Abstract
C/EBPα is implicated to regulate mouse amelogenin gene expression during tooth enamel formation, in vitro. Since enamel formation occurs during postnatal development and C/EBPα-deficient mice die at birth, we used the Cre/loxP recombination system to characterize amelogenin expression in C/EBPα conditional knock-out mice. Mice carrying the Cre-transgene under the control of the human keratin-14 (K14) promoter show robust Cre expression in the ameloblast cell lineage. Mating between mice bearing the floxed C/EBPα allele with K14-Cre mice generate C/EBPα conditional knock-out mice. Real-time PCR analysis shows that removal of one C/EBPα allele from the molar enamel epithelial organ of 3-day postnatal mice results in dramatic decrease in endogenous C/EBPα mRNA levels and coordinately altered amelogenin mRNA abundance. Conditional deletion of both C/EBPα alleles further diminishes C/EBPα mRNA levels, however, rather than ablating amelogenin expression, we observe wild-type amelogenin mRNA abundance levels. We examined C/EBPβ and NF-YA expression, two transcription factors that had previously been shown to modestly participate in amelogenin expression, in vitro, but found no significant changes in either of their mRNA abundance levels comparing conditional knock-out mice with wild-type counterparts. While the abundance of C/EBPδ is also unchanged in C/EBPα conditional knock-out mice, in vitro, we find that C/EBPδ activates the mouse amelogenin promoter and synergistically cooperates with NF-Y, suggesting that C/EBPδ can functionally substitute for C/EBPα to produce an enamel matrix competent to direct biomineralization.
In vitro, C/EBPα has been demonstrated to act as a strong transactivator for amelogenin and to synergize with NF-Y to further increase expression, while activation is opposed by Msx2 in a pathway working through protein to protein interactions (1–5). Mice homozygous for the C/EBPα gene deletion die within 8 hours of birth from either hypoglycemia (6) or impaired function of type II pneumocytes (7). This perinatal lethal phenotype has prevented our observation of the effects from the loss of C/EBPα on postnatal tooth formation as this is the precise developmental period when enamel formation is occurring and amelogenin mRNA expression is robust (8,9).
The Cre/LoxP recombination system offers a method to circumvent this problem and has been successfully applied to investigating the role of C/EBPα in energy metabolism in the liver and adipose tissues at later stages of postnatal development (10,11). The homozygous C/EBPα-loxP (C/EBPαfl/fl) mice are indistinguishable from their wild-type counterparts (10).
The search for a promoter useful for driving Cre recombinase in ectoderm-derived ameloblasts identified the keratin 14 (K14) promoter (12–15). Therefore, we used the K14 promoter to drive Cre mediated recombination of floxed C/EBPα loci predicted to result in the loss of amelogenin expression.
Here, we report on the generation of C/EBPα conditional knock-out mice created by crossing mice bearing the C/EBPαfl/fl with K14-Cre mice. This conditional ablation allowed us to investigate the relationship between C/EBPα and amelogenin expression in vivo utilizing real-time PCR technique to measure mRNA levels. We find that in the absence of mRNA for C/EBPα that C/EBPδ is redundant, serving to maintain amelogenin expression at wild-type levels in C/EBPα conditional knock-out mice.
Experimental Procedures
Transient transfection and luciferase assay
Transient transfection and luciferase assays were performed as described previously (3).
Animal Preparation
The K14-Cre transgenic line, the R26R reporter line, and the C/EBPαfl/fl (fl, flanked by loxP sites) mouse stain have been described previously (10,16,17). Mating K14-Cre+/− with R26R+/− mice generated “R26R;K14-Cre” mice (double transgenic). Mating K14-Cre+/− with fl/fl (C/EBPαfl/fl) mice generated “wt/fl;K14-Cre” mice with subsequent mating between “wt/fl;K14-Cre mice” generated “fl/fl;K14-Cre” homozygous conditional knock-out mice for C/EBPα. For ease of identification, we refer to wild-type animal as C/EBPα+/+, the “wt/fl;K14-Cre” animal as C/EBPα+/−, and the “fl/fl;K14-Cre” animal as C/EBPα−/− in order to allow the C/EBPα allele status to be easily tracked.
Genotyping of wild-type (+), loxP-targeted (fl), and Cre-mediated recombination (−) for C/EBPα alleles
Genomic DNA from mouse tails was isolated by digestion in a buffer containing 0.6 mg/ml proteinase-K, 50 mM Tris-Cl, pH 8.0, 100 mM EDTA, and 0.5 % SDS at 55°C overnight. The solution was subjected to extraction with phenol, phenol/chloroform, and chloroform. DNA in the aqueous phase was precipitated by the addition of 2 volumes of ethanol. An additional wash step in 70% ethanol was essential to remove traces of SDS and phenol prior to biochemical manipulation.
As shown in figure 1A, PCR primers F4, 5′-AACCTCCACC TCCCCTCG-3′; F6, 5′-TCTGATGCCG CCGTGTTC-3′; F7, 5′-CTCCAGTGTG GTCTGTGTTG G-3′; B4, 5′-GCCAAACCCC GTGTTCAC-3′; and B6, 5′-CCCCTGATGC TCTTCGTCCA G-3′, were used to differentiate the wild-type, floxed C/EBPα, and knock-out C/EBPα allele: the F7/B4 primer pairs were used to detect the 600 bp wild-type allele; the F6/B6 primer pairs were used to detect the 400 bp floxed C/EBPα allele; and the F4/B4 primer pairs were used to detect the 900 bp knock-out allele (Fig. 1A). The primer pair used to identify the 700 bp Cre allele was: forward primer, 5′-TGCTGTTTCA CTGGTTATGC GG-3′; and reverse primer, 5′-CCATTGCCCC TGTTTCACTA TCC-3′.
Figure 1.
Analysis of Cre mediated recombination. A, Solid triangles represent the loxP sites, and “E” represents the EcoRI restriction endonuclease site. Primers (arrows) F7 and B4 amplified the 600 bp wild-type allele; primers F6 and B6 amplified the 400 bp C/EBPαfl/fl allele; primers F4 and B4 amplified the 900 bp C/EBPα−/− allele. B, K14-Cre mice were mated with R26R transgenic mice and the first molars recovered by microdissection and subjected to β-galactosidase (lacZ) staining. Only epithelial cells (E) of the enamel organ, including the lingual enamel organ epithelia (open arrows) show positive reaction for lacZ staining while mesenchymal cells (M) are completely free of lacZ staining. C, Mandibles of newborn mice were used to produce frozen sections and subjected to lacZ staining to ascertain Cre recombination on a cell-by-cell basis. Occasional unstained ameloblast cells (arrow) were observed in the maturing region (d) and secretory region (e) of the incisor and in molars (f). M1, first molar; M2, second molar.
Detection of Cre-mediated recombination by β-galactosidase (lacZ) staining
Cells undergoing recombination were identified using the Cre/loxP system in which the K14-Cre transgene mediated DNA recombination after being crossed with the ROSA26 reporter transgene (17). As a consequence to recombination, β-gal expression was activated and restricted to the cells where the K14 promoter was expressed at levels sufficient to cause Cre-mediated activation of the ROSA26 marker. To assess K14 promoter activity in mouse teeth, first molars were dissected from newborn mice and stained for β-galactosidase activity. Molars were fixed overnight at 4°C in 0.2% glutaraldehyde in PBS (phosphate-buffered saline), washed three times in rinse solution (0.005% Nonidet P-40 and 0.01% sodium deoxycholate in PBS). Tissues were stained overnight at 37°C using a staining solution containing 2 mM MgCl2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 0.4% X-gal, rinsed twice in PBS and postfixed in 3.7% formaldehyde.
Cryostat sectioning
Mandibles of newborn mice were prepared for frozen sections and stained according to standard procedures. In brief, tissues were fixed in 0.2% glutaraldehyde solution overnight at 4°C, immersed in 10% sucrose in PBS containing 2mM MgCl2 for 30 min at 25°C, incubated in 30% sucrose, 50% OCT (optimum cutting temperature, Sakura Finetek USA, Inc., Torrance, CA), 2 mM MgCl2 in PBS for 1.5 h at 4°C and embedded in OCT by freezing on dry ice. Sections were cut at 10 μm thickness, mounted on gelatin-coated slides, fixed in 0.2% glutaraldehyde for 10 min on ice, and rinsed twice in PBS containing 2 mM MgCl2, followed by a 10 min wash in PBS with 2 mM MgCl2. Tissue sections were incubated in the detergent rinse solution (2 mM MgCl2, 0.005% NP-40 and 0.01% sodium deoxycholate in PBS) for 10 min at 4°C and chromogen developed in X-gal staining solution overnight at 37°C in the dark.
Epithelial-Mesenchyme Separation of Molars from 3-day Postnatal Mice
The first and second molars were dissected free of the mandible and incubated in 1% dispase (Invitrogen, Carlsbad, CA) in PBS for 1 h on ice. The sheets of epithelial cells were separated from the underling extracellular matrix with mesenchyme-derived cells and subjected to RNA extraction.
RNA Extraction and Reverse Transcription
Total RNA from enamel organ epithelial cells from the first and second molars of 3-day postnatal mice was extracted by using the RNA-Bee reagent (TEL-TEST, Inc., Friendswood, TX). First strand cDNA was synthesized with 100 ng random oligodeoxynucleotide decamers using a RETROscript reverse transcription kit (Ambion, Austin, TX).
Real-time PCR Analysis
PCR was carried out with the IQ SYBR green supermix kit (Bio-Rad, Hercules, CA) in a 20 μl final volume, 3–4 mM MgCl2 and 0.2–0.4 μM each primer (final concentration). Detailed primer sequences for each target gene are shown in supplemental table 1. For analyzing C/EBPα, amelogenin, NF-YA, and β-actin, PCR was performed using the iCycler iQ multicolor real-time PCR detection system (Bio-Rad, Hercules, CA) for 40 cycles at 95°C for 10 s and 55°C for 45 s. For C/EBPβ and C/EBPδ analysis, PCR was performed for 40 cycles at 95°C for 20 s, 59°C for 20 s and 72°C for 20 s. Amplification specificity was verified by analysis of the data derived from the melting curve for each sample following the manufacturer’s instructions. The iCycler iQ real-time PCR detection system software version 3.1 was used to analyze results, and the PCR baseline subtraction curve fit function was used to determine threshold cycle (CT) values. The threshold cycle value was converted to mRNA abundance, using a algorithm provided by the manufacturer.
Electrophoresis mobility shift assay (EMSA)
Nuclear extracts and EMSA assay were performed as described previously (3).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed according to the protocol for the EZ ChIP kit (17–371, Upstate Biotechnology, Temecula, CA). Cells were cross-linked with 1% formaldehyde for 10 min at 25°C. After washing with ice-cold PBS, cells were lysed in SDS lysis buffer (6×106 cells/0.4 ml of SDS lysis buffer) for 10 min on ice, sonicated at 4 watts for 4 pulses of 10 s each using a VirSonic 60 sonicator (Virtis Co., Gardiner, NY). Cell debris were removed by centrifugation for 10 min at 13,000 g. The soluble chromatin (100 μl per immunoprecipitation assay) was transferred to a new Eppendorf tube and diluted 10 times in ChIP dilution buffer, followed by pre-clearing with 60 μl of protein G agarose beads (16-201C, Upstate Biotechnology, Temecula, CA) for 60 min at 4°C with rotation. The pre-cleared lysates were immunoprecipitated using antibodies for either non-immune IgG (5 μg, PP64B, Upstate Biotechnology, Temecula, CA), C/EBPα (5 μg, sc-61x, Santa Cruz Biotechnology, Santa Cruz, CA) or C/EBPδ (5 μg, sc-636x, Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C with rotation. Immune complexes were collected by adding 50 μl of protein G agarose beads, and washed once with 1 ml of low salt wash buffer, once with 1 ml of high salt wash buffer, once with 1 ml of LiCl wash buffer, and twice with 1 ml of TE buffer. Immune complexes were eluted with elution buffer, cross-links reversed, and treated with proteinase-K. DNA was recovered using spin columns. The resulting ChIP DNA solution was diluted 100 times, and 2 μl of DNA was used for PCR detection. To amplify the 200 bp product, the primer set was 5′-GCTTCCCAAA CCTATTATTG CCTG-3′ and 5′-TTTCTTCCAA CTCTGTGCCC C-3′; to amplify the 584 bp product, the primer set was 5′-CTACTGTAAT AGTCTTGAGG TCGTGGC-3′ and 5′-CGATGGTTTC TTCCAACTCT GTGC-3′.
Statistical Analysis
Statistical analysis was performed using the one-way ANOVA test and statistical significance is defined as P < 0.05.
RESULTS
K14-Cre transgenic mice robustly express Cre recombinase in the ameloblast cell lineage
The K14-Cre transgenic mice were bred with R26R mice to generate transgenic animals, in which expression of the β-galactosidase occurs only upon Cre-mediated DNA excision. When newborn mandibles of “K14-Cre; R26R” mice were prepared for histologic examination, X-gal positive cells were restricted to the enamel organ of the incisor and molar teeth, including the ameloblasts (Fig. 1C). The first molar of newborn pups, with the genotype of “K14-Cre; R26R”, was isolated and subject to whole-mount X-Gal staining. As expected, the lacZ expression was uniformly distributed in enamel organ epithelial cells, while no lacZ stain was observed in mesenchymal cells (Fig. 1B, panels a, b, c; Fig 1C, panel f). In the lower incisor, the lacZ activity was detected only in enamel organ epithelial cells from the earliest stage of ameloblast differentiation within the stem cell compartment, continuously through the fully differentiated ameloblasts lining the incisor tip region (Fig. 1C). The enamel organ of the lingual surface of the incisor, an area that does not develop as secretory ameloblasts, was also identified as lacZ positive (Fig. 1C, open arrows). When examined in histologic sections, some unstained ameloblast cells were observed sporadically, suggesting an incomplete recombination event in some cells from the enamel organ (Fig. 1C, panel d and e; solid arrows).
K14-Cre-mediated C/EBPα ablation in mouse ameloblast cell lineage
To specifically disrupt C/EBPα gene expression in the ameloblast cell lineage (Fig. 1A), C/EBPαfl/fl (fl, flanked by loxP sites) mice were bred with K14-Cre transgenic mice to generate “C/EBPα+/−” heterozygous conditional knock-out mice. Subsequent mating between “C/EBPα +/−” mice produced homozygous conditional knock-out mice, “C/EBPα−/−”. RNA extracted from molar enamel organ epithelial cells of 3-day postnatal mice was reverse-transcribed, and the first strand cDNA was subject to real-time PCR analysis.
C/EBPα mRNA transcripts were reduced significantly upon removal of one C/EBPα allele and upon removal of the second C/EBPα allele (P<0.01, Fig. 2A), this finding suggesting successful K14-Cre-mediated C/EBPα gene excision to generate a conditional C/EBPα knockout in the enamel organ epithelium. The level of C/EBPα in “C/EBPα+/−” mice was reduced to 41–55% of levels compared to expression levels observed in wild-type control mice, while the level of C/EBPα in “C/EBPα−/−” mice was reduced to 2–32% of that in wild-type control mice (Fig. 2A). C/EBPα has been demonstrated as the strong transactivator of the amelogenin gene (1,2,5), and ablation of the C/EBPα gene is expected to affect amelogenin expression. Therefore, we examined the expression level of amelogenin mRNA from each of these C/EBPα conditional knock-out mice that varied only by the C/EBPα allele number. Amelogenin was reduced upon removal of one C/EBPα allele, with 61 to 85% of the amelogenin transcript abundance in “C/EBPα+/−” mice compared to the abundance found in wild-type control mice (P<0.05, Fig. 2E). Surprisingly, once two C/EBPα alleles were excised, we identified that the amelogenin mRNA transcript abundance for the “C/EBPα−/−” mice returned to the wild-type level (P>0.05, Fig. 2E). This finding suggests that amelogenin transcript abundance, in vivo, is independent of C/EBPα expression levels. Rather, amelogenin transcript abundance is compensated by an unknown mechanism(s) upon excision of two C/EBPα alleles.
Figure 2.
Real-time PCR analysis. The mRNA expression level for C/EBPα (A), C/EBPβ (B), C/EBPδ (C), NF-YA (D), and amelogenin (E) among wild-type or C/EBPα conditional knock-out mice was determined by real-time PCR. *P<0.05, **P<0.01. Shown in brackets is the mRNA abundance calculated from the threshold cycle (Ct) shown as a percent of wild type.
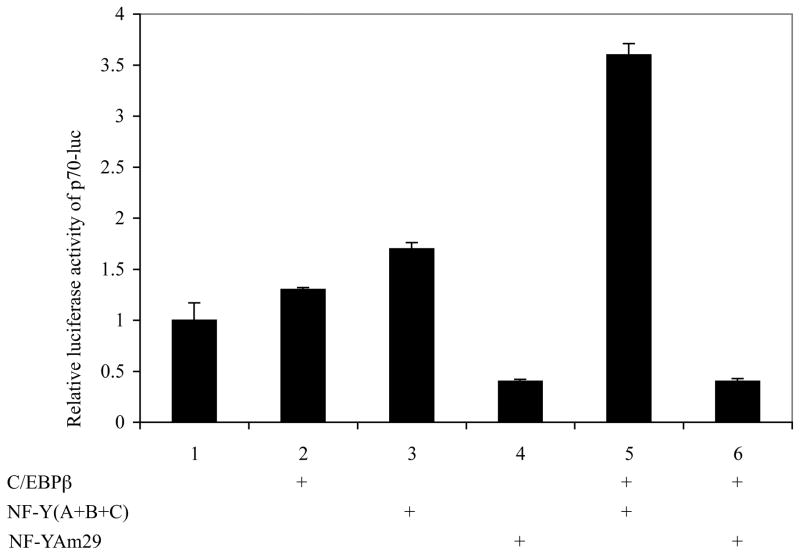
Several studies have suggested potential roles of C/EBPβ and NF-Y in facilitating C/EBPα-mediated transactivation (5,18). Functional assay were used to show that either C/EBPβ or NF-Y alone has only marginal effects on the amelogenin promoter (Fig. 3, lane 2 and 3). While NF-Y and C/EBPβ enhance the amelogenin promoter some 3.5 fold (Fig. 3, lane 5), this gain is ablated in the presence of a mutated NF-YA (NF-Yam29) (Fig. 3, lane 6).
Figure 3.
Effects of C/EBPβ and NF-Y on the amelogenin promoter. LS8 cells were transiently transfected with 250 ng of p70-luc reporter construct in the presence of 200 ng of an “empty” expression vector (lane 1), C/EBPβ (lane 2), NF-Y (lane 3), dominant negative form of NF-Y, NF-YAm 29 (lane 4), C/EBPβ and NF-Y (lane 5), and C/EBPβ and NF-YAm 29 (lane 6). A pCMV-lacZ plasmid (75 ng) was included in all experiment groups as an internal control for transfection efficiency. Data reflected the mean ± S.D. of three independent experiments, with the response level of p70-luc in the absence of exogenous C/EBPβ set arbitrarily as “1”.
Nonetheless, it is possible that increased levels of C/EBPβ and/or NF-Y mRNA level could lead to increased protein levels sufficient to activate the amelogenin promoter and produce enough amelogenin proteins to compensate for the loss of both C/EBPα alleles. Based upon this hypothesis, the level of C/EBPβ and NF-YA transcripts were ascertained in this study. We did not observe a significant change in the level of C/EBPβ among mice with either one or both C/EBPα alleles removed (P>0.05, Fig. 2B). The expression level of C/EBPβ in mice in which there was only one C/EBPα allele (“C/EBPα+/−”) was 75–134% of that observed in wild-type control mice, and the expression level of C/EBPβ in mice with no C/EBPα allele (“C/EBPα−/−”) was 100–189% of that observed in wild-type control mice (Fig. 2B). In addition, there was no significant difference in the level of NF-YA in either instance in which only the C/EBPα allele number varied (P>0.05, Fig. 2D). The level of NF-YA in heterozygous mice (“C/EBPα+/−”) was 70–101% of the expression levels observed in wild-type control mice. The level of NF-YA in conditional knockout mice (“C/EBPα−/−”) was 55–125% of that observed in wild-type control mice.
Standard histologic examination of the hematoxylin and eosin stained 3-day postnatal mouse mandibles showed that the enamel thickness of incisors (Fig. Supplemental 1A) and molars (Fig. S1B) was not grossly affected in mice with a complete absence of C/EBPα (“C/EBPα−/−”) when compared to enamel thickness of wild-type control mice. The absence of detectable diminution of the enamel thickness in the C/EBPα conditional knock-out animal is not surprising given the observed compensation that restored amelogenin mRNA levels to wild-type levels (Fig. 2E). However, it is unlikely that C/EBPβ and NF-Y, even working in combination are sufficient to restore amelogenin abundance to the near wild type levels that are observed (Fig. 2) especially in the absence of significant increases in C/EBPβ and NF-Y mRNA abundance in the teeth from C/EBPα conditional knock-out mice.
C/EBPδ is able to activate the mouse amelogenin promoter
The finding that the amelogenin mRNA level is restored upon excision of both CEBPα alleles prompted us to search for an alternative pathway in the regulation of the amelogenin gene. C/EBPδ was regarded as a candidate because both C/EBPδ and C/EBPα have similar nucleotide sequence preferences for promoter binding (19). To test whether C/EBPδ could function as a transactivator of the mouse amelogenin promoter, a C/EBPδ expression vector was co-transfected into LS8 cells with different amelogenin promoter reporter constructs: p2207-luc (the full-length amelogenin promoter), p70-luc (the minimal amelogenin promoter), mC/EBP-p2207-luc (the full-length amelogenin promoter with the mutated C/EBP site), and mC/EBP-p70-luc (the minimal amelogenin promoter with the mutated C/EBP site). Mutation of the C/EBP site abolished the basal promoter activity of mutant reporter constructs (mC/EBP-p2207-luc and mC/EBP-p70-luc) and their C/EBPδ-mediated transaction, whereas the reporter gene activity of wild-type constructs (p2207-luc, and p70-luc) was increased dramatically by co-transfection with C/EBPδ (Fig. 4A). This level of transactivation for the amelogenin promoter by C/EBPδ is similar to that observed for C/EBPα (5). Taken together, these data indicate that the C/EBP site is required to maintain basal amelogenin promoter activity and is responsive to either C/EBPδ or C/EBPα.
Figure 4.
In vitro analysis of the amelogenin promoter. A, the C/EBP site is required for maintaining the basal amelogenin promoter activity and C/EBPδ-mediated transactivation. Various reporter constructs (p2207-luc, p70-luc, mC/EBP-p2207-luc, and mC/EBP-p70-luc) each used at 250 ng were transiently transfected into LS8 cells with 200 ng of C/EBPα expression plasmid or empty vector pcDNA3. In all cases, pCMV-lacZ (75 ng) was included as an internal control for transfection efficiency. The relative luciferase activity was the normalization of luciferase activity with β-galactosidase activity. The mean ± S.D. from at least three independent experiments was represented, and the level of p70-luc in the absence of exogenous C/EBPδ was set arbitrarily as “1”. B, C/EBPδ and NF-Y synergize on the minimal amelogenin promoter. LS8 cells were transiently transfected with 250 ng of p70-luc reporter construct in the presence of 200 ng of empty “expression” vector (lane 1), C/EBPδ (lane 2), NF-Y (lane 3), dominant negative form of NF-Y, NF-YAm 29 (lane 4), C/EBPδ and NF-Y (lane 5), and C/EBPδ and NF-YAm 29 (lane 6). pCMV-lacZ plasmid (75 ng) was included in all experiment groups as an internal control for transfection efficiency. Data reflected the mean ± S.D. of three independent experiments, with the response level of p70-luc in the absence of exogenous C/EBPδ set arbitrarily as “1”.
Previous data has shown that NF-Y and C/EBPα synergistically activate the mouse amelogenin promoter (3). To investigate whether C/EBPδ has similar potency to that of C/EBPα, NF-Y and C/EBPδ expression plasmids were co-transfected with the p70-luc reporter construct into LS8 cells. As shown in figure 4B, C/EBPδ alone increased the promoter activity about 8-fold (lane 2), whereas exogenous expression of NF-Y in isolation had only marginal effects on the promoter (lane 3). Co-transfection of C/EBPδ with NF-Y served to synergistically increase the promoter activity to 16-fold (lane 5), a level that was two times more than that of C/EBPδ only. Furthermore, the presence of mutant NF-YA (NF-YAm29) greatly reduced the promoter activity, either in the absence (lane 4) or in the presence (lane 6) of exogenous C/EBPδ expression. These observations demonstrate that NF-Y facilitates C/EBPδ similar to the way it does for C/EBPα, to synergistically activate the mouse amelogenin promoter. However, NF-Y by itself exhibits only a marginal effect on the amelogenin promoter.
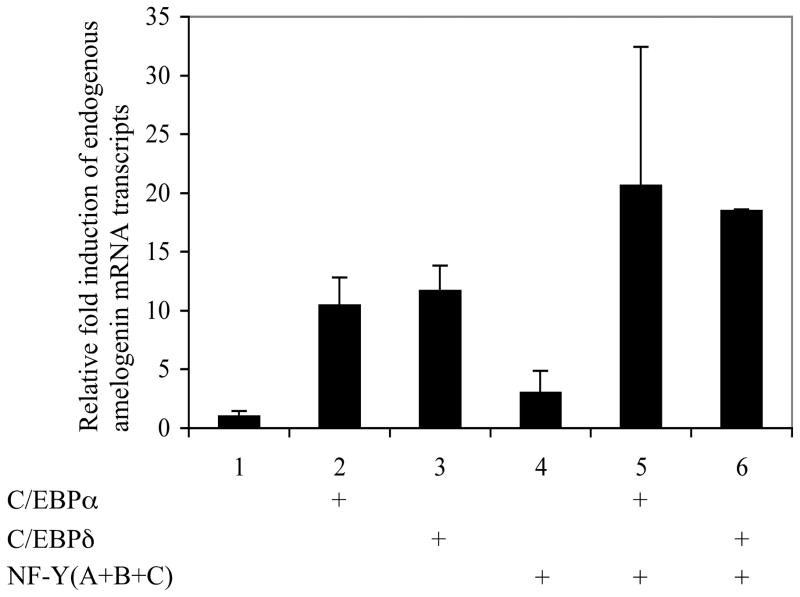
Nuclear extracts prepared from LS8 cells transfected with the C/EBPδ expression plasmid (NE/δ) were supershifted by the C/EBPδ antibody (Fig. 5, lane 4) as well as by the NF-Y antibody (Fig. 5, lane 6). ChIP assay was performed to assess the interaction of the mouse amelogenin promoter with its transcription factor C/EBPα or C/EBPδ in ameloblast-like LS8 cells transfected with the full-length mouse amelogenin promoter containing plasmid together with either the C/EBPα or the C/EBPδ expression vector. As a negative control, non-immune IgG did not show the enrichment for the amelogenin proximal promoter region, while both C/EBPα and C/EBPδ antibodies were able to enrich the promoter fragment containing the C/EBP consensus-binding motif (Fig. 6). Moreover, the capacity of the endogenous amelogenin promoter to respond to C/EBPα or C/EBPδ alone and in the presence of NF-Y was assessed in ameloblast-like LS8 cells. The endogenous amelogenin promoter is responsive to either C/EBPα or C/EBPδ alone (Fig. 7, lanes 2, 3, respectively) with each factor individually enhancing amelogenin mRNA greater than 10 fold. In the presence of NF-Y, either C/EBPα or C/EBPδ amelogenin mRNA transcript abundance nearly doubled (Fig. 7, lanes 5, 6, respectively).
Figure 5.
EMSA of the C/EBPδ binding site. Nuclear extracts were prepared from LS8 cells transfected with the C/EBPδ expression plasmid (NE/δ). Extracts were incubated with 32P-labeled probe, followed by electrophoretic separation, and visualized by autoradiography. For competition assay (lane 3), 50-fold molar excesses of unlabeled probe were incubated with 32P-labeled probe. For supershift assay, extracts were pre-incubated with a C/EBPδ antibody (lane 4, sc-636x, Santa Cruz Biotechnology, Santa Cruz, CA), normal rabbit serum (lane 5), NF-YA antibody (lane 6, sc-636x, Santa Cruz Biotechnology, Santa Cruz, CA), and normal goat serum (lane 7). The supershifted band is denoted by the symbol “*”.
Figure 6.
ChIP analysis for the binding of C/EBPα or C/EBPδ on the mouse amelogenin promoter. LS8 cells (100 mm plate) were transfected with p2207-luc (500 ng) with the expression vector for either C/EBPα (800 ng) or C/EBPδ (800 ng), and incubated for 24 h. Cells were fixed, sonicated and incubated with an non-immune IgG, or an anti-C/EBPα antibody (sc-61x, Santa Cruz Biotechnology, Santa Cruz, CA) or an anti-C/EBPδ antibody (sc-636x, Santa Cruz Biotechnology, Santa Cruz, CA), and immunoprecipitated with protein G-agarose beads (16-201C, Upstate Biotechnology, Temecula, CA).
Figure 7.
NF-Y facilitates C/EBPα or C/EBPδ and synergistically induces the endogenous amelogenin promoter as measured by mRNA transcripts. LS8 cells were transiently transfected with 200 ng of empty vector (lane 1), expression vector for C/EBPα (lane 2), C/EBPδ (lane 3), NF-Y (lane 4), C/EBPα and NF-Y (lane 5), C/EBPδ and NF-Y (lane 6). The mRNA expression level from the endogenous amelogenin gene was determined by real-time PCR analysis. Data represents three independent experiments, each done in triplicates. The level of amelogenin mRNA from cells treated with the empty vector was set arbitrarily as “1”.
In addition, immunohistochemistry detection of C/EBPδ protein signals were localized within nuclei of the ameloblast cell lineage in teeth (Fig. S2), that further suggests a role for C/EBPδ in the regulation of amelogenin gene expression in vivo. Thus, it is reasonable to propose a hypothesis that the synergism between NF-Y and C/EBPδ compensates for the absence of C/EBPα, restoring amelogenin expression levels to approximately wild-type status, in vivo.
DISCUSSION
Data from several in vitro experimental strategies have demonstrated that C/EBPα is a strong transactivator for amelogenin gene expression (1–3,5), however, no in vivo observation of the role for C/EBPα in amelogenin expression has been reported. As a step in exploring the function of C/EBPα in regulating amelogenin activation in vivo, and to circumvent the lethal neonatal phenotype in the conventional C/EBPα knock-out mice (6,7,20), we generated a conditional knock-out mouse strain in which the C/EBPα allele(s) was removed upon the expression of Cre recombinase under the control of K14 promoter. We used the R26R mouse to report on the cell specific expression of Cre recombinase. The K14 promoter drove Cre expression robustly as demonstrated by the lacZ staining in the enamel organ epithelia, specifically localizing to the ameloblast cell lineage from molars and incisors. However some unstained ameloblast cells were observed sporadically in the lower incisor. One explanation for this “sparing” is that expression of Cre recombinase in those cells unmarked by lacZ is below the threshold required for recombination and results in an incomplete gene excision indicated by the failure to express lacZ. To prevent potential contamination from mesenchymal cells, in which K14-Cre would not be active, molars of three-day postnatal mice were dissected, and the epithelial cells were mechanically separated from the mesenchyme.
We examined the expression level of amelogenin with regard to the C/EBPα allele number in these conditional knock-out mice. The amelogenin mRNA transcripts decreased in the C/EBPα heterozygous conditional knock-out mice (P<0.05, Fig. 2E). Surprisingly, in the C/EBPα homozygous conditional knock-out mice, amelogenin abundance was measured at essentially at the wild-type expression level (Fig. 2E). This finding strongly implies the existence of an alternative pathway for the activation of the amelogenin gene.
We examined the level of C/EBPβ and NF-YA in these C/EBPα conditional knock-out mice. Neither the level of C/EBPβ nor NF-YA was significantly affected in the mice with reduced C/EBPα allele numbers and consequently reduced C/EBPα mRNA levels. This observation is correlated with the finding that C/EBPβ and NF-Y exert only arithmetic accumulation effects in activating the amelogenin gene (Fig. 3), not the synergistic effect on the amelogenin gene as observed by over-expressing C/EBPα and NF-Y (3). Therefore, C/EBPβ and NF-Y are not likely to be capable of activating the amelogenin gene to the wild-type level in C/EBPα conditional knock-out mice. Hematoxylin and eosin stained 3-day postnatal mouse mandibles failed to show that enamel thickness was dramatically affected in C/EBPα conditional knock-out mice compared to the enamel thickness observed in wild-type control mice (Fig. S1).
The amelogenin expression level in the C/EBPα conditional knock-out mice suggested an alternative mechanism that circumvents the C/EBPα-mediated activation of amelogenin expression observed, in vitro (1–3,5). In searching for the potential candidate, C/EBPδ was analyzed because C/EBPα and C/EBPδ share the same binding site to the promoter (19). Data demonstrate that C/EBPδ, like C/EBPα, is able to bind to the amelogenin promoter (Figs. 5 and 6), and activates the mouse amelogenin promoter (Fig. 4). NF-Y could facilitate either C/EBPα or C/EBPδ to induce endogenous amelogenin expression in LS8 cells (Fig. 7). Immunohistochemistry for C/EBPδ protein revealed a nuclear localization in ameloblast cells (Fig. S2). However, we did not observe the increased C/EBPδ mRNA level in the enamel organ epithelia of the C/EBPα conditional knock-out mice. One explanation is that the mRNA level of a certain gene is not always proportional to its protein levels or to the functional activity of the protein. Although there is no significant change at the mRNA level of C/EBPδ, the activity of C/EBPδ protein may be increased, due to post-transcriptional and/or post-translational modifications, an outcome that is sufficient to produce enough amelogenin proteins in the C/EBPα conditional knock-out mice to build an apparently normal enamel matrix (Fig. S1).
Several studies have demonstrated a functional redundancy in the C/EBP family (21,22). Taken together, these data suggest that C/EBPδ and C/EBPα may have the functional redundancy in the regulation of mouse amelogenin gene.
Here, we report on the generation of C/EBPα conditional knock-out mice, in which C/EBPα alleles are specifically excised by Cre recombinase driven by the K14 promoter. Real-time PCR analysis demonstrated successful Cre-mediated C/EBPα ablation in mouse ameloblast cell lineages in vivo. Amelogenin mRNA levels were decreased in C/EBPα heterozygous conditional knock-out mice. However, amelogenin mRNA levels returned to the wild-type level in C/EBPα homozygous conditional knock-out mice. This finding implies an existence of an alternative pathway bypassing the C/EBPα pathway and compensating for the loss of C/EBPα. In search for other transcription factor(s) responsible for the induction of the amelogenin gene, we identified C/EBPδ. Like C/EBPα, C/EBPδ showed a similar synergistic potency with NF-Y to activate the mouse amelogenin promoter, a finding that suggests a functional redundancy between C/EBPα and C/EBPδ. We have successfully utilized ChIP assay to demonstrate the binding of C/EBPα or C/EBPδ on the amelogenin promoter in LS8 cells, in vitro. We are in the process of performing in vivo ChIP assay to investigate interactions of C/EBPα and C/EBPδ on the amelogenin promoter during the process of amelogenesis. The ultimate test will be to examine amelogenin expression and subsequent enamel biomineralization in C/EBPα and C/EBPδ double knock-out mice.
Supplementary Material
Acknowledgments
This work was supported by grant DE-06988 from the National Institute of Dental and Craniofacial Research, NIH.
We thank our colleagues at the University of Southern California, Center for Craniofacial Molecular Biology and Institute for Genetic Medicine for their support. We thank Dr. Sigal Gery (Cedars-Sinai Medical Center) for the C/EBPδ expression vector; Dr. Pierre Chambon (Institut de Genetique et de Biologie Moleculaire et Cellulaire, France) for the K14-Cre mice; Dr. Philippe Soriano (Fred Hutchinson Cancer Research Center) for the R26R mice; and Dr. Michael Wu (Millipore) for the technique support for the ChIP assay. We thank Dr. Baruch Frenkel (USC-Institute for Genetic Medicine) for his help in transferring the ChIP assay to our lab and Dr. Henry Sucov (USC-Institute for Genetic Medicine) for critical discussion during the execution of this work. We thank the two anonymous reviewers for their constructive criticisms.
The abbreviations used are
- C/EBPα
CCAAT/enhancer-binding protein alpha
- C/EBPβ
CCAAT/enhancer-binding protein beta
- C/EBPδ
CCAAT/enhancer-binding protein delta
- NF-Y
nuclear factor Y
- K14
keratin 14
- R26R
ROSA26 reporter
- PCR
polymerase chain reaction
- EDTA
ethylene diamine tetra acetic acid
- SDS
sodium dodecyl sulphate
- PBS
phosphate-buffered saline
- OCT
optimum cutting temperature
References
- 1.Xu Y, Zhou YL, Erickson RL, Macdougald OA, Snead ML. Biochem Biophys Res Commun. 2007;354(1):56–61. doi: 10.1016/j.bbrc.2006.12.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Y, Zhou YL, Ann DK, MacDougald OA, Shum L, Snead ML. Eur J Oral Sci. 2006;114(Suppl 1):169–177. doi: 10.1111/j.1600-0722.2006.00319.x. discussion 201–162, 381. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Zhou YL, Luo W, Zhu QS, Levy D, MacDougald OA, Snead ML. J Biol Chem. 2006;281(23):16090–16098. doi: 10.1074/jbc.M510514200. [DOI] [PubMed] [Google Scholar]
- 4.Zhou YL, Lei Y, Snead ML. J Biol Chem. 2000;275(37):29066–29075. doi: 10.1074/jbc.M002031200. [DOI] [PubMed] [Google Scholar]
- 5.Zhou YL, Snead ML. J Biol Chem. 2000;275(16):12273–12280. doi: 10.1074/jbc.275.16.12273. [DOI] [PubMed] [Google Scholar]
- 6.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Science. 1995;269(5227):1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 7.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. J Biol Chem. 1996;271(40):24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 8.Snead ML, Bringas P, Jr, Bessem C, Slavkin HC. Dev Biol. 1984;104(1):255–258. doi: 10.1016/0012-1606(84)90053-8. [DOI] [PubMed] [Google Scholar]
- 9.Snead ML, Luo W, Lau EC, Slavkin HC. Development. 1988;104(1):77–85. doi: 10.1242/dev.104.1.77. [DOI] [PubMed] [Google Scholar]
- 10.Lee YH, Sauer B, Johnson PF, Gonzalez FJ. Mol Cell Biol. 1997;17(10):6014–6022. doi: 10.1128/mcb.17.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ. J Biol Chem. 2004;279(43):44740–44748. doi: 10.1074/jbc.M405177200. [DOI] [PubMed] [Google Scholar]
- 12.Mustonen T, Ilmonen M, Pummila M, Kangas AT, Laurikkala J, Jaatinen R, Pispa J, Gaide O, Schneider P, Thesleff I, Mikkola ML. Development. 2004;131(20):4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 13.Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Wankell M, Werner S, Thesleff I. Dev Dyn. 2004;231(1):98–108. doi: 10.1002/dvdy.20118. [DOI] [PubMed] [Google Scholar]
- 14.Plikus MV, Zeichner-David M, Mayer JA, Reyna J, Bringas P, Thewissen JG, Snead ML, Chai Y, Chuong CM. Evol Dev. 2005;7(5):440–457. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabata MJ, Matsumura T, Liu JG, Wakisaka S, Kurisu K. Arch Oral Biol. 1996;41(11):1019–1027. doi: 10.1016/s0003-9969(96)00087-8. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Chiba H, Warot X, Messaddeq N, Gerard C, Chambon P, Metzger D. Development. 2001;128(5):675–688. doi: 10.1242/dev.128.5.675. [DOI] [PubMed] [Google Scholar]
- 17.Soriano P. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Zhu QS, Qian B, Levy D. J Biol Chem. 2004;279(29):29902–29910. doi: 10.1074/jbc.M400438200. [DOI] [PubMed] [Google Scholar]
- 19.Osada S, Yamamoto H, Nishihara T, Imagawa M. J Biol Chem. 1996;271(7):3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 20.Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ. Proc Natl Acad Sci U S A. 2001;98(22):12532–12537. doi: 10.1073/pnas.211416898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SS, Chen JF, Johnson PF, Muppala V, Lee YH. Mol Cell Biol. 2000;20(19):7292–7299. doi: 10.1128/mcb.20.19.7292-7299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones LC, Lin ML, Chen SS, Krug U, Hofmann WK, Lee S, Lee YH, Koeffler HP. Blood. 2002;99(6):2032–2036. doi: 10.1182/blood.v99.6.2032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.