Abstract
The successful adoption by clinicians of evidence-based clinical practice guidelines (CPGs) contained in clinical information systems requires efficient translation of free-text guidelines into computable formats. Natural language processing (NLP) has the potential to improve the efficiency of such translation. However, it is laborious to develop NLP to structure free-text CPGs using existing formal knowledge representations (KR). In response to this challenge, this vision paper discusses the value and feasibility of supporting symbiosis in text-based knowledge acquisition (KA) and KR. We compare two ontologies: (1) an ontology manually created by domain experts for CPG eligibility criteria and (2) an upper-level ontology derived from a semantic pattern-based approach for automatic KA from CPG eligibility criteria text. Then we discuss the strengths and limitations of interweaving KA and NLP for KR purposes and important considerations for achieving the symbiosis of KR and NLP for structuring CPGs to achieve evidence-based clinical practice.
Keywords: Knowledge Representation, Natural Language Processing, Clinical Trial, Practice Guidelines
Introduction
It takes about 17 years for new medical evidence to be routinely applied in patient care, and on average patients receive 54.9% of recommended care in the United States (US). Furthermore, the fast growing literature of clinical evidence has exceeded human cognitive capacity. Simultaneously, clinical decision-making processes for diagnosis and treatment have become so complex as to require clinical decision support (CDS) to promote evidence-based practice – a key concern of nurses and other practitioners. In this context, the Institute of Medicine requested that practice guideline developers structure the format, vocabulary, and content of computer-based practice guidelines to facilitate implementation of CDS.
A major barrier to evidence-based care is the difficulty of translating of free practice guidelines into a format that is actionable in the context of clinical practice. Many formal representations for clinical practice guidelines (CPGs) have been developed to generate computable rules to provide CDS (examples can be found at www.openclinical.org). However, most representations face two major obstacles to wide implementation and adoption in real clinical care settings. First, such computerized guidelines often take significant time and domain expertise to formalize. An experienced knowledge engineer often must manually extract knowledge from free-text guidelines and map it into a logic-based formalism or ontology with the assistance of domain experts. Moreover, this labor-intensive practice often causes variations in guideline interpretation and introduces potential biases and errors of omission. Second, execution of computerized CPGs requires data triggers, but many existing guideline ontologies face the fundamental challenge of the “semantic gap”: the difference between the coarse-grained concepts in free-text guidelines and the fine-grained data representations in electronic health records (EHR). Moreover, the requisite data might not even be available in the EHR in a discrete and computable format.
To overcome these barriers, researchers have recently explored knowledge representations (KR) that are pragmatic, tolerant of natural language, and data-interoperable. For example, Shiffman et al. used controlled natural language to write CPGs [1]. Similarly, rather than fully capturing the semantics of clinical research eligibility criteria directly in a formal language, Sim et al. used an annotation approach to leverage NLP to convert free-text eligibility criteria into a computable format [2]. To integrate a comprehensive model of clinical semantics with language processing types, Wu et al. developed a common type system for various clinical NLP uses to improve the interoperability of different NLP systems [3], while Peleg created a knowledge-data ontology mapper for guideline representations [4].
These approaches share the common idea of maximizing the support for text-based knowledge engineering for guideline KRs through the use of NLP. Semantic KR is a prerequisite for developing a symbolic NLP system. It is also referred to as sublanguage analysis for identifying controlled vocabularies or information structure in textual information. Unfortunately, such linguistic knowledge is rarely considered in KR efforts for guideline automation, largely because KR and NLP have evolved as two separate domains in biomedical informatics research so that researchers in the two domains often perform NLP and KR tasks independently of each other in separate silos. An exception to this trend can be found in recent work by Serban, who used Unified Medical Language System (UMLS) knowledge and linguistic patterns to manually formalize guidelines [5, 6].
There is potentially a strong connection between KR and natural language, but a collaborative approach has not been well explored by the KR, standardization, or formal methods research communities. In order to structure free-text guidelines, the KR community should provide not only a logical form but also a “natural” KR that preserves the information structure from the text. For example, a free-text guideline itself is an expressive natural KR. The process for structuring guidelines essentially decreases expressivity without losing the meaning yet increases tractability of the KR. In this process, preservation of the information structure in text can allow NLP to support text-based knowledge engineering. The advances in NLP research have generated many tools for accurate syntactic and semantic parsing. Applying text mining and pattern recognition to parsed text can make feasible the identification of the sublanguage, statement templates, and upper-level ontology [7], which defines very general classes and relationships, for free-text guidelines.
1. The Proposed Model
We argue that a synergistic model that incorporates both NLP and KR, with a formal KR informed by semantic KR analysis for NLP purposes, can ease text-based knowledge acquisition (KA) and improve KR efficiency. In this context, we define such a model as one that employs techniques adhering to the definition of a hybrid KA and KR methodology, wherein both the conceptual and procedural knowledge necessary to inform the parsing and codification of a CPG are generated in an integrated manner. Based on this premise, we provide our perspective on the aforementioned issues, propose an approach to achieve the symbiosis using the principles of upper-level ontology and structured narrative [8], present our preliminary results, and discuss needed future work.
2. Methods
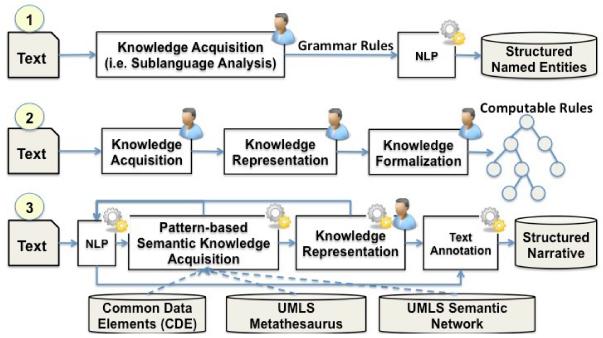
Figure 1 contrasts the two existing approaches to structuring free text CPGs (1 and 2) with our proposed approach (3).
Figure 1.
A comparison of three approaches to structuring free-text CPG documents: (1) NLP-based information extraction; (2) Manual knowledge acquisition and formalization; (3) NLP-based knowledge acquisition, knowledge representation, and text annotation. The person icon indicates a domain expert. The gear icon indicates an automated process.
Approach 1 performs named entity or semantic relationship extraction on guideline text. Manual knowledge acquisition for sublanguage analysis of the text usually occurs before automatic symbolic NLP, but this knowledge is rarely included in any KR for CPGs. This approach identifies instances of named entities but usually does not generate reusable KRs.
Approach 2 is most widely used by the biomedical KR community. It heavily relies on manual efforts for text-based KA, KR, and formalization. Occasionally, the third step is assisted by limited NLP. Approaches 1 and 2 both involve expert-driven, laborious KR efforts. However, due to the lack of coordination and exchange between KR and NLP, the ontologies created by domain experts are often not informed by the conceptual knowledge incumbent to the information structure in the text identified during sublanguage analysis. A typical result is that additional — and potentially error-prone — translation between the two constructs is required, which can include the instantiation of mappings from textual information to ontology-based concepts by domain experts or by NLP engineers respectively.
Approach 3 uses a hybrid KA and KR technique to achieve the symbiosis in KR and NLP through four steps: (1) NLP, (2) pattern-based KA, (3) NLP-assisted KR, and (4) NLP-assisted text annotation. It uses existing conceptual knowledge such as those found in the UMLS to link atomic information units to each other, and combines syntactic and semantic data standards for common data elements (CDEs) with text mining to perform NLP and pattern recognition from text before constructing a KR. This approach identifies the information structure in text and builds an upper-ontology [7]. An upper ontology differs from a full ontology by defining general concepts and concept relationships at a high level for a selected domain. Examples include BIOTOP for molecular biology [9] and Basic Formal Ontology (BFO) [10]. An upper-ontology for practice guidelines can inform NLP-based KR. This approach annotates text with concepts and semantic relationships (i.e., conceptual knowledge) and eases the process of more fine-grained manual knowledge engineering or NLP. Such a systematic conceptual knowledge-based representation of a given CPG provides the foundational basis for the induction of procedural knowledge at the time of guideline execution.
The insight gained from this comparison is that the semantic representation of knowledge expressed in natural language can play a central role in connecting all components of NLP systems, such as the automatic understanding of natural language, the rational reasoning over knowledge bases, or the generation of natural language expressions from formal KRs. Approach 3 leverages the UMLS conceptual knowledge resource to analyze the controlled vocabulary and semantic patterns in free text to assist with sublanguage analysis of the text, using the formalism of conceptual graphs, which can be further used to guide NLP of the text to support automated knowledge formalization. This method provides an abstracted and unified conceptual overview of a selected domain by integrating KR and NLP.
We believe that approach 3 has several advantages over those that use NLP or KR separately. First and most important, the results of sublanguage analysis inform KR, which subsequently supports NLP. Second, the process can be automated to some degree using conceptual knowledge resources such as UMLS and text mining tools, thereby improving the efficiency of knowledge acquisition and formulation. Third, the use of conceptual knowledge resources such as UMLS can improve the interoperability of the KR. Finally such conceptual knowledge can be used to induce procedural knowledge related to CPG execution.
This approach has an inherent limitation, in that the output is not fully computable but is instead semi-structured text or a structured narrative [8] (e.g., partially formatted text such as a nursing progress note). However, an upper-level ontology provides a foundation on which domain experts can build to further increase its computability.
3. A Case Study of Clinical Eligibility Criteria
We use clinical eligibility criteria as an example to contrast the representative KR schemata resulting from the three approaches and to illustrate the importance and feasibility of supporting the symbiosis of KR and NLP. Clinical eligibility criteria define characteristics a patient must possess to qualify for a CPG. An example is “children aged 2 months through 5 years with acute gastroenteritis.”
3.1. Output for Approach 1: Instances but not Necessarily Knowledge
Many biomedical NLP systems have been created to parse clinical text, notably MedLEE, MetaMap, and cTAKES. These systems can be applied to extract named entities and their properties, such as certainty, degree, and quantity. The NLP output contains discrete entities and properties, such as “children” and “acute gastroenteritis”. However, it does not generate reusable knowledge and hence falls short of a KR.
3.2. Output for Approach 2: Ontologies, e.g., ERGO
Tu et al. defined the Eligibility Rule Grammar and Ontology (ERGO). A criterion is categorized as a simple, compound, or complex clinical statement. It defines temporal constraints and quantifiers and uses terminology standards to encode biomedical concepts. Although ERGO comes with an annotation-based NLP algorithm for formalizing free-text criteria into this formalism, many decisions, such as ascertaining whether a criterion is simple or complex and whether a constraint is temporal or quantitative, must be made manually. Therefore, the knowledge in this NLP algorithm is about rules for processing text to fit the target ontology. Such knowledge is procedural knowledge for NLP purposes and is not linguistic knowledge that characterizes how domain experts compose eligibility criteria, which is critical for deducing the semantic patterns in eligibility criteria. A KR that is created independently from NLP considerations, as are many existing biomedical ontologies created to represent the universal truth by philosophers or engineers, is likely to require additional KR effort to support NLP. In this way, users who want to use ontologies to structure guidelines have to use two KRs, one for representing the domain and the other for translating the textual description to the first KR. An expressive ontology such as ERGO can pose significant difficulties for the knowledge formulation step because it requires significant NLP support for knowledge formalization, which often is unavailable in practice and mandates manual work, thereby impeding the process.
3.3. Output for Approach 3: Upper-Ontology, e.g., EliXR
Unlike the previous two approaches, in which KR remains independent from NLP, we propose a three-step process to structure free-text CPGs and translate them into computable rules: (1) perform NLP using conceptual knowledge resources such as UMLS; (2) identify the semantic patterns in text using a dependency parser; and (3) semi-automatically construct a KR based on merged semantic patterns.
This approach is advantageous over others in that the KR is informed by the NLP requirements so that no additional KR is required when extracting text to use the KR as the target structure to populate the knowledge base. We first tested the feasibility of manually using the semantic types and semantic relationships from the UMLS Semantic Network to represent eligibility criteria. We found that by adding five new semantic types and semantic predicates, we were able to construct an UMLS-like semantic network representation for eligibility criteria. Motivated by these promising results, we automated the above process using a dependency parser called AQUA. Rather than using a top-down approach for KR, we used a bottom-up, data-driven approach. The technical details for pattern mining and upper-level ontology construction have been previously reported [11].
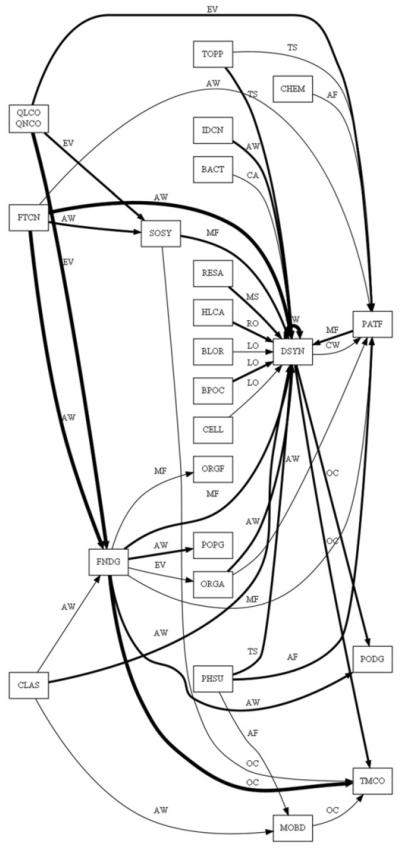
Figure 2 shows the partial network: each node is a UMLS semantic type and each edge is a UMLS semantic relationship. The core semantic patterns cover 86.2% of the 943 eligibility criteria that contained more than one medical concept. Our preliminary study of another 10,000 randomly selected eligibility criterion sentences confirmed that the number of patterns stabilizes at a manageable number as the sample size increases.
Figure 2.
A partial view of the automatically constructed upper-ontology, EliXR, for cancer clinical eligibility criteria. Each UMLS semantic type or semantic relationship is represented by its abbreviation. For example, DSYN represents “diseases or syndromes” and AW represents “associated with”.
4. Discussion
A literature review by Payne showed that text-based KA has been neglected in the informatics community, in terms of rigorous methods for both discovery and of validation [12]. Because KA is so labor intensive, there is a need for a rigorous, data-driven, and reproducible method. Given the significant amount of work to translate free-text CPGs to a structured format, the biomedical informatics research community needs scalable and explicit KA.
Approaches to CPG automation present tradeoffs between labor and accuracy, and the degree of automation affects the kind of language used to express the CPGs. For example, a human author could describe a CPG using a formal language. This facilitates automation of subsequent steps but places a huge burden on authors while restricting the richness of expression. Or, the author could express the CPGs in natural language. This transfers the burden of formalization to someone else, such as a knowledge engineer who must encode the guideline by hand. If we choose to automate the translation of free-text to a formal representation, the burden is transferred to the NLP developer.
Formalization too early in the process could result in simplistic procedures that do not capture the complexity of clinical practice. Translation into a formal KR at a later stage may be hindered by a mismatch between the concepts and processes described by the author and the limited structures afforded by a formal language, leading to errors in translation. A better balance of the distribution of efforts is needed. A semi-structured, iterative process offers such a balance between these extremes. Automated recognition of small-scale semantic units early in the process can benefit later, more complex KR tasks. By examining the ways in which these elements combine in the original text, knowledge engineers may be able to induce concepts and relationships that had not previously been considered. By basing the final KR on real data using a “bottom up” approach, it becomes easier and more scalable to automate the conversion of the text into formal structures. In this type of model, work is distributed between the knowledge engineer and the developer of NLP software, which will likely reduce the total amount of labor by making their processes more harmonious. Such a synergistic model can also facilitate more effective interaction between KA and NLP for guideline formalization. Tools and methods are needed to make text-based KA easier and more integrated with the KA and KR processes.
Researchers have surveyed the common methods of, and problems with, text-based ontology learning. Ontology learning can be considered different tasks, such as concept clustering, lexicon construction, or template generation for information extraction. Many problems remain to be solved in this area. For example, definitions for ontologies differ between the field of philosophy and the fields of computer science or engineering. Our proposed method may not necessarily be able to create an ontology acceptable to certain philosophies for KA for eligibility criteria but it does offer efficiency, scalability, and flexibility from an engineering perspective.
Moreover, there is a wide divide between statistical and symbolic NLP research communities. Neither approach is perfect on its own. An integration of the two is needed to support text-based KA and KR. To enable a feedback loop between iterative KA and NLP, we can take the following 3-step approach: (1) identification of CDE; (2) syntactic and semantic annotation of CDEs in text; and (3) the use of annotations to train CDE extraction and classification. NLP may assist with CDE identification. We can first identify basic CDEs of CPGs, such as population and recommendation. An annotator tool could then use NLP to derive syntactic structure. The user would indicate which text fragments are associated with the CDEs in the CPG text.
5. Conclusions
In summary, KA for KR should be made explicit, scalable, elastic, iterative, and “just expressive enough” to allow NLP-assisted knowledge engineering and increase the facility by which clinical practice guidelines are translated from research into practice. We present a highly efficient and systematic hybrid approach to KA and KR and provide a solid basis for the induction of requisite procedural knowledge from the encoded guidelines and their incumbent conceptual knowledge at the time of execution.
Acknowledgments
This research was funded by grants R01LM009886, R01LM010815, R01HS022961, and UL1TR000040.
References
- [1].Shiffman R, Miche l.G., Krauthammer M, Fuchs N, Kaljurand K, Kuhn T. Writing clinical practice guidelines in controlled natural language. In: Fuchs NE, editor. Conrolled Natural Language. Springer; Heidelberg: 2010. pp. 264–280. 2010. [Google Scholar]
- [2].Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, Sim I. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2011;44(2):239–250. doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wu ST, Kaggal VC, Savova GK, Liu H, Dligach D, Zheng J, Chapman WW, Chute CG. Generality and Reuse in a Common Type System for Clinical Natural Language Processing; Managing Interoperability and Complexity in Health Systems (MIXHS’11); Glasgow, Scotland, United Kingdom. 2011.pp. 27–34. [Google Scholar]
- [4].Peleg M, Keren S, Denekamp Y. Mapping computerized clinical guidelines to electronic medical records: Knowledge-data ontological mapper (KDOM) J Biomed Inform. 2008;41(1):180–201. doi: 10.1016/j.jbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- [5].Serban R, ten Teije A. Exploiting thesauri knowledge in medical guideline formalization. Methods Inf Med. 2009;48(5):468–74. doi: 10.3414/ME0629. [DOI] [PubMed] [Google Scholar]
- [6].Serban R, ten Teije A, van Harmelen F, Marcos M, Polo-Conde C. Extraction and use of linguistic patterns for modelling medical guidelines. Artif Intell Med. 2007;39(2):137–49. doi: 10.1016/j.artmed.2006.07.012. [DOI] [PubMed] [Google Scholar]
- [7].McCray AT. An Upper-Level Ontology for the Biomedical Domain. Comp Funct Genomics. 2003;4(1):80–4. doi: 10.1002/cfg.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Johnson SB, Bakken S, Dine D, Hyun S, Mendonça E, Morrison F, Bright T, Van Vleck T, Wrenn J, Stetson P. An Electronic Health Record Based on Structured Narrative. JAMIA. 2008;15(1):54–64. doi: 10.1197/jamia.M2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Beißwanger E, Schulz S, Stenzhorn H, Hahn U. BioTop: An Upper Domain Ontology for the Life Sciences - A Description of its Current Structure, Contents, and Interfaces to OBO Ontologies. Applied Ontology. 2008;3(4):205–212. [Google Scholar]
- [10].Arp R, Smith B. Function, role, and disposition in Basic Formal Ontology. Nature Precedings. 2008 http://precedings.nature.com/documents/1941/version/1. [Google Scholar]
- [11].Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach toeligibility criteria extraction and representation. JAMIA. 2011;18(Suppl 1):i116–i124. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Payne PRO, Mendonça EA, Johnson SB, Starren JB. Conceptual knowledge acquisition in biomedicine: A methodological review. Journal of Biomedical Informatics. 2007;40(5):582. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]