Abstract
We summarize 12 experimental methods that have been developed for profiling gene expression by focusing on the 3′-end of poly(A+) mRNA, distilling both common and unique features. Of this family of methods, we provide detailed protocol for MAPS, a method we believe is the simplest and most cost-effective for profiling gene expression and quantifying alternative polyadenylation events by oligo-dT priming followed by random priming and deep sequencing. This method also enables library multiplexing by using a set of bar coding primers during PCR amplification. We also provide a general guideline for analysis of the data generated by MAPS by using the software package maps3end.
Keywords: Gene expression profiling, RNA-seq, Alternative polyadenylation, Multiplexing strategies
1 Introduction
Gene expression profiling is a key strategy to study regulated gene expression, to classify disease samples based on gene expression signatures, and to discover potential drug targets. Full-length RNA-seq is now a widely used technology to measure gene expression and detect structural variations in processed transcripts genome-wide. However, the cost is still prohibitive in cases where there are lots of samples and multiple replicates to be analyzed, emphasizing the need for efficient and simplified methods.
Polyadenylation is a conserved process in eukaryotic cells, which proceeds in two essential steps: cleavage at near the 3′-end of the primary transcripts and addition of a polyA tail of 200–250 nt in length [1]. Importantly, more than 40 % genes have been shown to undergo alternative polyadenylation (APA), as detected in zebrafish [2], mice, and humans [3], indicating APA as a critical regulatory step in gene expression. APA can cause 3′ UTR lengthening or shortening, resulting in different platforms for regulation by miRNAs, nonsense-mediated RNA decay (NMD), mRNA localization, and translational control [4, 5]. APA is dynamically regulated during cell proliferation and development via cis-elements, including PAS (Poly(A) Signal), USE (Upstream Sequence Element) and DSE (Downstream Sequence Element), and trans-factors, such as core 3′-end processing factors like CPSF and CstF. APA is coupled with other layers of gene expression and is subject to modulation by RNA-binding proteins like HuR and Nova2 [5]. Mis-regulated APA has been linked to various human diseases, particularly cancers [6]. Researchers are still early in deciphering the polyadenylation code, similar to the splicing code, an effort that began 10 years ago [7].
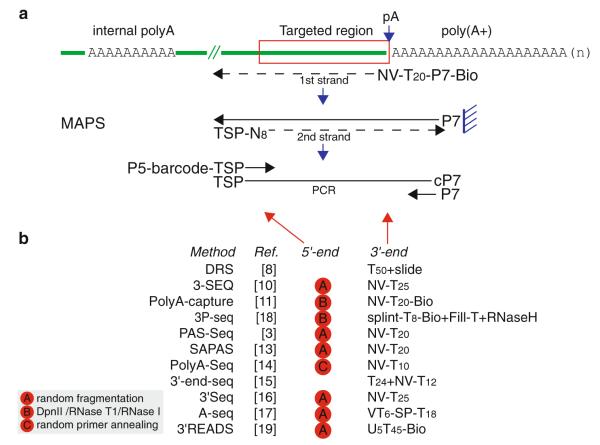
Motivated by the requirement for gene expression profiling and pA site usage quantification, a family of RNA-seq methods named 3′-end RNA-seq has been developed with the basic idea to only count reads in the proximity of pA sites. Sequencing reads mapped at the end of genes reflect the levels of gene expression and pA usage, but do not provide information on other differences in the transcripts, such as alternative promoter usage and alternative splicing. Up to now, there are at least 12 deep sequencing-based methods that have been described in literature, as summarized in Fig. 1. All these methods utilize the unique signature of the long polyA tail for pulling down mRNAs with complementary oligo-dT. One major confounding factor is the internal A-rich sequences, which may be annealed to oligo-dT to generate unwanted signals. Some methods have been specifically designed to address this problem.
Fig. 1.
MAPS and other strategies in 3′-end RNA-seq. (a) Schematic of MAPS procedure. Green line represents an mRNA with polyadenylated tail and one internal polyA stretch. Red box highlights the targeted region where the sequencing reads are supposed to be mapped. (b) Summary of other strategies. The methods are sorted by the publication date, with abbreviated description of experimental designs. TSP target sequencing primer, cP7 reverse complement of P7 primer, Bio biotin, Ref. reference number, SP sequencing primer in a stem-loop, N A/C/T/G, V A/C/G
DRS [8] permits direct sequencing of the captured mRNAs from 50 oligo-dT coated on the slide, which is exemplified with several successful applications [6, 9]. However, this method is only amenable on the Helicos platform, which has not been widely available to the research community. All other methods generally involve several common steps that include extraction of small RNA fragments close to pA sites, conversion of RNA to cDNA by RT, ligation to sequencing adaptors and amplification by PCR, and finally, sequencing on Illumina or 454 sequencers. The main differences lie at the treatment at the 5′-end and 3′-end of short RNA fragments. To remove most upstream sequences from the pA site, three strategies are used: random fragmentation by heating, enzymatic cleavage with DpnII/RNase T1/RNase I, or random priming with octamer. Several creative strategies have been developed to treat the 3′-end. NV-anchor oligo-dT are used in 3-SEQ [10], PolyA capture [11], PAS-Seq [3], MAPS [12], SAPAS [13], PolyA-Seq [14], 3′-end-seq [15], 3′Seq [16], and A-seq [17]. All of these methods require bioinformatics treatment to remove reads from potential internal priming [4]. In contrast, 3P-seq [18] is designed to remove internal polyA priming experimentally by attaching splint oligos to the end of polyA tail followed by selection with biotin. 3′READS [19] uses a composed oligo (U5T45) coupled with stringent wash conditions to enrich for polyadenylated RNA fragments.
We believe that the MAPS method we have developed is the simplest and most cost-effective for profiling the 3′-end of mRNA by RNA-seq, as illustrated in Fig. 1. In this method, oligo-dT linked to the 3′ adaptor is first used to prime reverse transcription from mRNAs. After removing free priming oligos and biotin selection, the second-strand synthesis is initiated on beads with random octamers directly anchored to the 5′ adaptor. The released second-strand products are PCR amplified, which takes advantage of PCR bias in combination with size selection to enrich small fragments near the 3′-end. This method provides strand-specific information. A set of bar-coded primers is individually used in the PCR reactions for multiplexing libraries to be sequenced, permitting genome-wide gene expression profiling of up to 12 libraries per lane on the Illumina HiSeq 2000 platform [12]. Here we describe a detailed protocol for this Multiplex Analysis of PolyA-linked Sequences (MAPS) method and provide a general guide to data analysis by using the software package maps3end.
2 Materials
SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies, cat. no. 18080-051), including 10× RT buffer, 25 mM MgCl2, 0.1 M DTT, 10 mM dNTP mix, 200 U/μL SuperScript III RT, and 40 U/μL RNaseOUT.
NucleoSpin gel and PCR clean-up (Macherey-Nagel, cat. no. 470609).
Terminal transferase (NEB, cat. no. M0315S), including 10× buffer, 2.5 mM CoCl2, and 20 U/μL Terminal Transferase.
Dynabeads Myone Streptavidin C1 (Life Technologies, cat. no. 650.01).
Taq DNA polymerase (NEB, cat. no. M0237L).
Amplitaq Gold DNA polymerase (Life Technology, cat. no. N8080241).
10 mM dNTP mix and 10 mM ddNTP mix.
Washing buffer: 20 mM Tris–Cl, pH 7.6, 0.1 M NaCl, 1 mM disodium EDTA, 0.1 % Tween-80.
Library construction primers – first-strand RT primer: biotin-CAAGCAGAAGACGGCATACGAGT(20) VN; second-strand primer: GCTGATGCTACGACCACAGGN(8).
PCR amplification primers – bar coding primers: AATGATACGGCGACCACCGAGATN(4)GCTGATGCTACGACCACAGG; P7 primer: CAAGCAGAAGACGGCATACGAG.
Sequencing primers – P5 primer (for sequencing barcodes): AATGATACGGCGACCACCGAGAT; target sequencing primer: GCTGATGCTACGACCACAGG.
3 Methods
3.1 Reverse Transcription and Blocking
- Add the following components to denature RNA:
- 1 μg of Trizol-isolated total RNA.
- 1 μL of 50 μM first-strand RT primer.
- 1 μL of 10 mM dNTP mix.
- RNase-free H2O to 10 μL.
Incubate at 65 °C for 5 min and then place on ice for at least 1 min.
- Add the following components to the above mixture to initiate the RT reaction:
- 2 μL of 10× RT buffer.
- 4 μL of 25 mM MgCl2.
- 2 μL of 0.1 M DTT.
- 1 μL of 40 U/μL RNaseOUT.
- 1 μL of 200 U/μL SuperScript III.
Incubate at 50 °C for 50 min and terminate the reaction at 85 °C for 5 min.
Purify the first-strand cDNA with NucleoSpin gel and PCR clean-up kit following manufacturer’s instruction to remove free RT primer.
- Add the following components to block the 3′-end of the cDNA:
- 19 μL of purified cDNA (supplement with water if necessary).
- 2.5 μL of 10× terminal transferase buffer.
- 2.5 μL of 2.5 mM CoCl2.
- 0.25 μL of 10 mM ddNTP.
- 0.25 μL of 20 U/μL terminal transferase.
Incubate at 37 °C for 30 min and terminate reaction at 70 °C for 10 min.
Wash magnetic beads (5 μL per reaction) twice with washing buffer and resuspend in washing buffer (equivalent to the original volume).
Add the beads in washing buffer to the above mixture and incubate at room temperature for 30 min, agitating a few times during the incubation.
Collect beads on a magnetic stand and discard the supernatant.
Briefly denature the cDNA by suspending beads in 100 μL of 0.1 N NaOH, followed by incubation at room temperature for 5 min, and washing the beads twice with H2O.
3.2 Synthesis of the Second-Strand cDNA and PCR
- Add the following components to washed beads to synthesize the second strand:
- 1 μL of 100 μM AD primer.
- 5 μL of 10× Taq DNA Polymerase Buffer.
- 1 μL of 10 mM dNTP mix.
- 1 μL of 5 U/μL Taq DNA polymerase (NEB).
- Add H2O to 50 μL.
Incubate at 25 °C for 60 min, heat at 68 °C for 30 s, 75 °C for 5 min, and immediately put the tubes on magnetic stand and wash the beads twice with washing buffer.
Resuspend beads with 20 μL of H2O.
Heat at 95 °C for 5 min and immediately put the tubes on magnetic stand to collect supernatant containing the released second-strand cDNA.
- Add the following components for PCR amplification (see Note 1):
- 5–20 μL of the second-strand cDNA.
- 5 μL of 10× PCR buffer.
- 3 μL of 25 mM MgCl2.
- 1 μL of 10 mM dNTP mix.
- 1.5 μL of 20 μM P7 primer.
- 1.5 μL of 20 μM bar coding primer.
- 0.3 μL of 5 U/μL Amplitaq Gold DNA polymerase (Life Technologies).
- Add H2O to 50 μL.
Run PCR 20–25 cycles in the thermal cycler using the following program: 94 °C for 10 min (initial denaturation and polymerase activation), 94 °C for 30 s (denaturation), 58 °C for 30 s (annealing), and 72 °C for 30 s (extension).
Examine PCR products in 2 % agarose gel.
Cut band corresponding to 200–400 nt and purify PCR product with NucleoSpin gel and PCR clean-up kit following manufacturer’s instruction.
Quantify the amount of purified PCR products by using a Qubit fluorometer, and pool purified PCR products for multiplex sequencing (see Note 2).
3.3 Sequencing
Use a Qubit fluorometer to quantify the pooled PCR products to make an estimate for loading onto individual lanes of Illumina sequencer (see Note 3).
Load 10 pM of multiplexed libraries to individual lanes of flow cell.
Use the target sequencing primer to sequence a desired length (35–75 nt).
Strip off the sequenced products (using the procedure described in the Illumina sequencing manual) and then prime the flow cell with the P5 primer to sequence the barcode region.
3.4 Data Analysis
Here we describe a basic pipeline for processing MAPS data and also provide some example scripts at https://code.google.com/p/maps3end/
-
Decoding of multiplexed samples.
The output from a MAPS sequencing run is a FASTQ file containing the reads for all samples. The first step is to decode the samples according to specific barcodes incorporated in different libraries. The program decode.py in the maps3end package is designed for this task, which can be run in the following way: decode.py [--startpos=37 –mismatch=0 –outprefix lane1] f_fastq f_barcode. The f_barcode is a tab-delimited text file and contains bar coding information for all libraries sequenced in the given lane. In this text file, each line represents a library where the first column is the barcode sequence and the second column is the unique library name. The option startpos is for setting the start position of the barcode in the read sequence. The option mismatch is the maximum number of mismatches allowed in assigning read to the corresponding barcode (we normally allow 0 or 1 mismatch, as long as the read can be assigned to a single barcode). The decoded reads are written into individual files for each library and any reads that failed to be decoded are saved in the file starting with outprefix for diagnosis of potential sequencing problems.
-
Mapping of sequencing reads.
MAPS generates strand-specific reads from mRNAs, which should be only mapped to the sense strand of genes. Generally we map reads directly to the reference genome (e.g., mm9 for mouse, hg18 for human, see Note 4), which could be downloaded from the UCSC genome browser FTP site [20]. There are multiple programs, such as Bowtie [21] and BWA [22], for mapping the reads to genomes. We normally use Bowtie with the following parameters: bowtie -l25 -n2 -e 200 -m1 –best –strata –trim5 4 EBWT sample.fastq (see Note 5). EBWT is the reference index built from genome sequence in FASTA format by the bowtie-build program. As the beginning nucleotides from random primers are not required for perfect matching with primed sequences, we skip the first 4 nt of reads in mapping. Two mismatches are allowed in the remaining first 25 nt seed sequences. Only the reads uniquely mapped to the genome are kept for further analysis.
-
Filtering reads resulting from internal priming.
Potential internal priming is checked for each read against the downstream sequence up to 300 nt to determine the presence of one or more polyA stretches (consecutive 8 As or 9 As in a 10 nt window) [23]. The polyA stretches can be scanned by the script scan4astretch.py across the sequences of all annotated genes or the genome and saved for later use. The output BED file can be compared with the mapped reads by the intersectBed program in BEDTools [24] to filter out those potential internal priming events.
-
Quantification of gene expression and APA usage.
Even though the current MAPS protocol cannot accurately assign the pA position in each gene (see Note 6 for an alternative sequencing strategy to assign pA sites), the data can be used to quantify gene expression without confounding effect of gene length. For the purpose of gene expression profiling, we sum up all reads mapped to the targeted regions (300 nt upstream of the 3′-end, see Note 7) in annotated genes with more than one 3′-ends. To calculate differential APA usage, we use reads mapped to individual targeted regions that belong to the same gene. We take two steps in computing the quantitative information at each pA site. We first count uniquely assignable reads at individual pA sites and then use the frequency at each site to assign a specific fraction of reads that are mapped to multiple 3′-ends. We provide the scripts gene2land.py and land2exp.py to assist such computation, which report the quantitative information in terms of reads per kb per million mapped reads (RPKM) for each gene or pA site.
Acknowledgments
We acknowledge early contribution of Kristi Fox-Walsh to this method. This work was supported by NIH grants (HG004659) to XDF.
4 Notes
A single bar coding primer from a synthetic set of primers each containing a specific sequence in the barcode region is used in combination with the common primer (P7) to amplify each library. We normally take a small aliquot (2 μL) of the second-strand cDNA to perform preliminary amplification to estimate the yield, based on which to use the right amount of the second-strand cDNA for final PCR amplification in order to obtain detectable products with a minimal PCR cycle number to minimize PCR biases.
For transcriptome profiling in mammalian genomes, we take an equal amount of PCR-amplified materials from each library to make a pool for multiplex sequencing. We normally pool 12 libraries for sequencing on a single HiSeq 2000 lane, which will yield at least ten million reads per library.
Before sequencing, it is important to make an estimation of the materials for loading, which can be determined by various quantitative methods, such as Qubit or real-time PCR. Although 10 pM is recommended for loading into each lane, careful adjustment is required based on previous sequencing results to load the right amount to obtain the optimal density of ~200 millions per lane on HiSeq 2000.
The genome version choice, like mm9 or mm10 both for mouse, depends on user’s preference, because of other existing data for comparative purposes.
Alternatively, to increase the number of assignable reads, we can first map the reads to the mRNA reference sequences and assign remaining reads to the genome reference. We also use TopHat [25] to map reads that span exon-exon junctions.
The library can be sequenced from the 3′-end by using a different sequencing primer to determine the accurate pA sites as described [26].
The 300 nt interval is a rough choice based on the size selection during the preparation of sequencing library. The sequencing data can be used to estimate the average distance between the positions of mapped reads and the actual 3′-ends of transcripts that contain a single pA site.
References
- 1.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Sun Y, Fu Y, et al. Dynamic landscape of tandem 3′ UTRs during zebrafish development. Genome Res. 2012;22(10):1899–1906. doi: 10.1101/gr.128488.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepard PJ, Choi EA, Lu J, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17(4):761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Dowell RD, Yi R. Genome-wide maps of polyadenylation reveal dynamic mRNA 3′-end formation in mammalian cell lineages. RNA. 2013;19(3):413–425. doi: 10.1261/rna.035360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18(12):2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y, Li Z, Ozsolak F, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40(17):8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu XD. Towards a splicing code. Cell. 2004;119(6):736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 8.Ozsolak F, Platt AR, Jones DR, et al. Direct RNA sequencing. Nature. 2009;461(7265):814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 9.Ozsolak F, Kapranov P, Foissac S, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143(6):1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck AH, Weng Z, Witten DM, et al. 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PloS One. 2010;5(1):e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangone M, Manoharan AP, Thierry-Mieg D, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329(5990):432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox-Walsh K, Davis-Turak J, Zhou Y, et al. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics. 2011;98(4):266–271. doi: 10.1016/j.ygeno.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Sun Y, Li Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21(5):741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derti A, Garrett-Engele P, Macisaac KD, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haenni S, Ji Z, Hoque M, et al. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3′-end-seq. Nucleic Acids Res. 2012;40(13):6304–6318. doi: 10.1093/nar/gks282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenal M, Elkon R, Loayza-Puch F, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149(3):538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Martin G, Gruber AR, Keller W, et al. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1(6):753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Jan CH, Friedman RC, Ruby JG, et al. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469(7328):97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoque M, Ji Z, Zheng D, Luo W, et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10(2):133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer LR, Zweig AS, Hinrichs AS, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11(9):1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkening S, Pelechano V, Jarvelin AI, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41(5):e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]