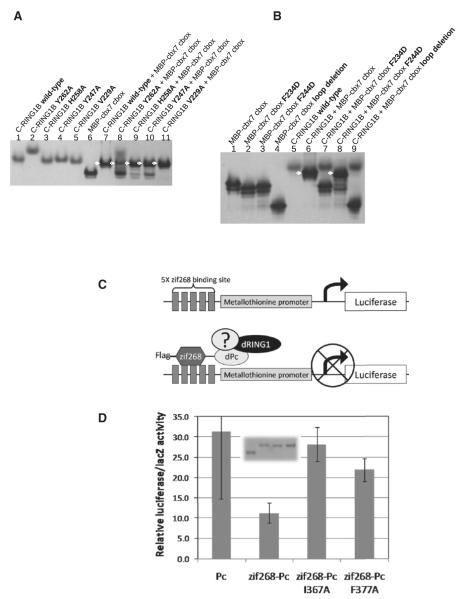
We next tested whether both the beta sheet and the loop structure of the cbox domain are necessary for binding. We introduced mutations into C-RING1B that alter either the beta sheet binding site (Tyr262Ala and His258Ala) or the cbox loop binding site (Val227Ala and Tyr247Ala) and then tested each of these mutant proteins for binding to the cbx7 cbox domain using a native gel binding assay (Figure 2A). These mutations are of surface residues and do not disrupt the tertiary fold (Figure S2). The mutant C-RING1B proteins showed diminished ability to bind cbx7 cbox, as indicated by the significant amounts of residual unbound cbx7 cbox that was observed for all the mutants except for Val229Ala (compare the bands for faster migrating unbound cbx7 cbox in Figure 2A lane 7 for wild-type to the corresponding bands in lanes 8–11). We also performed the complementary experiment of testing the binding of cbx7 cbox proteins mutated at the beta sheet and loop regions. When cbx7 cbox was mutated in the beta sheet, Phe234Asp, there was no binding to C-RING1B (Figure 2B, lane 7). The cbx7 loop mutation Phe244Asp was found to be capable of associating with C-RING1B, but with reduced affinity, as indicated by the presence of a substantial quantity of unbound cbx7 protein (Figure 2B, compare the bands for cbx7 in lanes 6 and 8). We further tested the role of the cbox loop interaction with C-RING1B by deleting cbx7 loop residues altogether. Deleting the loop of cbx7 cbox completely disrupted binding (Figure 2B, lane 9). The results of these mutagenesis studies are consistent with our NMR data which showed that Pc cbox domains were unstructured in the absence of C-RING1B (Wang et al., 2008). NMR conformational dynamics measurements revealed that upon binding C-RING1B, both the beta sheet and the loop structure of the cbx7 cbox domain become ordered (Figure S1A).
Figure 2. Beta Sheet and Loop Interactions Are Both Required for Binding and Repression.
(A) Native gel binding assay testing C-RING1B wild-type and mutant proteins binding to maltose binding protein (MBP) fused cbx7 cbox. Arrows indicate the 1:1 complex between the C-RING1B protein and its binding partner.
(B) Binding reactions using MBP-cbx7 cbox mutants. The MBP-cbx7 loop deletion terminates at cbx7 residue 240. The C-RING1B Tyr262Ala mutant likely disrupts the homodimerization of C-RING1B which would alter its mobility in a native gel compared to the other C-RING1B proteins.
(C) Transcription assay. Five repeats of the zif268 binding site are placed upstream of the metallothionine promoter (MTp). The zinc finger DNA binding domain of zif268 fused to dPc allows targeting to the MTp. Binding of endogenous dRING1 and other endogenous components to the MTp can lead to repression.
(D) Transcription assay results. Error bars are the standard deviations from three independent transfections. (inset) Anti-FLAG immunoblot of the exogenous dPc proteins expressed in cells. Equivalent lysate volumes used in the transcription assay were used for the immunoblot. Unfortunately, a similar experiment using dRING1 could not be performed because overexpression of dRING1 alone and one that is targeted to the MTp resulted in similar levels of luciferase activity.
Previously published transcription assays using recombinant Pc proteins targeted to reporter genes had shown that the cbox domain is required for repression (Bardos et al., 2000; Bunker and Kingston, 1994; Muller, 1995). We wondered whether specific, structure-guided point mutations within the cbox domain designed to disrupt binding to C-RING1B would hinder the ability of Pc to repress transcription. To this end, we used a transcription assay carried out in Drosophila S2 cells using the full-length Drosophila Pc (dPc) protein (Figure 2C). Using this assay, we found that the dPc that was targeted to an exogenous metallothionine promoter (MTp) was able to repress transcription of the luciferase reporter gene. In contrast, there was elevated luciferase activity with a dPc that was similarly expressed but not targeted to the MTp (Figure 2D). We also introduced mutations into dPc cbox that are equivalent to the cbx7 cbox mutations that hinder cbx7 cbox binding to RING1B (discussed above). The cbox beta sheet mutation Ile367Ala (equivalent to cbx7 Phe 234) and the loop residue mutant Phe377Ala (Phe 244 in cbx7) were both found to be incapable of repressing the expression of the reporter gene as compared to wild-type. Taking all of the in vitro and in vivo results together, we conclude that both the cbox beta sheet and loop structures are required to form a stable complex with C-RING1B, which in turn is required for full transcription repression.
RYBP/YAF2 Binding to C-RING1B Is Similar to the C-RING1B/Pc cbox Interaction
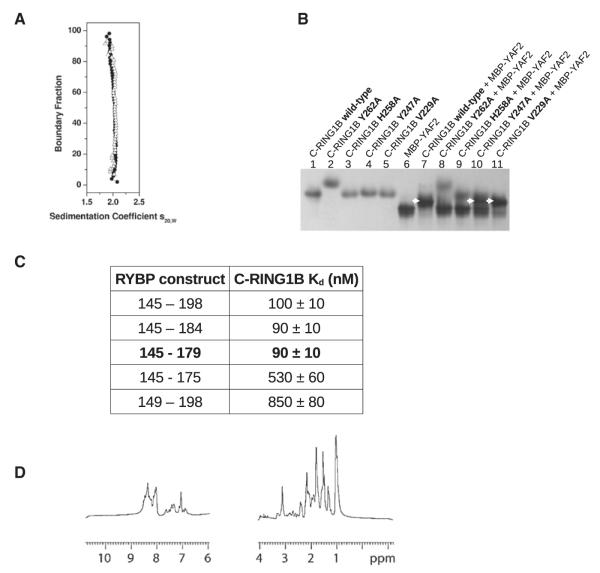
Evidence indicates a strong likelihood that RING1B targeting is dependent on the identity of the binding partner(s) of C-RING1B. For example, multiprotein complexes that include RING1B and RYBP (or its homolog YAF2) also contain proteins like E2F6, E2F2, E2F3, hGABPβ, and YY1, all of which have DNA binding domains (Garcia et al., 1999; Ogawa et al., 2002; Sawa et al., 2002; Schlisio et al., 2002; Trimarchi et al., 2001; Zheng et al., 2001). If the binding target of RING1B is a specific DNA sequence and not methylated chromatin, then C-RING1B would have to bind RYBP with 1:1 stoichiometry while not allowing C-RING1B to bind the Pc cbox domains. If the stoichiometry of the interactions is greater than 1:1 utilizing multiple binding surfaces on C-RING1B, then a single RING1B protein could simultaneously bind to two different sites. Since there is little sequence similarity between RYBP and the Pc cbox domains (Figure 1A), it is difficult to predict how RYBP binds to C-RING1B. We performed a series of experiments to investigate the C-RING1B/RYBP interaction and compared the results to those for the C-RING1B/Pc cbox complex. Using velocity sedimentation, we assessed the stoichiometry of the complex between C-RING1B and RYBP residues 145–198, a previously identified C-terminal region of RYBP that is capable of binding C-RING1B (Garcia et al., 1999) (Figure 3A). The sedimentation velocity van Holde-Weischet distribution plot of the C-RING1B/RYBP 145–198 complex revealed the presence of a homogeneous species with an S-value of 2. Using a genetic algorithm (Brookes and Demeler, 2007) and a nonlinear least-squares finite element analysis (Demeler and Saber, 1998), we determined the molecular weight of the complex to be 21.6 kDa, matching the calculated molecular weight for a 1:1 stoichiometry. In order to determine if the same C-RING1B surface binds both the Pc cbox domain and RYBP, we used a native gel binding assay and tested the association between the structure-guided C-RING1B mutants and YAF2, a close homolog to RYBP (Figure 3B). (We used YAF2 in lieu of RYBP because the migration of C-RING1B/RYBP complex in the native gel could not be distinguished from the individual components.) Our finding that the C-RING1B mutants that hinder Pc cbox binding also disrupted YAF2 binding suggested that the same surface is used to bind both proteins. Next, we determined the minimum region within RYBP required to bind C-RING1B. We made a series of RYBP constructs (145–184, 145–179, 145–175, 149–198) and measured their affinity to C-RING1B using surface plasmon resonance (SPR) (Figure 3C; Figure S3A–S3E). The equilibrium dissociation constants (Kd) for C-RING1B binding to RYBP 145–198 and RYBP 145–179 were similar (100 and 90 nM, respectively) while RYBP constructs shorter than 145–179 exhibited significantly lower affinity. This suggests that the 35 residues in RYBP 145–179 represent the minimum region required to bind C-RING1B. Like the Pc cbox domains which are unfolded in the absence of C-RING1B (Wang et al., 2008), RYBP 145–179 shows no upfield or downfield shifts in its 1D NMR spectrum, indicating the absence of a tertiary fold (Figure 3D). This is consistent with previous observations from analysis of a larger region of RYBP (Neira et al., 2009). Also similar to the Pc cbox domains, the 2D HSQC spectrum of C-RING1B shows a greater dispersion of chemical shifts in the presence of RYBP 145–179 compared to C-RING1B alone (Figure S3B). Conformational tightening within C-RING1B when in complex with RYBP or the Pc cbox domain would be expected to result in a more dispersed spectrum.
Figure 3. C-RING1B/RYBP Interaction and Structure.
(A) The van Holde-Weischet integral distribution plot of the C-RING1B/RYBP 145–198 complex at two different concentrations (Δ: 16 μM; ●: 56 μM).
(B) Native gel binding assay between C-RING1B (wild-type and mutant proteins) and MBP-YAF2 103–150.
(C) Table of the equilibrium dissociation constants between the indicated RYBP construct and C-RING1B measured using SPR (Figure S3). (D) 1D 1H NMR of RYBP 145–179.
RYBP Shares the Same Binding Site on C-RING1B as the cbx7 cbox Domain
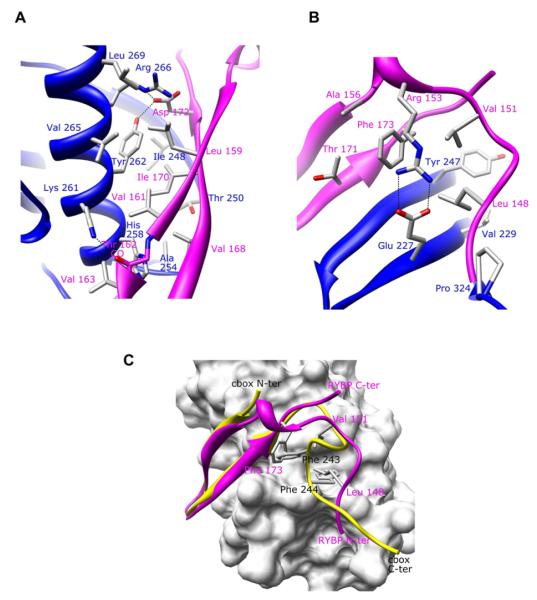
To view the binding mode between RYBP and C-RING1B, we determined the 1.7 Å crystal structure of C-RING1B bound to RYBP 145–179 (Figure 4). The structure revealed that despite significant differences in their sequences (Figure 1A), RYBP 145–179 binds to the same hydrophobic patches on the surface of C-RING1B as the cbx7 cbox domain. RYBP residues 158–172 folds into an antiparallel beta sheet that is nearly identical to the beta sheet formed by the cbx7 cbox domain (Figure 4A). Similar to cbx7 cbox, RYBP also has a loop structure which binds to the same C-RING1B surface that the cbx7 loop structure binds to. However, there are significant differences between the loop structures from cbx7 cbox and RYBP. For example, while the cbx7 loop is formed by residues that are C-terminal to the cbx7 cbox beta sheet, it is residues that are N-terminal to the RYBP beta sheet that form the RYBP loop. Because of this, it is not surprising that the RYBP backbone conformation is completely different from that of the cbx7 cbox loop (Figures 4B and 4C). A notable way that RYBP can adopt a conformation differently than the Pc cbox is demonstrated by RYBP residues 173–175. In the ClustalW alignment (Figure 1A), RYBP residues 173–175 (FKE) align with cbx7 residue 234–236 (FRE). Cbx7 Phe 234 is part of the intermolecular beta sheet and participates in a key hydrophobic stacking interaction with C-RING1B Tyr 262, while cbx7 Glu 236 forms a key salt bridge interaction (Figure 1D). RYBP Phe 173 also plays a key role in binding C-RING1B, but rather than participating in a stacking interaction with C-RING1B Tyr 262 in the beta sheet region as cbx7 Phe 234 does, RYBP Phe 173 binds to the loop binding surface on C-RING1B while Glu 175 does not contact C-RING1B at all (Figure 4B). These results demonstrate the conformational diversity that is present among C-RING1B binding partners that are capable of using a single, shared binding surface on C-RING1B. The consequence of the shared binding site would result in C-RING1B having a single binding partner at one time.
Figure 4. C-RING1B/RYBP Structure (A and B) Close up of the.
(A) beta sheet interaction and (B) the loop interaction. RING1B and RYBP are drawn in blue and magenta, respectively.
(C) Overlay of the C-RING1B/RYBP 145–179 on the C-RING1B/cbx7 cbox. The C-RING1B is shown as a surface representation, cbx7 cbox is colored yellow and RYBP is in magenta. Key hydrophobic residues that share the same C-RING1B loop binding surface are highlighted on both cbx7 and RYBP. RYBP residues are labeled in magenta and cbx7 residues are labeled in black.
Mutation of the Shared Binding Site of C-RING1B Disrupts Chromosome Localization
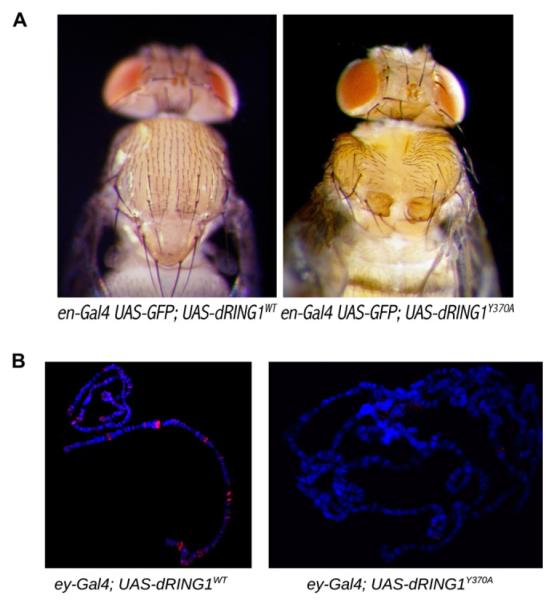
We next determined the consequences of disrupting the shared binding site of RING1B in vivo. To do so, we performed experiments using transgenic Drosophila and introduced a mutation to the Drosophila RING1B ortholog, dRING1. Tyr 370 of dRING1 is equivalent to Tyr 262 of human RING1B; as discussed above, Tyr 262 is a key residue that is required for stable complex formation with the Pc cbox domain and RYBP (Figures 1D, 2A, 2B, and 4A). For this experiment, we expressed both wild-type and a Tyr370Ala mutant form of FLAG epitope-tagged dRING1 in the posterior compartment of the wing disc (Figure S5). Whereas overexpression of wild-type dRING1 failed to affect development, expression of dRING1Y370A was pupal lethal. A few escapers were observed, all of which exhibited defects in thorax closure (Figure 5A). Interestingly, a similar thorax closure phenotype was previously observed in Drosophila Pc/pnrGal4 transheterozygous mutants (Pena-Rangel et al., 2002). Given that we designed the mutant with the intent of disrupting the binding function of dRING1, it was perhaps not surprising that the Tyr370Ala mutant functioned as a dominant negative. When the dRING1 transgenes were expressed in the salivary glands (Figure 5B), the exogenous wild-type dRING1 showed a banding pattern similar to that previously observed for endogenous dRING1 (Gorfinkiel et al., 2004). The ability to associate with chromosomes was found to be severely compromised for the Try370Ala dRING1 mutant, reflecting the key targeting role played by C-dRING1.
Figure 5. Transgenic Drosophila melanogaster.
(A) Expression of the mutant from the en-Gal4 driver results in a dominant negative mutant phenotype. The images are of the dorsal side of the flies showing a deficiency in thorax closure.
(B) Polytene chromosome from ey-Gal4; dRING1 transgenes were stained with Rb-anti antibodies against the FLAG epitope to monitor transgene expression (red) while DNA was counterstained with Hoescht dye (blue).
DISCUSSION
We have shown that two C-RING1B binding partners that differ in amino acid sequence and attain different conformations bind to the same site on the surface of C-RING1B. Our finding has important implications for PRC1 assembly and targeting. If a PRC1 complex contains a single RING1B protein and a single Pc protein, the likely target would be methylated chromatin. RYBP binds C-RING1B with affinity (Kd = 90 nM) (Figure 3C; Figure S3A) that is comparable to the binding affinity between C-RING1B and the Pc cbox domains (Kd = 9.2–180 nM (Wang et al., 2008). Because both RYBP and Pc cbox bind to the same surface on C-RING1B, C-RING1B would be able to bind only one of these proteins at a time. Therefore, rather than being recruited to methylated chromatin when RING1B binds Pc, the genomic location to which RING1B is targeted would depend on the binding partner of RYBP that houses a DNA binding domain.
It is worth noting that all PRC1-like complexes isolated from vertebrate cells contain both RING1B and its close homolog RING1A (Elderkin et al., 2007; Levine et al., 2002; Ogawa et al., 2002; Sanchez et al., 2007; Wang et al., 2004a). How both RING1A and RING1B can be present within a single PRC1 is not clear, though several protein-protein interaction domains exist within PRC1 that may facilitate higher order stoichiometries. Having both RING1A and RING1B in the same complex could affect PRC1 targeting. If the C-terminal domain of RING1A (C-RING1A) and C-RING1B bind different proteins, e.g., RYBP and Pc, then the PRC1 complex could use a combinatorial approach utilizing both a specific DNA sequence through RYBP and also methylated chromatin that is recognized by the Pc chromo domain, to determine the specific location for PRC1 to bind to. Alternatively, two different genomic sites targeted by the RING1A and RING1B binding partners could be used to gather genomic elements that are separated by large intervening sequences creating higher order chromatin structures that are part of the coordinated repression mediated by PRC1.
In addition to the Pc cbox domain and RYBP/YAF2, C-RING1B has also been shown to bind methylated DNA binding protein 1 (MBD1) (Sakamoto et al., 2007). Furthermore, a proteomic analysis of RING1B-associated proteins has identified many other binding partners (Sanchez et al., 2007). While the Psc orthologs (e.g., Bmi-1 and Mel-18) can form the heterodimeric RING finger ubiquitin ligase with the N-terminal RING finger domain of RING1B, it remains to be seen if any of the other proteins identified in the proteomics study directly bind C-RING1B. Our data indicate that regions as short as 30 residues can bind C-RING1B with strong affinity. Even among the cbox domains of the different Pc orthologs, there is sufficient sequence diversity to predict that there will be alternative structures of the cbox domains when they are bound to C-RING1B. As seen in the sequence alignment of the Pc cbox domains (Figure 1A), the cbox domain of cbx4 lacks residues that are equivalent to cbx7 Phe 243 and Phe 244. These two Phe residues in cbx7 are required for stable complex formation (Figures 1E and 2B). They are also conserved in the Drosophila Pc protein and are required for repression of transcription (Figure 2D). Despite the absence of these critical residues, cbx4 cbox is still capable of forming a stable complex with C-RING1B (Wang et al., 2008). We have determined the structure of the cbox domain of cbx4 (also called Pc2) bound to C-RING1B and found that cbx4 cbox utilizes the same binding surface on C-RING1B as cbx7 cbox and RYBP (our unpublished data). While the C-RING1B/cbx4 cbox structure further confirms that sequence variations can be accommodated in an association with the same binding surface on C-RING1B, it also provides insight into a possible reason why cbx4 evolved a different mode of interaction with C-RING1B because cbx4 cbox may be unique in its ability to perform an oligomerization function. We are currently investigating this possibility. The ability of C-RING1B to bind to different proteins that have different functions, even among the different Pc orthologs (Bernstein et al., 2006), may allow RING1B to not only bind different sites but also to act as repressors in a variety of pathways in different cell types. These and other future studies with other C-RING1B binding partners will be needed to help clarify the role of C-RING1B and provide new insights into PcG assembly and function.
EXPERIMENTAL PROCEDURES
Protein Preparations
All proteins used for this study are summarized in Table S1. C-RING1B complexes used for NMR and X-ray crystallography were obtained by coexpressing the C-RING1B construct in pET-30a and the hexahistidine-tagged binding partner in pET-3c while maintaining both plasmids in the presence of kanamycin and ampicillin, respectively. Proteins using the T7 promoter were expressed in BL21-Gold (DE3) (Stratagene) that had been pretransformed with the pRARE plasmid (Novagen). MBP fusion proteins were obtained by cloning the target gene into pBADM-41+ (EMBL) and were expressed in ARI814 (Schatz et al., 1996) cells. One millimolar IPTG or 0.2% arabinose was used to induce protein expression for the pET (T7 promoter) and pBADM-41+ (araBAD promoter) clones, respectively. Typical protein purification involved resuspending bacterial cells from a 1 liter culture in 10 ml of 50 mM Tris (pH 8.0), 100 mM NaCl, 25 mM imidazole (pH 7.5), 1 mM PMSF, and 5% glycerol, cell lysis by sonication, and extraction of the protein using Ni affinity chromatography, followed by ion exchange chromatography. The protein complexes used for crystallization were first purified on a Ni affinity column, digested with TEV to remove the tag sequence on the cbx7 219–248 and RYBP 145–179 constructs, followed by a second Ni affinity chromatography where the nonbinding fractions were collected. The complexes were further purified by ion exchange chromatography.
NMR Spectroscopy
All samples were labeled with 15N or with both 15N and 13C using minimal media containing isotopically labeled 15N NH4Cl and 13C glucose. The purified complexes were all prepared in 10 mM Na PO4 buffer (pH 6.0), 50 mM NaCl, and 5% D2O. Spectra were recorded on a Bruker Av700 spectrometer equipped with an actively shielded z-gradient triple resonance probe at a temperature of 310 K. Spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRView (Johnson and Blevins, 1994). Backbone amide relaxation data sets were recorded using standard Bruker pulse programs.
X-Ray Crystallography
C-RING1B/cbx7 219–248
The C-RING1B construct used for crystallization encompassed human RING1B residues 223–333. Additionally, we mutated C-RING1B residue Asn 306 to Asp to prevent deamidation of the Asn residue. Asn 306 is not important for cbox binding as mutating this residue to an Asp does not hinder cbox binding (Figure S1B). The C-RING1B 223–333 N306D/cbx7 219–248 copurified complex was prepared to a concentration of 26 mg/ml and screened for crystals using hanging drop vapor diffusion. See Supplemental Material for all crystallization conditions. Data sets from the first native, iodide, and Se-Met derivative crystals were collected on a Rigaku MicroMax 007HF rotating anode X-ray generator equipped with R-AXIS HTC imaging plate system (structure II), processed using HKL2000 (Otwinowski et al., 2003), and input into SHARP (de La Fortelle and Bricogne 1997). SHELX-D (Sheldrick, 2008) identified the heavy atom positions from which phases were calculated. Maps calculated following density modification (Collaborative Computational Project, Number 4, 1994) allowed automatic model building of the initial chain using ARP/wARP (Langer et al., 2008) and manual rebuilding was completed using Coot (Emsley and Cowtan, 2004). The structure was refined using PHENIX (Adams et al., 2002) against a second native data set collected to 1.7 Å. A portion of the electron density map is shown in Figure S1C. Crystallographic statistics are shown in Table S1.
C-RING1B/RYBP 145–179
Crystals of both the native and Se-Met C-RING1B Asn306Asp/RYBP complex were grown by hanging drop vapor diffusion set up at 16°C using a protein concentration of 20 mg/ml which was mixed with equal volume of well buffer. Crystals were flash-cooled in liquid nitrogen prior to data collection. All data were collected at the Advanced Light Source (ALS) Beamline 4.4.2 and processed using d*TREK (Pflugrath, 1999). Fifteen selenium atom positions were determined using SHELX (Sheldrick, 2008), and phases were calculated and refined using autoSHARP (Vonrhein et al., 2007). Density modification, model building, and refinement were carried out as described above.
Native Gel Electrophoresis Binding Assay
All binding reactions were carried out in 15 μl using equimolar concentrations of C-RING1B and its binding partner. The protein concentration used for all the reactions was 0.14 nmol. The binding reactions were equilibrated at room temperature for at least 30 min in 10 mM Tris-HCl (pH 7.8), 50 mM NaCl, and 5 mM β-mercaptoethanol prior to loading the samples onto a 10% native polyacrylamide gel.
Transcription Assay
All exogenous Pc proteins were constitutively expressed using the actin 5c promoter (plasmid was a kind gift from Dr. Albert J. Courey). The vector containing the lacZ gene, also under control of the actin 5c promoter, was a kind gift from Dr. Yuzuru Shiio. On day 1 of the assay, the plasmid bearing the Pc gene, pGL2-Basic, lacZ plasmids (100, 7.5, and 7.5 ng, respectively) were transiently transfected into 1 × 105 Drosophila S2 cells in 100 μl of media using Fugene HD transfection reagent (Roche Applied Science). On day 3, the expression of the luciferase gene was induced by adding CuSO4 (100 μM). Cells were harvested on day 4. LacZ activity was first measured using various amounts of lysate in order to calculate volumes required to provide equivalent amounts of lacZ activity for all of the individual transfections. Lysate volumes calculated from this initial experiment were used in a second experiment measuring both luciferase and lacZ activities. The Dual-Light Combine Reporter Gene Assay System (Applied Biosystems) was used to measure both enzyme activities using a microplate luminometer (Veritas). The data are presented as the ratio of luciferase to β-galactosidase activity.
Analytical Ultracentrifugation
Protein samples were prepared in 10 mM Na PO4 buffer (pH 6.0) and 50 mM NaCl. All sedimentation experiments were performed with a Beckman Optima XL-I at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies (CAUMA) at the University of Texas Health Science Center at San Antonio. The sedimentation experiments were performed at 55,000 rpm. The sedimentation velocity data were analyzed by UltraScan version 9.9 (Demeler, 2005). Additional methods are described in the Supplemental Experimental Procedures.
Transgenic Drosophila melanogaster
FLAG epitope-tagged dRING1 wild-type and Tyr370Ala genes under control of the Gal4 dependent UAS promoter were cloned into a vector required for PhiC31 integration (Bischof et al., 2007). The DNA was injected into early syncytial-stage blastoderm embryos (Rainbow Transgenics) that carries both a source of the PhiC31 integrase on the X chromosome and attP target site at chromosomal position 86Fa (yw; M{eGFP.vas-int.Dm}2A;M {RFP.attP}86Fa). Successful transformants were selected on the basis of red eye color associated with stable integration of the white gene into white mutant background. Multiple independent transformant lines for each construct were derived. To promote the expression of Gal4 in the posterior half of the developing wing imaginal disc and the salivary glands, the transgenic flies were mated with the engrailed Gal4 (en-Gal4) and the eyeless (ey-Gal4) lines, respectively. Detailed descriptions of Gal4 driver lines can be found at http://flybase.org.
Polytene Chromosome Staining
Polytene chromosome squashes were prepared as described (Schwartz et al., 2004). Recombinant proteins were labeled with a rabbit anti-DDDDK sera (Abcam) and visualized with a goat anti-rabbit Alexa 568 secondary antibody (Molecular Probes), whereas DNA was counterstained with Hoechst dye.
Supplementary Material
Figure S1. C-RING1B/cbx7 cbox conformational dynamics and structure
(A) Backbone conformational dynamics of the C-RING1B/cbx7 cbox complex. T1, T2, and 15N-{1H} NOEs for cbx7 211 – 251 and for RING1B residues 222 – 336. The error bars for the T1 and T2 relaxation times are calculated by Monte Carlo sampling methods. Using an 15N and 13C labeled 1 mM complex (see Table S1 for a list of all proteins used in this study), we first assigned backbone atoms (1HN, 15N, 13CO, 13Cβ, 13Cα) and then proceeded to measure the 15N HSQC based longitudinal (T1) and transverse (T2) relaxation times along with the 15N-{1H} heteronuclear Overhauser effects (15N-{1H} NOE) on the 15N labeled sample. The relaxation measurements show a stretch of approximately 30 residues within cbx7 that is highly ordered when in complex with C-RING1B. The long T1 and short T2 measurements along with the high 15N-{1H} NOE values within cbx7 residues 219 – 248 compared to the residues at each termini indicate a well-ordered structure. The regions of disorder within C-RING1B precisely correspond to gaps in the sequence alignment between the RING orthologs (Fig. 1B). The areas of disorder observed in these NMR relaxation measurements are consistent with the structure of the complex (Fig. 1C – E).
(B) C-RING1B Asn 306 mutants bind the cbox domain. A native electrophoresis binding assay is shown. The difference in the two wild-type C-RING1B proteins (lanes 4 and 5) shows the heterogeneity that can stem from Asn deamidation. The lane 4 wild-type differs from the lane 5 wild-type in that lane 4 is a more recent prep and additional cation exchange chromatography was performed. Lane 5 wild-type shows a significant fraction of the protein sample migrating faster in the gel and similar to the rate of the Asn306Asp mutant. This band likely corresponds to a deamidated protein because a single Asn deamindated protein and the Asn306Asp mutant would have the same charge and be nearly identical in molecular weight.
(C) Sample electron density of the C-RING1B/cbx7 219 – 248 crystal structure. Stereoview of the 1.7 Å, 2Fo – Fc map contoured at 1σ. The map is centered at the cbx7 Phe 234 base stacking interaction with C-RING1B Tyr 262.
Figure S2. 1D 1H NMR spectra of wild-type C-RING1B and the structure guided C-RING1B mutants used in the binding studies
The spectra aligned from top to bottom are Val229Ala, Tyr247Ala, His258Ala, Tyr262Ala, and wild-type.
Figure S3. C-RING1B/RYBP interaction
(A - E) Determination of the minimal binding region of RYBP required to bind C-RING1B using surface plasmon resonance (SPR). Data for each variant are shown in the indicated panel (A - E). In each panel, the plot on the left is an overlay of the sensorgrams for a representative concentration series. The black bar marks when the indicated RYBP variant was injected. The plot on the right shows the response at equilibrium as a function of MBP-RYBP protein concentration. The color of each data point in the plot on the right, corresponds to the trace of identical color in the plot on the left. The data were fit to a hyperbolic equation (red lines), and the Kd values indicated are the mean value from experiments performed on two different surfaces, while the standard deviation was calculated by descriptive statistics using Origin 7 software. The SPR data clearly identifies RYBP residues 145 – 179 as the minimum binding region for maintaining the high affinity with C-RING1B (Kd = 90 nM). These residues include both a beta sheet that is similar to the cbx7 cbox domain, and a loop structure that is completely different than the cbx7 cbox loop (Fig. 3E). Thus, in addition to identifying the minimum binding region, the SPR data also shows that both the beta sheet and loop structures are required to maintain a stable complex with C-RING1B.
(F) 5N-1H HSQC of 15N labeled C-RING1B alone (150 μM) (left) and with 1.5 molar equivalents of non-labeled RYBP 145 – 179 added to the same sample (right).
Figure S4. Stereo view of the 1.7 Å, 2Fo – Fc map contoured at 1σ of the RYBP loop interaction There are six C-RING1B/RYBP complexes in the asymmetric unit. All descriptions in the text were of a C-RING1B/RYBP complex with the best electron density
Figure S5. Wing disc images of the transgenic flies
Expression of the dRING1 exogenous gene products in the posterior compartment of the wing disc is accomplished through the engrailed Gal4 driver (en-Gal4) which results in the expression of both GFP and dRING1 genes that are inserted into two different chromosomal locations and are under control of the Gal4 upstream activating sequence (UAS). Blue hoechst dye stains DNA and is used to highlight the entire wing disc.
Table S1. Protein constructs used for in vitro experiments.
ACKNOWLEDGMENTS
We thank Paul F. Fitzpatrick and Susan T. Weintraub for their comments on the manuscript; we also thank Weintraub and her staff at the UTHSCSA Institutional Mass Spectrometry Laboratory for mass spectrometry analysis of the Se-Met proteins and Jay Nix for the data collection at the Advanced Light Source (ALS). This work was supported by the American Heart Association (0830111N), the American Cancer Society (RSG-08-285-01-GMC) and the Department of Defense Breast Cancer Research Program (BC075278) (CAK). Support for the X-ray Crystallography Laboratory, NMR facility, the Center for Macromolecular Interactions (CAUMA and SPR) and the Mass Spectrometry Laboratory was provided by UTHSCSA and NIH-NCI P30 CA54174 (CTRC at UTHSCSA). The development of the UltraScan software is supported by the National Institutes of Health (RR022200 to BD), supercomputer allocations were provided through National Science Foundation (TG-MCB070038 to BD). Structure figures were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (NIH P41 RR-01081).
Footnotes
ACCESSION NUMBERS
The resonance assignments have been deposited to the Biological Magnetic Resonance Bank (accession number 16229). Coordinates have been deposited in the RCSB PDB with PDB IDs 3GS2 (C-RING1B/cbx7 219–248) and 3ISX (C-RING1B/RYBP 145–179).
Supplemental Information includes Experimental Procedures, five figures, and one table and can be found with this article online at doi:10.1016/j.str.2010.04.013.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bardos JI, Saurin AJ, Tissot C, Duprez E, Freemont PS. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J. Biol. Chem. 2000;275:28785–28792. doi: 10.1074/jbc.M001835200. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezsonova I, Walker JR, Bacik JP, Duan S, Dhe-Paganon S, Arrowsmith CH. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry. 2009;48:10542–10548. doi: 10.1021/bi901131u. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brookes E, Demeler B. Paper presented at GECCO Proceedings ACM 978-1-59593-69-4/07/0007.2007. [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker CA, Kingston RE. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Czypionka A, de los Panos OR, Mateu MG, Barrera FN, Hurtado-Gomez E, Gomez J, Vidal M, Neira JL. The isolated C-terminal domain of Ring1B is a dimer made of stable, well-structured monomers. Biochemistry. 2007;46:12764–12776. doi: 10.1021/bi701343q. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwave-length anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Demeler B. UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. Royal Society of Chemistry; London: 2005. pp. 210–229. [Google Scholar]
- Demeler B, Saber H. Determination of molecular parameters by fitting sedimentation data to finite-element solutions of the Lamm equation. Biophys. J. 1998;74:444–454. doi: 10.1016/S0006-3495(98)77802-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B, Brookes E, Wang R, Schirf V, Kim CA. Characterization of reversible associations by sedimentation velocity with ultrascan. Macromolec. Biosci. 2010;10:755–782. doi: 10.1002/mabi.200900481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderkin S, Maertens GN, Endoh M, Mallery DL, Morrice N, Koseki H, Peters G, Brockdorff N, Hiom K. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell. 2007;28:107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol. Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Fanti L, Melgar T, Garcia E, Pimpinelli S, Guerrero I, Vidal M. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 2004;121:449–462. doi: 10.1016/j.mod.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Muller J, Muller CW. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a BMI-1-ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Mol. Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Muller J. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira JL, Roman-Trufero M, Contreras LM, Prieto J, Singh G, Barrera FN, Renart ML, Vidal M. The transcriptional repressor RYBP is a natively unfolded protein which folds upon binding to DNA. Biochemistry. 2009;48:1348–1360. doi: 10.1021/bi801933c. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- Pena-Rangel MT, Rodriguez I, Riesgo-Escovar JR. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics. 2002;160:1035–1050. doi: 10.1093/genetics/160.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura JM, Paro R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Watanabe S, Ichimura T, Kawasuji M, Koseki H, Baba H, Nakao M. Overlapping roles of the methylated DNA-binding protein MBD1 and polycomb group proteins in transcriptional repression of HOXA genes and heterochromatin foci formation. J. Biol. Chem. 2007;282:16391–16400. doi: 10.1074/jbc.M700011200. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell. Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M. RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics. 2008;9:308. doi: 10.1186/1471-2164-9-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa C, Yoshikawa T, Matsuda-Suzuki F, Delehouzee S, Goto M, Watanabe H, Sawada J, Kataoka K, Handa H. YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J. Biol. Chem. 2002;277:22484–22490. doi: 10.1074/jbc.M203060200. [DOI] [PubMed] [Google Scholar]
- Schatz PJ, Cull MG, Martin EL, Gates CM. Screening of peptide libraries linked to lac repressor. Methods Enzymol. 1996;267:171–191. doi: 10.1016/s0076-6879(96)67012-8. [DOI] [PubMed] [Google Scholar]
- Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, Marcos-Gutierrez C, Were F, Martinez R, Garcia E, Satijn DP, Otte AP, Vidal M. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Schwartz BE, Werner JK, Lis JT. Indirect immunofluorescent labeling of Drosophila polytene chromosomes: visualizing protein interactions with chromatin in vivo. Methods Enzymol. 2004;376:393–404. doi: 10.1016/S0076-6879(03)76026-1. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Muijrers I, de Wit E, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Wen J, Lees JA. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA. 2001;98:1519–1524. doi: 10.1073/pnas.041597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004a;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 2004b;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wang R, Ilangovan U, Robinson AK, Schirf V, Schwarz PM, Lafer EM, Demeler B, Hinck AP, Kim CA. Structural transitions of the RING1B C-terminal region upon binding the polycomb cbox domain. Biochemistry. 2008;47:8007–8015. doi: 10.1021/bi800857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Schickling O, Peter ME, Lenardo MJ. The death effector domain-associated factor plays distinct regulatory roles in the nucleus and cytoplasm. J. Biol. Chem. 2001;276:31945–31952. doi: 10.1074/jbc.M102799200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. C-RING1B/cbx7 cbox conformational dynamics and structure
(A) Backbone conformational dynamics of the C-RING1B/cbx7 cbox complex. T1, T2, and 15N-{1H} NOEs for cbx7 211 – 251 and for RING1B residues 222 – 336. The error bars for the T1 and T2 relaxation times are calculated by Monte Carlo sampling methods. Using an 15N and 13C labeled 1 mM complex (see Table S1 for a list of all proteins used in this study), we first assigned backbone atoms (1HN, 15N, 13CO, 13Cβ, 13Cα) and then proceeded to measure the 15N HSQC based longitudinal (T1) and transverse (T2) relaxation times along with the 15N-{1H} heteronuclear Overhauser effects (15N-{1H} NOE) on the 15N labeled sample. The relaxation measurements show a stretch of approximately 30 residues within cbx7 that is highly ordered when in complex with C-RING1B. The long T1 and short T2 measurements along with the high 15N-{1H} NOE values within cbx7 residues 219 – 248 compared to the residues at each termini indicate a well-ordered structure. The regions of disorder within C-RING1B precisely correspond to gaps in the sequence alignment between the RING orthologs (Fig. 1B). The areas of disorder observed in these NMR relaxation measurements are consistent with the structure of the complex (Fig. 1C – E).
(B) C-RING1B Asn 306 mutants bind the cbox domain. A native electrophoresis binding assay is shown. The difference in the two wild-type C-RING1B proteins (lanes 4 and 5) shows the heterogeneity that can stem from Asn deamidation. The lane 4 wild-type differs from the lane 5 wild-type in that lane 4 is a more recent prep and additional cation exchange chromatography was performed. Lane 5 wild-type shows a significant fraction of the protein sample migrating faster in the gel and similar to the rate of the Asn306Asp mutant. This band likely corresponds to a deamidated protein because a single Asn deamindated protein and the Asn306Asp mutant would have the same charge and be nearly identical in molecular weight.
(C) Sample electron density of the C-RING1B/cbx7 219 – 248 crystal structure. Stereoview of the 1.7 Å, 2Fo – Fc map contoured at 1σ. The map is centered at the cbx7 Phe 234 base stacking interaction with C-RING1B Tyr 262.
Figure S2. 1D 1H NMR spectra of wild-type C-RING1B and the structure guided C-RING1B mutants used in the binding studies
The spectra aligned from top to bottom are Val229Ala, Tyr247Ala, His258Ala, Tyr262Ala, and wild-type.
Figure S3. C-RING1B/RYBP interaction
(A - E) Determination of the minimal binding region of RYBP required to bind C-RING1B using surface plasmon resonance (SPR). Data for each variant are shown in the indicated panel (A - E). In each panel, the plot on the left is an overlay of the sensorgrams for a representative concentration series. The black bar marks when the indicated RYBP variant was injected. The plot on the right shows the response at equilibrium as a function of MBP-RYBP protein concentration. The color of each data point in the plot on the right, corresponds to the trace of identical color in the plot on the left. The data were fit to a hyperbolic equation (red lines), and the Kd values indicated are the mean value from experiments performed on two different surfaces, while the standard deviation was calculated by descriptive statistics using Origin 7 software. The SPR data clearly identifies RYBP residues 145 – 179 as the minimum binding region for maintaining the high affinity with C-RING1B (Kd = 90 nM). These residues include both a beta sheet that is similar to the cbx7 cbox domain, and a loop structure that is completely different than the cbx7 cbox loop (Fig. 3E). Thus, in addition to identifying the minimum binding region, the SPR data also shows that both the beta sheet and loop structures are required to maintain a stable complex with C-RING1B.
(F) 5N-1H HSQC of 15N labeled C-RING1B alone (150 μM) (left) and with 1.5 molar equivalents of non-labeled RYBP 145 – 179 added to the same sample (right).
Figure S4. Stereo view of the 1.7 Å, 2Fo – Fc map contoured at 1σ of the RYBP loop interaction There are six C-RING1B/RYBP complexes in the asymmetric unit. All descriptions in the text were of a C-RING1B/RYBP complex with the best electron density
Figure S5. Wing disc images of the transgenic flies
Expression of the dRING1 exogenous gene products in the posterior compartment of the wing disc is accomplished through the engrailed Gal4 driver (en-Gal4) which results in the expression of both GFP and dRING1 genes that are inserted into two different chromosomal locations and are under control of the Gal4 upstream activating sequence (UAS). Blue hoechst dye stains DNA and is used to highlight the entire wing disc.
Table S1. Protein constructs used for in vitro experiments.