Abstract
The availability of complete genome sequences of diverse bacteria and archaea makes comparative sequence analysis a powerful tool for analyzing signal transduction systems encoded in these genomes. However, most signal transduction proteins consist of two or more individual protein domains, which significantly complicates their functional annotation and makes automated annotation of these proteins in the course of large-scale genome sequencing projects particularly unreliable. We describe here certain common-sense protocols for sequence analysis of two-component histidine kinases and response regulators, as well as other components of the prokaryotic signal transduction machinery: Ser/Thr/Tyr protein kinases and protein phosphatases, adenylate and diguanylate cyclases and c-di-GMP phosphodiesterases. These protocols rely on publicly available computational tools and databases and can be utilized by anyone with an Internet access.
Introduction
Sequence analysis of regulatory proteins played a key role in the discovery of the two-component signal transduction. Indeed, it were the sequence alignments of the chemotaxis response regulator CheY and transcriptional regulators OmpR and ArcA from Escherichia coli with Bacillus subtilis sporulation proteins Spo0F and Spo0A by James Hoch and colleagues (Trach et al., 1985) and with the N-terminal fragment of the chemotaxis methylesterase CheB by Ann and Jeffry Stock and Daniel Koshland (Stock et al., 1985) that convinced them that all these protein fragments were homologous. This homology, in turn, suggested “an evolutionary and functional relationship between the chemotaxis system and systems that are thought to regulate gene expression in response to changing environmental conditions” (Stock et al., 1985). This prescient conclusion has been verified in subsequent studies that described phosphorylation of these proteins and identified their common CheY-like receiver (REC) domain as an evolutionarily stable compact structural unit (Stock et al., 1989; 1993; Volz and Matsumura, 1991) that undergoes a distinctive change upon phosphorylation (Kern et al., 1999; Lee et al., 2001).
The identification of the receiver domain was followed by sequence analysis of histidine kinases, most importantly by Parkinson and Kofoid (1992), who described five conserved sequence motifs (H, N, G1, F and G2 boxes), and by Grebe and Stock (1999), who classified histidine kinases into 11 families based on sequence similarity in their kinase domains. These papers provided a solid basis for recognition of histidine kinases in genomic sequences and analysis of the diversity in their domain organization (Dutta et al., 1999).
The importance of sequence analysis in studies of bacterial and archaeal signal transduction systems has received an additional boost from the genome sequencing projects, which provided virtually unlimited material for comparative studies. However, these studies revealed a stunning complexity and diversity of signal transduction systems in various microorganisms. The total number of sensory histidine kinases encoded in the genomes of E. coli K12 and B. subtilis, 30 and 36, respectively, proved to be quite modest compared to the sets of histidine kinases encoded by such environmental organisms as Pseudomonas aeruginosa (62 proteins), Streptomyces coelicolor (95 proteins) or Myxococcus xanthus (138 proteins), see http://www.ncbi.nlm.nih.gov/Complete_Genomes/SignalCensus.html (Galperin, 2005). Furthermore, the list of microbial environmental receptors has been expanded and now, in addition to histidine kinases and methyl-accepting chemotaxis proteins, includes Ser/Thr protein kinases and protein phosphatases, as well as adenylate and diguanylate cyclases and c-di-GMP phosphodiesterases (Galperin, 2004, 2005; Kennelly, 2002; Kennelly and Potts, 1996; Römling et al., 2005). All these environmental receptors share a pool of sensory domains, which can be extracytoplasmic (periplasmic or - in gram-positive bacteria - extracellular), membrane-embedded or cytoplasmic (Galperin et al., 2001; Nikolskaya et al., 2003; Zhulin et al., 2003), see reviews by Taylor and Zhulin (1999) and Galperin (2004). Another important development was characterization of a complex system of “one-component” intracellular signaling proteins (Galperin, 2004; Ulrich et al., 2005), such as the anaerobic nitric oxide reductase transcription regulator NorR, which combines a sensor GAF domain with an enhancer binding ATPase and a DNA binding domain (Gardner et al., 2003; Pohlmann et al., 2000). To complicate the picture even further, certain receptors contain more than one sensory domain and/or more than one output domain and participate in the cross-talk between different signal transduction pathways (Galperin, 2004). However, this very complexity makes case-by-case sequence analysis of signal transduction proteins so effective. In the following paragraphs we discuss the computational tools and databases that are most commonly used in sequence analysis of sensory and signal transduction proteins and described analytical methods used for recognizing histidine kinases, response regulators and other bacterial signaling components in genomic sequences and for delineating their constituent domains.
Computational tools for domain identification
Identification of the CheY-like receiver (REC) domain (Stock et al., 1985; Trach et al., 1985) as a common phosphoacceptor domain in various two-component systems demonstrated the power of comparative sequence analysis in studies of the prokaryotic signal transduction systems. In subsequent studies, many other conserved protein domains that are involved in signal transduction were identified and included in public domain databases, such as Pfam, SMART, InterPro and CDD (Table 1). Each of these databases comes with a search tool that allows comparing any given protein sequence against the domain library to identify the (known) domains that this protein consists of. In addition, these databases contain pre-computed profiles for previously sequenced proteins and provide graphical views of their domain architectures (Fig. 1). As new genomic data are added, deduced proteins are automatically analyzed for domain content. Therefore, unless the protein to be analyzed is a newly sequenced one that is still absent from the NCBI protein database and/or UniProt, its domain architecture should be available in protein domain databases. Importantly, position-specific scoring matrices (PSSMs) and hidden Markov models (HMMs) that are used for sequence searches in domain databases already reflect sequence divergence within each protein family. This makes comparing a protein sequence against a domain database (a library of PSSMs or HMMs) much more sensitive than any pairwise comparisons, used, e.g. in the BLAST algorithm. However, domain recognition and functional annotation of multi-domain proteins is a tedious process that cannot be readily automated (see below). Furthermore, the standard methodology of assigning protein function based on the function of its closest experimentally characterized homolog is not readily applicable to signal transduction components, as proteins with very similar sequences (e.g. B. subtilis response regulators PhoP and ResD) may have dramatically different biological functions. As a result, many signal transduction proteins have incomplete, biased, or even erroneous annotation. Given that most protein annotations these days are made in an automated high-throughput fashion, it would be unrealistic to put too much trust into these annotations, especially when planning long-term experimental research. For many experimentally uncharacterized proteins, an imprecise annotation, such as “response regulator, OmpR type”) in the PIRSF protein family classification system (Nikolskaya et al., 2006) or “COG0745: Response regulators consisting of a CheY-like receiver domain and a winged-helix DNA-binding domain” in the COG database (Tatusov et al., 2000) would actually be far more accurate than a more precise, but likely erroneous, functional assignment. Protein annotations in specialized databases, dedicated to signal transduction, such as Sentra and MiST, are much more reliable but even these might need to be verified.
Table 1.
Computational resources for sequence analysis of signal transduction proteins
| Name | URL | Comment | Ref. |
|---|---|---|---|
| Specialized databases of signal transduction proteins | |||
| KEGG | http://www.genome.jp/kegg-bin/show_pathway?ko02020 | Graphical representation of two-component systems in bacteria with sequenced genomes | Kanehisa et al., 2004 |
| MiST | http://mistdb.com | Predicted signal transduction proteins from all completely sequenced prokaryotic genomes | Ulrich and Zhulin, 2007 |
| Signaling Census | http://www.ncbi.nlm.nih.gov/Complete_Genomes/SignalCensus.html | Total counts of signal transduction proteins in completely sequenced prokaryotic genomes | Galperin, 2005 |
| KinG | http://megha.garudaindia.in/king/index.jsp | Kinases in Genomes, a listing of Ser/Thr/Tyr-specific protein kinases encoded in complete genomes of prokaryotes and eukaryotes | Krupa et al., 2004 |
| Sequence motif databases | |||
| PROSITE | http://www.expasy.org/prosite/ | Protein sequence patterns and profiles that define protein domains | Hulo et al., 2006 |
| BLOCKS | http://blocks.fhcrc.org/ | Protein sequence motifs represented as Blocks, multiply aligned ungapped sequence segments | Henikoff et al., 2000 |
| PRINTS | http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS/ | Protein fingerprints groups of conserved motifs used to characterize each protein family | Attwood et al., 2003 |
| Protein domain databases | |||
| Pfam | http://pfam.xfam.org/ | An extensive collection of protein domains, including those with unknown functions | Finn et al., 2006 |
| SMART | http://smart.embl.de/ | Prokaryotic and eukaryotic signaling domains and domain architectures | Letunic et al., 2006 |
| ProDom | http://prodom.prabi.fr/ | An automatically generated listing of protein domains | Servant et al., 2002 |
| CDD | http://www.ncbi.nlm.nih.gov/cdd | Conserved domains with curated alignments that are based on available 3D structures | Marchler-Bauer et al., 2005 |
| Protein family databases (full-length proteins) | |||
| COG | http://www.ncbi.nlm.nih.gov/COG/ | Clusters of orthologous groups that represent either whole proteins or individual domains | Tatusov et al., 2000 |
| PIRSF | http://pir.georgetown.edu/pirwww/dbinfo/pirsf.shtml | Families of proteins with shared domain architecture and full-length similarity | Wu et al., 2004 |
| Integrated motif, domain and family database | |||
| InterPro | http://www.ebi.ac.uk/interpro/ | An umbrella database combining results from several of the above databases | Mulder et al., 2005 |
| Protein structure database | |||
| PDB | http://www.rcsb.org/pdb | 3D structures of proteins and other molecules | Berman et al., 2000 |
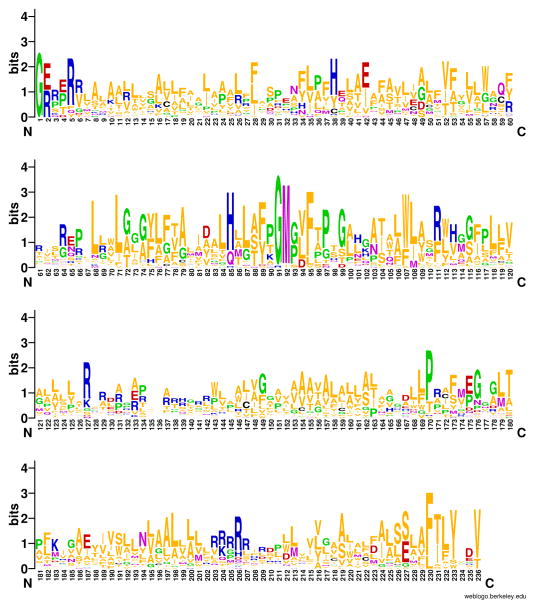
Figure 1.
Sequence logo of the MASE3 domain, generated using the WebLogo (http://weblogo.berkeley.edu) tool from a multiple alignment of 35 different sequences of the MASE3 domain aligned to the Methanosarcina acetivorans histidine kinase MA_3481. Residue numbering starts from Gly-4 of the MA_3481 sequence.
As a starting point in sequence analysis of a putative signal transduction protein, it is often useful to compare it against several (or even all) domain databases that are listed in Table 1. Each of these databases uses its own search tool, so the results are likely to be non-uniform, both in terms of domain recognition and in terms of domain boundaries for the same domain. A careful analysis of all meaningful annotations from these different sources, taking into account the similarity scores, the underlying experimental evidence and the available references, is the best way to avoid costly mistakes. We provide several examples of such an analysis further in this chapter.
Sequence analysis of histidine kinases
Overview
A typical sensory histidine kinase consists of at least three distinct domains: a sensor (signal input) domain, a His-containing phosphoacceptor (dimerization) HisKA domain and an ATP-binding HATPase domain (Dutta et al., 1999; Grebe and Stock, 1999; Hoch, 2000; Stock et al., 2000). There are numerous variations on this common theme. Sensor domains can be periplasmic, membrane-embedded or cytoplasmic, and a single histidine kinase can contain two or more sensory domains. Extracytoplasmic sensor domains are connected to the intracellular HisKA domains by one or more transmembrane segments and, sometimes, the cytoplasmic helical linker (HAMP) domain (Aravind and Ponting, 1999; Williams and Stewart, 1999). In addition, certain histidine kinases contain C-terminal CheY-like phosphoacceptor (receiver, REC) domains (these enzymes are commonly referred to as hybrid histidine kinases) and/or histidine phosphotransfer (HPt) domains (Dutta et al., 1999; Matsushika and Mizuno, 1998; Mizuno, 1997).
The diversity of histidine kinases makes recognizing them in genomic sequences a formidable task. In 1992, Parkinson and Kofoid described conserved sequence motifs (H, N, G1, F and G2 boxes), common for most histidine kinases (Parkinson and Kofoid, 1992). These motifs were subsequently used in numerous papers, most significantly in the histidine kinase classification by Grebe and Stock (1999). However, proteins that lack one or more such motifs can still function as histidine kinases. Examples include proteins that belong to the HPK8 and HPK10 in classification by Grebe and Stock (COG2972 and COG3275 in the COG database), such as Pseudomonas aeruginosa sensor protein FimS (Yu et al., 1997) and many others. While these motifs can be captured by such databases as Blocks, PRINTS, or PROSITE (Table 1), in the past several years the motif-based approach to identification of histidine kinases has been largely replaced by an approach based on domain analysis. Sensory domains are by far the most diverse ones of the three core domains in histidine kinases; many of them are unique or have a narrow phylogenetic representation. Phosphoacceptor (dimerization) HisKA domains, which contain the H box with the phosphoryl-accepting His residue, are less diverse and have similar three-dimensional structures, consisting of long alpha-helices (Stock et al., 2000; Wolanin et al., 2002). Still, recognizing these domains through sequence similarity alone may be complicated. In the latest version of the Pfam database, HisKA domains are divided into four separate domain families, HisKA (PF00512), HisKA_2 (PF07568), HisKA_3 (PF07730), and HWE_HK (PF07536), which are unified into the His Kinase A (phosphoacceptor) domain clan (Finn et al., 2006). It should be noted that the four HisKA domains currently in Pfam do not cover the full diversity of these domains: some experimentally characterized sensory histidine kinases, such as Clostridium perfringens VirS (Cheung and Rood, 2000), as well as many archaeal histidine kinases, contain dimerization domains that are not recognized by either of the Pfam profiles, at least at standard confidence levels (E<10−3). The ATPase domain of histidine kinases, referred to as HATPase_c domain (PF02518) in the Pfam database, contains the N, G1, F and G2 boxes of Parkinson and Kofoid. It is by far the most conserved domain in histidine kinases and the easiest one to recognize in sequence similarity searches. However, very similar ATPase domains of the GHKL family can be found in a stand-alone form in the DNA gyrase (gyrB gene product) and DNA repair protein MutL, or as a component of the heat-shock protein HSP90 (Ban and Yang, 1998; Dutta and Inouye, 2000). Therefore, recognition of a histidine kinase by sequence analysis relies on finding a (usually C-terminal) ATPase domain of the GKHL superfamily that does not belong to GyrB, MutL, or HtpG family. This domain should be preceded by a histidine kinase A (phosphoacceptor) domain: either one of the HisKA domains listed in Pfam or a poorly conserved domain of ~60 amino acid residues that consists of predicted alpha-helices and contains an invariant His residue. Finally, there should be an N-terminal fragment, corresponding to a sensory domain, which may or may not have close homologs in the existing protein databases. The presence of these three domains would qualify the protein in question as a two-component sensory histidine kinase. Determining its exact substrate (ligand) specificity would require further analysis and may be impossible without additional experimental data. A significant fraction of histidine kinases are encoded in conserved operons with their cognate response regulators that can be easily recognized by their highly conserved REC domain. Although not a universal trait, presence of an adjacent gene encoding a response regulator could strengthen the case for the analyzed protein being a histidine kinase. Thus, sequence analysis of potential sensory kinases should always include examination of their gene neighborhoods.
Identification of MA_3481 as a sensory histidine kinase
Find the entry for the Methanosarcina acetivorans protein MA_3481 in GenBank (accession no. AE010299) or directly in one of the protein databases: UniProtKB (accession no. Q8TKC7) or the NCBI protein database (AAM06847)1.
-
Inspect the annotation of MA_3481 and its constituent domains in each of these databases. Note that both the NCBI protein database (a non-curated database) and in UniProt (a curated database) annotate MA_3481 as a “hypothetical protein”, in keeping with its original annotation by the scientists at the Whitehead Institute Center for Genome Research (Cambridge, MA). Nevertheless, both NCBI and UniProt entries include the list of the domains that are recognized in the MA_3481 sequence by various tools, which provide numerous hints that MA_3481 might be a histidine kinase.
In the NCBI entry, these domains come from the CDD databases and are linked to the CDD entries for the PAS domain and the HATPase_c domain. The exact borders of each domain vary depending on the source of the domain entry: in the COG database the PAS domain (COG2202) occupies amino acid residues 243 to 446, whereas the CDD’s own cd00130 entry recognizes a much smaller region, 339 to 439, as the PAS domain. Likewise, COG3920 “Signal transduction histidine kinase” covers amino acid residues 436 to 702, whereas the SMART entry SM00387 recognizes only the C-terminal HATPase_c domain (amino acid residues 557 to 702) with a somewhat unreliable expectation value of 0.004. The complete domain organization can be viewed on the NCBI web site by clicking the “Conserved Domains” link or by entering the link http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=19917534 where the last 8 digits correspond to the gi number. When the protein in question is not in the database, one would need to compare its sequence against the CDD using RPS-BLAST (see below).
The UniProt entry for MA_3481 contains even more hints that MA_3481 is a histidine kinase. For example, this entry contains links to the Gene Ontology (GO) “Molecular function: two-component sensor activity (GO: 0000155)” and “Biological process: two-component signal transduction system (GO: 0000160)”. Still, these annotations should be treated with caution. For example, one of them says “Biological process: regulation of transcription, DNA-dependent (GO:0006355), which is probably not true for MA_3481, as DNA-binding response regulators are very rare in archaea (Galperin, 2006) and, like most archaeal histidine kinases, MA_3481 does not appear to regulate transcription. The UniProt entry for MA_3481 also contains links to the PROSITE database and several domain databases, such as InterPro, Pfam, SMART and TIGRFAMs. A very useful link to the “Graphical view of the domain structure” in InterPro (http://www.ebi.ac.uk/interpro/protein/Q8TKC7) allows one to take a birds-eye view at all the domain recognized by individual tools used in InterPro. Again, all of them recognize the PAS (or PAS/PAC) domain in the 320–449 region of the protein and the ATPase domain in the 550–706 region of the protein. Two InterPro tools also cover the intermediate 450–550 region: Pfam recognizes it as HisKA_2 (PF07568) domain (corresponding to the InterPro entry IPR011495), whereas PROSITE unifies it with the ATPase domain into the single HIS_KIN entry spanning the entire C-terminal half of MA_3481 from Glu-459 to its C-terminus. Pfam graphical view, which can be obtained by following a link from the UniProt entry or by going directly to http://pfam.xfam.org/protein/Q8TKC7 will show the presence of all three required domains, i.e. the sensory PAS domain, the phosphorylation/dimerization HisKA_2 domain, and the C-terminal HATPase_c domain. In addition, Pfam shows that the N-terminus of MA_3481 consists of 7 transmembrane segments, which might form an additional sensory domain (see below).
Although most sequence analysis tools recognize the C-terminal region of MA_3481 as the HATPase domain, it is necessary to check conservation of the key (active site) residues to make sure that this protein actually can function as an ATPase (or kinase). The easiest way to do that is to use the CDD tool that compares the sequence in question against the consensus sequence for the given domain. In the current version this can be done by clicking the ‘plus’ sign in the CDD output. Although the current CDD entry does not have any information about the active site residues, this alignment allows one to recognize the G1 box and the less obvious N and G2 boxes (the F box is poorly conserved in the MA_3481 sequence). Therefore, it is necessary to verify conservation of the key residues in the HATPase_c domain of MA_3481 by comparing it against the active site residues of a well-characterized ATPase domain, e.g. with the nucleotide binding domain of Thermotoga maritima CheA (TM0702, gi|15643465), whose structure (PDB entry: 1I5A and others) has been solved in a complex with an ATP analog (Bilwes et al., 1999; Bilwes et al., 2001).
The quickest way to compare two closely related sequences is by aligning them using the Blast 2 sequences tool (Tatusova and Madden, 1999), available on the NCBI BLAST web page http://www.ncbi.nlm.nih.gov/BLAST/ in the “Special” category. (Other on-line tools for aligning two sequences, such as EMBOSS, http://www.ebi.ac.uk/emboss/align/, or LALIGN, http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi?rm=lalign, can be used as well). Clicking on “bl2sec” opens two sequence windows; paste the gi number of MA_3481, 19917534, into one of them and the gi number of the Thermotoga maritima CheA, 15643465, into the other, change the program to be run from “blastn” to “blastp”, and press the “Align” key”. Although the resulting alignment shows only a relatively low sequence similarity (21% identity in the 279 amino acid overlap; expect value E = 0.71), comparing it with the article by Bilwes and colleagues (Bilwes et al., 2001) shows that the key nucleotide-binding residues of CheA, Asn-409, His-413, and Gly-506 are all conserved in MA_3481. Hence, MA_3481 appears to have a functional ATPase domain.
According to the Pfam database, MA_3481 contains a dimerization and phosphoacceptor domain HisKA_2 (PF07568). The only thing that needs to be checked here is the presence of the phosphoryl-accepting His residue. A BLAST search using the MA_3481 residues 430–550 as a query reveals a large number of sequences with a conserved (HNQDR)HR motif, characteristic of the recently described HWE family of histidine kinases, where the conserved His residue serves as the phosphorylation site (Karniol and Vierstra, 2004). A similar result can be obtained by comparing MA_3481 with Agrobacterium fabrum protein Atu2165 (AGR_C_3927, gi|15157306), which was experimentally characterized by Karniol and Vierstra (2004). These comparisons show that MA_3481 indeed contains a functional dimerization/phosphoacceptor domain with a phosphoryl-accepting His residue.
Summing up, the above analysis shows that MA_3481 contains a C-terminal HATPase domain, preceded by a HisKA domain and a PAS domain. In addition, it contains an N-terminal 7TM region, which could be another sensor domain (see below). Although the MA_3481 gene neighborhood does not contain a gene for a potential response regulator, MA_3481 satisfies all the key criteria listed above and can be confidently annotated as sensory histidine kinase.
Analysis of sensory domains in histidine kinases
The sensory domains of histidine kinases are extremely diverse, reflecting the diversity of the signals they perceive. However, as far as we can judge, members of the same domain family typically recognize the same (or very close) substrates. Therefore, functional characterization of a sensory domain in one organism often serves as a basis for functional annotation of all proteins that contain the same domain. Still, functions of many periplasmic, membrane-embedded or intracellular sensory domains such domains are still unknown. Furthermore, not every N-terminal domain in every histidine kinases necessarily serves as a sensor. For example, the N-terminal membrane domain of the E. coli UhpB, a histidine kinase that regulates transport and metabolism of glucose-6-phosphate and related sugars, does not appear to work as a sensor. Instead, its function appears to be limited to the interaction with UhpC, another membrane protein that is evolutionarily related to sugar phosphate transport proteins but actually works as a sensor of sugar phosphate in the UhpB-UhbC complex (Island and Kadner, 1993). Therefore, describing new sensory domains requires certain caution. Still, in most cases one can safely assume that the (periplasmic) N-terminal region of a histidine kinase is its sensory domain. This assumption is definitely justified for those domains that are found in combination with more than one type of membrane sensors, for example, histidine kinases and chemotaxis methyl-accepting proteins, adenylate or diguanylate cyclases (Galperin et al., 2001; Nikolskaya et al., 2003; Zhulin et al., 2003).
Analysis of the putative sensory domain of MA_3481
-
1
Extract the sequence of MA_3481 from UniProt (accession no. Q8TKC7) or the NCBI protein database (accession no. AAM06847 or gi|19917534). Note that its N-terminal region is not covered by any known domain in CDD, whereas in Pfam it is represented by seven predicted transmembrane segments.
-
2
Select the first 240 amino acid residues of the MA_3481 sequence, copy them and paste into the PSI-BLAST window on the NCBI web site (http://www.ncbi.nlm.nih.gov/BLAST/) and press the “Run BLAST” and “Format” keys. Upon receiving the results, press “Run PSI-BLAST iteration 2” and then “Format” keys again. Continue this procedure until convergence, i.e. until PSI-BLAST reports “No new sequences were found above the 0.005 threshold!” The search should converge after 6 or 7 iterations, resulting in a list of ~90 database hits.
-
4
Visually inspect the degree of sequence conservation by scrolling down from the highest-scoring to the lowest-scoring proteins. Note that none of them has been experimentally characterized so far. In addition, although most proteins in the hit list are annotated as “sensory transduction histidine kinase”, several of them (including Thermotoga maritima protein TM0972 and Thiobacillus denitrificans protein Tbd_2578) are annotated as “GGDEF domain” or “diguanylate cyclase”, while others are annotated as “Protein phosphatase 2C-like” (Clostridium thermocellum protein ABN51324, gi|125712832 or “Stage II sporulation E” (Alkaliphilus metalliredigenes protein ABR47247, gi|149948719). This diversity of annotations deserves a further investigation to check if the N-terminal region of MA_3481 can be found in signaling proteins other than histidine kinases.
-
5
By pressing the “Taxonomy reports” link on top of the BLAST output, generate the listing of database hits, sorted by their taxonomic representation, and save to your disk in HTML and/or plain text format.
-
6
Using formatting options for the BLAST results, remove the low scoring hits by changing the “Expect value range” parameter to the range from 0 to 1e-10 and press the “Format” key. Save the resulting BLAST output file to your disk in HTML and/or plain text format.
-
7
Manually inspect each of the non-histidine kinase hits in the output by following the link to the source protein and then checking out the “Conserved domains” link. This should confirm that the N-terminal region of MA_3481, used in the PSI-BLAST search indeed can be fused to a variety of signal output domains and, hence, comprises a novel sensory domain. By analogy to the previously described membrane-associated sensory domains of unknown function, MASE1 and MASE2 (Nikolskaya et al., 2003), we can tentatively name it MASE3.
-
8
To create a multiple alignment of the MASE3 domain, return to formatting options and change the “Alignment view” from “Pairwise” to “flat-query-anchored without identities”. In addition, unselect the Graphical Overview, Linkout, and Sequence Retrieval boxes in the “Show” menu and again press “Format”. Save the resulting multiple alignment to your disk in HTML and/or plain text format (the plain text file can also be obtained by selecting the “Alignment in Plain text” option in the “Show” menu). After manual curation (trimming, removal of the unjustified gaps, etc.), such an alignment can be colored using Microsoft Word or a dedicate software program such as GeneDoc (http://www.nrbsc.org/gfx/genedoc/ebinet.htm), BoxShade (http://www.ch.embnet.org/software/BOX_form.html), or Cinema (http://aig.cs.man.ac.uk/research/utopia/utopia.php), and used for publication.
-
9
In certain cases, it might make sense to remove from the alignment the sequences coming from the unfinished genome sequences. This can be done using the “Limit results by entrez query” option and selecting the limit “srcdb_refseq_provisional[prop]” or a similar one.
-
10
The most conserved residues in the domain can be visualized using the WebLogo tool (Crooks et al., 2004). For a first look, change the BLAST results formatting options to the “query-anchored without identities” alignment view and save the resulting alignment as a text file. Remove the unaligned amino acid residues by deleting all lines that do not start with a gi number, fill the empty spaces with dots or dashes, and submit the resulting alignment to http://weblogo.berkeley.edu/. The formatted logo (Fig. 1) shows several well-conserved residues and groups of positively-charged residues that could be used to determine the membrane topology of the domain, using the “positive-inside” rule (von Heijne, 1992). For a publishable sequence logo, use the manually curated alignment from step 8.
-
11
It would be helpful to check whether all identified instances of MASE3 domain indeed consist of 7 transmembrane segments. This could be done using a variety of software tools, listed in Table 2. As always, it is recommended that at least 3 different methods were used, their results compared, and any discrepancies in the outcomes analyzed on case-by-case basis.
-
12
In conclusion, the described procedure identified a new integral membrane sensory domain (MASE3) found in histidine kinases, diguanylate cyclases, c-di-GMP phosphodiesterases, and PP2C-type protein phosphatases from a variety of bacteria (alpha-, beta-, gamma-, and deltaproteobacteria, firmicutes and Thermotoga spp.) and archaea. The signal sensed by this domain is currently unknown and can only be identified through experimental studies of the respective signaling proteins.
Table 2.
Computational resources for prediction of signal peptides and transmembrane segments in proteins
The actual limitation for PSORT is 600,000 characters, for SVMtm - 10 KB; N/A means that the server accept multiple sequences but the exact limit is not specified.
Sequence analysis of response regulators
Overview
All response regulators of the two-component signal transduction system contain the CheY-like phosphoacceptor (receiver, REC) domain (Stock et al., 2000; West and Stock, 2001), either in a stand-alone form (for example, the chemotaxis response regulator CheY or the sporulation regulator Spo0F) or fused to an effector, or output, domain, which is usually located at the C-terminus of the polypeptide chain (Grebe and Stock, 1999; Stock et al., 2000). Two-domain response regulators are typically thought of as transcriptional regulators that combine the REC domain with a DNA-binding output domain. Indeed, recent studies have shown that the great majority of output domains are involved in DNA binding (Galperin, 2006; Ulrich et al., 2005). However, a substantial fraction of response regulators have RNA-binding, enzymatic or ligand-binding (noncatalytic) output domains, or uncharacterized output domains whose function is unknown (Galperin, 2006; Ulrich et al., 2005). Phylogenetic analysis of the receiver and output domains in various response regulators has shown that receiver domains typically co-evolve with the corresponding effector domains, although some of them show signs of relatively recent domain shuffling (Pao and Saier, 1995).
The mechanism of two-component signal transduction includes phosphoryl transfer from the His residue in the HisKA domain of the sensor histidine kinase to an Asp residue in the REC domain of its cognate response regulator (Stock et al., 2000; West and Stock, 2001). Phosphorylation induces conformational changes in the REC domain (Kern et al., 1999), which affects its binding properties, including its association with the output domain (if any). In different response regulators, there appear to be several different mechanisms of signal transmission. These include dimerization of the REC domain (in OmpR/PhoB family and potentially in all DNA-binding response regulators (Toro-Roman et al., 2005a, 2005b)), direct protein-protein interaction with a variety of target proteins (in stand-alone receiver domains, such as CheY or Spo0F), and a relief-of-inhibition mechanism (and potentially also a stimulatory effect on catalysis) in case of enzymatically active output domains (Anand and Stock, 2002).
The most common response regulators are the DNA-binding transcriptional regulators that belong to two largest families: 1) the OmpR/PhoB family regulators that contain winged helix-turn-helix (HTH) output domains (Martinez-Hackert and Stock, 1997a, 1997b) and 2) the NarL/FixJ family regulators that contain a single HTH motif in the middle of a four-helix bundle (Baikalov et al., 1996). Other, less common, DNA-binding response regulators contain DNA-binding output domains of the Fis type (NtrC and PrrA families), AraC type (YesN family), LytTR type (AgrA/LytR family), Spo0A type (Spo0A family), and several others (Galperin, 2006). Within each family of response regulators, the signaling specificity is determined by minute details of the interactions of the REC domains with the cognate histidine kinases and of the HTH domains with the target sites on the DNA. As a result, response regulators within each particular family typically show high levels of sequence similarity, even when they regulate dramatically different biological processes. This circumstance makes it almost impossible to assign function to newly sequenced response regulators based solely on sequence similarity. Therefore, the goals of sequence analysis have to be far more modest: 1) identification of protein in question as a response regulator, based on the presence of the REC domain; 2) identification of the output domain of the given response regulator (if any) and its function (if known); and 3) assignment of this response regulator to a particular family, followed by assignment of a general function, such as DNA binding, RNA binding, small-molecule ligand binding, or an enzymatic activity.
Identification of Spo0A as a response regulator
By definition, almost any protein containing the receiver (REC) domain can be considered a response regulator. Exceptions include hybrid histidine kinases that contain a C-terminal REC domain and other multidomain signal transduction proteins that combine the REC domain with various sensory and/or output domains [see, for example, Fig. 2 in ref. (Galperin, 2006)]. The relatively high sequence conservation of the REC domain makes its identification relatively straightforward. Comparing the protein in question against any of the domain databases, such as CDD, Pfam, InterPro, or ProDom, using their default parameters, is usually sufficient to find out whether this protein contains the REC domain and, if it does, what are the domain boundaries. The output of this comparison will also show if this protein also contains a recognized output domain [see Galperin (2006) for a recently compiled listing]. Consider the following example of the well characterized DNA-binding response regulator Spo0A, which controls initiation of sporulation in Gram-positive bacteria (Stephenson and Lewis, 2005).
Retrieve the sequence of Bacillus anthracis Spo0A (BA_4394) from UniProt (SP0A_BACAN, accession no. P52928) or the NCBI protein database (accession no. AAP28110; gi|30258892). Inspect the annotation of BA_4394 in these databases. Note that this protein is uniformly annotated as “Stage 0 sporulation protein A”.
Inspect the domain architecture of BA_4394 as represented in Pfam. To do that, go to the Pfam search page at http://pfam.xfam.org/search and enter the UniProt name, accession number or the protein sequence in the appropriate windows. Results will show at http://pfam.xfam.org/search/sequence). Also, look at the domain representation in CDD (click the “Conserved Domains” link from the NCBI protein entry or go to http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml and then enter the NCBI accession number, gi number, or sequence. The results will also show up at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=30258892). Both CDD and Pfam recognize in the BA_4394 sequence an N-terminal REC domain with convincing similarity scores. Furthermore, inspection of domain alignment shows conservation of the phosphoacceptor Asp residue, confirming that BA_4394 contains a functional REC domain and, hence, is a genuine response regulator.
Identification of the output domain of Spo0A
In some cases, including the one above, comparing a sequence of a response regulator against protein domain databases shows that 1) the REC domain is the only one recognized in the given sequence and 2) it occupies only a certain part of the protein, leaving 50 or even more amino acid residues not assigned to any domain. Given that some protein domains can be as short as 25 amino acid residues [(e.g. ATP-hook, (Aravind and Landsman, 1998)] and many HTH domains are not much longer (Aravind et al., 2005), these unassigned regions could well belong to as yet unrecognized protein domains. On the other hand, some of such unassigned regions appear to lack any (predicted) secondary structure and therefore should not be considered separate domains. The following example continues sequence analysis of the Bacillus anthracis protein Spo0A (BA_4394).
Note that the C-terminal region of BA_4394 (amino acid residues 130–264) is now covered in CDD by the Pfam domain PF08759.
Copy the 134-aa C-terminal sequence fragment of BA_4394 (residues 131–264) into the PSI-BLAST search page at the NCBI web site, http://www.ncbi.nlm.nih.gov/BLAST/. Run PSI-BLAST until convergence using the default parameters on the web page. The search should converge after 3 or 4 iterations, resulting in a list of ~90 database hits. Visually inspect the degree of sequence conservation by scrolling down from the highest-scoring to the lowest-scoring proteins to confirm that all the hits are genuine homologs.
By pressing the “Taxonomy reports” link on top of the BLAST output, generate and save the listing of database hits, sorted by their taxonomic representation. You will see that, with a single exception, all high-scoring hits (bitscore of > 133, which corresponds to the expectation value E < 2×10−30) belong to the Firmicutes (low G+C Gram-positive bacteria). The only exception is Symbiobacterium thermophilum, which, owing to its relatively high G+C content, has been initially assigned to the phylum Actinobacteria but obviously belongs to the Firmicutes.
In the BLAST output, find hits to the known 3D structures (marked by red squares with the letter “S”). Follow the links to the Protein Data Base (PDB) entry 1FC3 to see a detailed description of this domain (Lewis et al., 2000) and then the link “Structure” to view its six-helix structure. Alternatively, follow the link to the PDB entry 1LQ1 to see this domain bound to the DNA (Zhao et al., 2002). Note that the C-terminal DNA-binding fragment of Spo0A forms a compact and stable 3D structure and thus comprises a separate well-defined protein domain.
Sequence analysis of prokaryotic signal transducers
Overview
Analysis of the rapidly accumulating genome sequences from diverse bacteria and archaea revealed the great variety of sensory proteins. The characteristic architecture of histidine kinases and MCPs, which includes a periplasmic sensory domain, a transmembrane segment with one or more transmembrane helices, and a cytoplasmically located output domain, was predicted for many proteins encoded in the newly sequenced genomes (Galperin, 2004; Galperin et al., 2001). However, while their N-terminal sensory domains were shared with histidine kinases and/or MCPs (Zhulin et al., 2003), their C-terminal output domains could be adenylate cyclases, diguanylate cyclases, c-di-GMP phosphodiesterases of EAL or HD-GYP type, as well as Ser/Thr protein kinases and protein phosphatases (in Ser/Thr protein kinases, the protein kinase domain is typically at the N-terminus and the sensory domains at the C-terminus). The computational predictions were followed by experimental data detailing participation of these new (predicted) receptors in bacterial signal transduction. Thus, activities of cyanobacterial membrane-bound adenylate cyclase and Rhodobacter sphaeroides bacteriophytochrome BphG1 (diguanylate cyclase/phosphodiesterase) were shown to respond to the red and blue light (Ohmori and Okamoto, 2004; Tarutina et al., 2006). Activities of many other bacterial receptor-type proteins appear to be regulated by environmental factors as well; however, the nature of these factors still remains obscure (Galperin, 2004, 2005; Kennelly, 2002; Lory et al., 2004; Römling et al., 2005; Zhang and Shi, 2004). Still, recognition of a potential receptor protein in a given piece of DNA sequence could be an important step towards understanding the function of that protein and/or its neighbors. We briefly describe here the domain-based approaches to the identification of diguanylate cyclases (the GGDEF domain), c-di-GMP-specific phosphodiesterases (EAL and HD-GYP domains), class III adenylate cyclases (the CyaA domain), eukaryotic-type Ser/Thr/Tyr-specific protein kinases (the STYK domain), and PP2C-family Ser/Thr/Tyr-specific protein phosphatases (the PP2C-SIG domain) and mention several caveats of such searches that may lead to false-positive hits.
Identification of diguanylate cyclases (the GGDEF domain)
The diguanylate cyclase activity, synthesis of the bacterial second messenger c-di-GMP from two molecules of GTP, is a property of the so-called GGDEF domain, named after its most conserved sequence motif, Gly-Gly-Asp/Glu-Glu-Phe (Galperin et al., 2001; Jenal and Malone, 2006; Römling et al., 2005). The GGDEF domain is structurally related to the eukaryotic adenylate cyclase domains (Chan et al., 2004) and has a number of well-conserved residues (see the sequence logo of this domain at http://mmbr.asm.org/content/77/1/1/F3.expansion.html), which makes its identification fairly straightforward. The key problem with its sequence analysis is recognition of inactivated and/or truncated GGDEF domains, which has to be done by meticulously checking the active site residues (Christen et al., 2006; Paul et al., 2004) and deciding whether if any given mutation allows correct folding of the protein and is compatible with its activity. Unfortunately, few residues outside the GGDEF loop and the allosteric I-site (Christen et al., 2006) have been mutated so far (see http://mmbr.asm.org/content/77/1/1/F4.expansion.html, and their contribution to activity remains unknown. The listings of the GGDEF-containing proteins encoded in completely sequenced bacterial genomes are available on the SignalCensus web site http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html and in the MiST database.
Identification of type I c-di-GMP-specific phosphodiesterases (the EAL domain)
Just like the GGDEF domain, the type I c-di-GMP-specific phosphodiesterase (the EAL domain) is very well conserved and easily identified through comparison with any of the protein domain databases. In addition to the conserved EAL motif, two acidic residues required for the activity have been identified (see the sequence logo of this domain at http://mmbr.asm.org/content/77/1/1/F3.expansion.html). An alignment of active and inactive EAL domains (http://mmbr.asm.org/content/77/1/1/F4.expansion.html and Schmidt et al., 2005) could provide further clues to which amino acid residues are required for activity.
Identification of type II c-di-GMP-specific phosphodiesterases (the HD-GYP domain)
The HD-GYP domain (Galperin et al., 1999), recently proven to function as a c-di-GMP-specific phosphodiesterase (Ryan et al., 2006), is an extended variant of the widespread HD-type phosphohydrolase domain (Aravind and Koonin, 1998) that contains extra conserved residues at its C-terminus. Because of that, Pfam and SMART databases do not recognize HD-GYP as a separate domain and list its N-terminal 110 amino acid residues as HD catalytic domain. In contrast, PIRSF and COGs list HD-GYP as a separate domain, while InterPro has a separate entry for RpfG-like response regulators that combine REC and HD-GYP domains (Ryan et al., 2006). In sequence similarity searches of HD-GYP domains, generic HD domains often show up with higher similarity scores than genuine HD-GYP domains. Here, the listing of HD-GYP-containing proteins on the SignalCensus web site could be used as a guide.
Identification of adenylate cyclases
Several unrelated (analogous) forms (classes) of the adenylate cyclase (EC 4.6.1.1) have been described but only one of them, usually referred to as class III, is widespread in bacteria and has been shown to function as environmental sensor (Lory et al., 2004; Ohmori and Okamoto, 2004). Class III adenylate cyclase domain (designated the Guanylate_cyc domain in Pfam and PROSITE and A/G cyclase in InterPro) is well conserved and can be easily recognized through protein sequence comparison against any of the domain databases. It should be noted that the eukaryotic form of this domain can use both ATP and GTP as substrates, producing, respectively, cAMP and cGMP, while in most bacteria it appears to be specific for ATP and have little, if any guanylate cyclase activity (Baker and Kelly, 2004; Shenoy and Visweswariah, 2004). Therefore, for bacteria, the name “adenylate cyclase domain” appears to be more appropriate than any other. Many class III adenylate cyclases are cytoplasmic enzymes and only a small fraction of them are membrane-bound and can be considered genuine environmental sensors.
Identification of Ser/Thr/Tyr-specific protein kinases
Curiously, the vast majority of prokaryotic Ser/Thr/Tyr-specific protein kinases belong to the so-called eukaryotic-type protein kinase superfamily (Kennelly and Potts, 1996). This superfamily includes several other kinase families, such as choline kinases, lipopolysaccharide kinases, aminoglycoside 3’-phosphotransferases. This fact makes correct identification of Ser/Thr/Tyr protein kinases fairly complicated, particularly because Ser/Thr protein kinases from different families sometimes show less similarity to each other than to 3-deoxy-D-manno-octulosonic acid (KDO) kinase (the product of the waaP gene, assigned in Pfam to a separate domain PF06293). In addition, there is a long-standing controversy regarding the functions of the proteins of ABC1/AarF/UbiB family, which are required for (Poon et al., 2000). These widespread proteins, which belong to the Pfam family PF03109 and COG0661 and most likely function as protein kinases that regulate ubiquinone biosynthesis pathway, are sometimes misannotated as ABC transporters or even as ABC transporter substrate binding proteins. Identification of an unknown protein as a Ser/Thr/Tyr protein kinase should take into account domain assignments in several domain databases, presence or absence of additional – sensory or signal output – domains, and genomic context, e.g. the operon structure of the adjacent genes. Functional assignments for completely sequenced genomes are available on MiST, Sentra, and SignalCensus web sites, as well as in KinG, a database dedicated specifically to Ser/Thr/Tyr protein kinases (Krupa et al., 2004).
Identification of Ser/Thr/Tyr protein phosphatases
Prokaryotic Ser/Thr/Tyr-specific protein phosphatases of the PP2C family are reasonably well conserved and can be easily recognized through protein sequence comparison against any of the domain databases. However, only a small fraction of these enzymes have an attached sensory domain and can be considered genuine environmental sensors.
Functional annotation of multidomain proteins
The complexity of microbial signal transduction machinery and the paucity of experimentally characterized proteins make annotating signaling proteins even in well-studied organisms an arduous task. For example, of the 30 histidine kinases encoded by E. coli K12, functions of seven (AtoS, RstB, YehU, YpdA, YfhK, YedV, YjoN) are unknown and several others have poorly defined substrates. For (predicted) signal transduction proteins encoded in the newly sequenced genomes this task becomes even more daunting. Although assigning the signal transduction protein to a general class, such as “histidine kinase”, “response regulator”, “methyl-accepting chemotaxis protein” or “diguanylate cyclase”, is usually easy (see above), it is practically impossible to identify the signal that this protein responds to. Indeed, standard methods of protein sequence analysis, where the function of newly sequenced protein is assigned based on the function of its experimentally characterized (close) homolog, are rarely applicable to signal transduction proteins. Thus, the degree of sequence similarity of the response regulators of the NtrC family is determined more by their central ATPase domains than by their N-terminal REC or C-terminal DNA-binding Fis-like domains, which are in fact responsible for the specificity of these transcriptional regulators. As a result, proteins that show the highest similarity scores are very likely to regulate entirely different biological processes. Therefore, functional annotation of newly sequenced signal transduction proteins should rely on the following simple rules:
If at all possible, the protein should be classified as a signaling, signal transduction, or signal output protein.
For signaling proteins, the enzymatic activity of the signal transduction domain (histidine kinase, Ser/Thr/Tyr kinase, protein phosphatase, adenylate cyclase, diguanylate cyclase, c-di-GMP phosphodiesterase) can be used as a basis for protein annotation.
Any protein that contains more than one enzymatic output domain should be annotated as “multidomain signaling protein containing such and such domain”. The only exceptions to this rule are the proteins combining the GGDEF and EAL domains, which, unless one of the domains is known (or predicted) to be inactive, can be annotated as “diguanylate cyclase/phosphodiesterase”.
It is also useful to identify the transmembrane segments, if any, and decide whether the respective signaling protein contains a periplasmic (extracytoplasmic) or membrane-embedded sensory domain, or it is sensing the cytoplasmic milieu.
If the signaling protein contains a known sensory domain, its name (or, better yet, ligand specificity) should be included in the annotation, e.g. “Citrate-sensing histidine kinase”, “Histidine kinase with two PAS and one GAF sensory domains”, “Adenylate cyclase with a PAS sensory domain”, or “Diguanylate cyclase with MASE2 sensory domain”.
Any protein containing the REC domain is annotated either as a response regulator, or as a hybrid histidine kinase, or as a multidomain signal transduction protein.
Response regulators are classified into families and named according to the specificities of their output domains, e.g. “DNA-binding response regulator, OmpR family” or a “Transcriptional regulator, OmpR/PhoB family”. In the rare cases when the transmitted signal is known, this information has to be reflected in the name, whereas the family designation can be omitted, for example, NarL can be annotated either as “Nitrate/nitrite response regulator NarL” or as “Transcriptional regulator of nitrate/nitrite response, NarL family”.
Response regulators with enzymatic output domains can have family-based or domain-based names. For consistency, it would be appropriate to annotate them based on their enzymatic activities, just as it is being done for signaling proteins. For example, the response regulator of the REC-GGDEF domain architecture can be called a “Response regulator, WspR family” or a “Response regulator with a diguanylate cyclase output domain”. Finally, response regulators with ligand-binding output domains have to be named based on their domain architectures, e.g. “Response regulator with a PAS output domain, REC-PAS”.
Finally, other components of the signal transduction machinery, unless their function has been established experimentally, should be annotated based on their domain composition. For example, we would recommend that uncharacterized homologs of the nitric oxide reductase transcription regulator NorR be annotated as “Transcriptional regulator containing GAF, AAA-type ATPase, and Fis-like DNA binding domains, NorR/HyfR family”, as some of them may turn out to respond to other signals than nitric oxide.
Most importantly, all signal transduction proteins should be annotated based on their total domain composition, not just based on the database hits that might cover the whole protein.
In conclusion, it is worth noting that sequence analyses and functional annotations described in this chapter cannot be readily be automated. Although analysis of protein domain organization is usually fairly straightforward, proper utilization of proper databases and software tools requires a human with certain biological education. While this may be considered a nuisance by some, this reflects a more general principle that to recognize novelty, one has to be aware of the state of the art. If the latest developments are any indication, signal transduction proteins with complex domain architectures would not be amenable to fully automated analysis for years to come.
Footnotes
All URLs and database references in this chapter were correct at the time of writing (Oct. 2006). We apologize for any confusion that might arise from subsequent changes in database content, sequence and/or annotation updates.
References
- Anand GS, Stock AM. Kinetic basis for the stimulatory effect of phosphorylation on the methylesterase activity of CheB. Biochemistry. 2002;41:6752–6760. doi: 10.1021/bi012102n. [DOI] [PubMed] [Google Scholar]
- Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–110561. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- Baker DA, Kelly JM. Structure, function and evolution of microbial adenylyl and guanylyl cyclases. Mol Microbiol. 2004;52:1229–1242. doi: 10.1111/j.1365-2958.2004.04067.x. [DOI] [PubMed] [Google Scholar]
- Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung JK, Rood JI. Glutamate residues in the putative transmembrane region are required for the function of the VirS sensor histidine kinase from Clostridium perfringens. Microbiology. 2000;146:517–525. doi: 10.1099/00221287-146-2-517. [DOI] [PubMed] [Google Scholar]
- Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics. 2004;20:136–137. doi: 10.1093/bioinformatics/btg394. [DOI] [PubMed] [Google Scholar]
- D’Souza M, Glass EM, Syed MH, Zhang Y, Rodriguez A, Maltsev N, Galperin MY. Sentra, a database of signal transduction proteins for comparative genome analysis. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkl949. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Gaidenko TA, Mulkidjanian AY, Nakano M, Price CW. MHYT, a new integral membrane sensor domain. FEMS Microbiol Lett. 2001;205:17–23. doi: 10.1111/j.1574-6968.2001.tb10919.x. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM, Gessner CR, Gardner PR. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and σ54 in the nitric oxide stress response. J Biol Chem. 2003;278:10081–10086. doi: 10.1074/jbc.M212462200. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Gomi M, Sonoyama M, Mitaku S. High performance system for signal peptide prediction: SOSUIsignal. Chem-Bio Info J. 2004;4:142–147. [Google Scholar]
- Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Greene EA, Pietrokovski S, Henikoff S. Increased coverage of protein families with the blocks database servers. Nucleic Acids Res. 2000;28:228–230. doi: 10.1093/nar/28.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Island MD, Kadner RJ. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175:5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ, Potts M. Fancy meeting you here! A fresh look at “prokaryotic” protein phosphorylation. J Bacteriol. 1996;178:4759–4764. doi: 10.1128/jb.178.16.4759-4764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol Lett. 2002;206:1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Krupa A, Abhinandan KR, Srinivasan N. KinG: a database of protein kinases in genomes. Nucleic Acids Res. 2004;32:D153–D155. doi: 10.1093/nar/gkh019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA, Wemmer DE. Crystal structure of an activated response regulator bound to its target. Nat Struct Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ, Krzywda S, Brannigan JA, Turkenburg JP, Muchova K, Dodson EJ, Barak I, Wilkinson AJ. The trans-activation domain of the sporulation response regulator Spo0A revealed by X-ray crystallography. Mol Microbiol. 2000;38:198–212. doi: 10.1046/j.1365-2958.2000.02134.x. [DOI] [PubMed] [Google Scholar]
- Lory S, Wolfgang M, Lee V, Smith R. The multi-talented bacterial adenylate cyclases. Int J Med Microbiol. 2004;293:479–482. doi: 10.1078/1438-4221-00297. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E, Stock AM. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure. 1997a;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- Martinez-Hackert E, Stock AM. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997b;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Mizuno T. The structure and function of the histidine-containing phosphotransfer (HPt) signaling domain of the Escherichia coli ArcB sensor. J Biochem (Tokyo) 1998;124:440–445. doi: 10.1093/oxfordjournals.jbchem.a022132. [DOI] [PubMed] [Google Scholar]
- Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya AN, Mulkidjanian AY, Beech IB, Galperin MY. MASE1 and MASE2: two novel integral membrane sensory domains. J Mol Microbiol Biotechnol. 2003;5:11–16. doi: 10.1159/000068720. [DOI] [PubMed] [Google Scholar]
- Nikolskaya AN, Arighi CN, Huang H, Barker WC, Wu CH. PIRSF family classification system for protein functional and evolutionary analysis. Evolutionary Bioinformatics Online. 2006;2:209–221. [PMC free article] [PubMed] [Google Scholar]
- Ohmori M, Okamoto S. Photoresponsive cAMP signal transduction in cyanobacteria. Photochem Photobiol Sci. 2004;3:503–511. doi: 10.1039/b401623h. [DOI] [PubMed] [Google Scholar]
- Pao GM, Saier MH., Jr Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J Mol Evol. 1995;40:136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B, Argos P. Prediction of membrane protein topology utilizing multiple sequence alignments. J Protein Chem. 1997;16:453–457. doi: 10.1023/a:1026353225758. [DOI] [PubMed] [Google Scholar]
- Pohlmann A, Cramm R, Schmelz K, Friedrich B. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol Microbiol. 2000;38:626–638. doi: 10.1046/j.1365-2958.2000.02157.x. [DOI] [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Gomelsky M, Galperin MY. C-di-GMP: The dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. Ubiquitous protein domain EAL encodes cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant F, Bru C, Carrere S, Courcelle E, Gouzy J, Peyruc D, Kahn D. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- Shenoy AR, Visweswariah SS. Class III nucleotide cyclases in bacteria and archaebacteria: lineage-spefic expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 2004;561:11–21. doi: 10.1016/s0014-5793(04)00128-0. [DOI] [PubMed] [Google Scholar]
- Stephenson K, Lewis RJ. Molecular insights into the initiation of sporulation in Gram-positive bacteria: new technologies for an old phenomenon. FEMS Microbiol Rev. 2005;29:281–301. doi: 10.1016/j.femsre.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Stock A, Koshland DE, Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci USA. 1985;82:7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Mottonen JM, Stock JB, Schutt CE. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Tarutina M, Ryjenkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem. 2006;281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the a4-β5-α5 face. J Mol Biol. 2005a;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro-Roman A, Wu T, Stock AM. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 2005b;14:3077–3088. doi: 10.1110/ps.051722805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach KA, Chapman JW, Piggot PJ, Hoch JA. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci USA. 1985;82:7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Zhulin IB. MiST: a microbial signal transduction database. Nucleic Acids Res. 2007;35:D386–D390. doi: 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Williams SB, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol. 1999;33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 2002;3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mudd M, Boucher JC, Schurr MJ, Deretic V. Identification of the algZ gene upstream of the response regulator algR and its participation in control of alginate production in Pseudomonas aeruginosa. J Bacteriol. 1997;179:187–193. doi: 10.1128/jb.179.1.187-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Mattick JS, Teasdale RD. SVMtm: support vector machines to predict transmembrane segments. J Comput Chem. 2004;25:632–636. doi: 10.1002/jcc.10411. [DOI] [PubMed] [Google Scholar]
- Zhang W, Shi L. Evolution of the PPM-family protein phosphatases in Streptomyces: duplication of catalytic domain and lateral recruitment of additional sensory domains. Microbiology. 2004;150:4189–4197. doi: 10.1099/mic.0.27480-0. [DOI] [PubMed] [Google Scholar]
- Zhao H, Msadek T, Zapf J, Madhusudan, Hoch JA, Varughese KI. DNA complexed structure of the key transcription factor initiating development in sporulating bacteria. Structure. 2002;10:1041–1050. doi: 10.1016/s0969-2126(02)00803-1. [DOI] [PubMed] [Google Scholar]
- Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J Bacteriol. 2003;185:285–294. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]