Abstract
Background
Asparaginyl-β-hydroxylase (AAH) promotes cell adhesion, migration, and invasion via Notch activation. AAH’s expression is up-regulated by insulin/IGF signaling through PI3K-Akt, but its protein is independently regulated by GSK-3β. The multiple predicted GSK-3β phosphorylation sites suggest post-translational mechanisms may regulate AAH protein expression.
Methods
Human Huh7 hepatoma cells were transfected with recombinant plasmids that expressed full-length N-terminal Myc-tagged (N-Myc-AAH) or C-terminal HA-tagged (C-HA-AAH) cDNA. Effects of IGF-1 on AAH protein were examined using cellular ELISAs, immunofluorescence, and Western blotting. Effects of kinase inhibitors relevant to AAH’s predicted phosphorylation sites were studied.
Results
IGF-1 stimulation increased AAH protein expression and shifted AAH’s localization from the perinuclear zone to the cell periphery, including podocytes. Subsequently, Notch-1 intracellular domain was translocated to the nucleus, which is critical for Notch- modulated gene expression. Besides GSK-3β, inhibition of PKC, PKA, and CK2, which could potentially phosphorylate AAH, increased IGF-1 stimulated AAH protein. Finally, insulin and LiCl independently and additively increased long-term AAH protein expression.
Conclusion
Insulin/IGF-1 stimulation of AAH and Notch are enhanced by inhibiting kinases that could phosphorylate AAH protein. Targeted manipulation of AAH’s phosphorylation state may have therapeutic value for reducing AAH-Notch activation and attendant infiltrative growth of hepatocellular carcinomas.
Keywords: Hepatocellular carcinoma, insulin, IGF, aspartyl-asparaginyl-β-hydroxylase, Notch
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide [1, 2], and among males in the United States [3]. Molecular mechanisms of HCC carcinogenesis include dysregulation of cell cycle checkpoints, apoptosis [4–6], and growth factor signaling [7–9]. Insulin/insulin-like growth factor (IGF) signaling are up-regulated in HCC [7, 9, 10]. Mechanisms include, gain-of-function mutations in phoshoinositide 3-kinase (PI3K) [11], impaired expression/function of phosphatase and tensin homolog [12], and over-expression of IGF-1 and IGF-1 receptor [9].
Aspartyl-asparaginyl-β-hydroxylase (AAH) is an important target of insulin/IGF signaling, and promotes cell migration and invasion [13–15] necessary for HCC cell infiltration and metastatic spread [16]. AAH is an ~86 kD [13] Type 2 transmembrane protein located in the endoplasmic reticulum (ER) [17, 18]. AAH is physiologically cleaved into a ~30–34 kD N-terminal fragment that is identical to Humbug (Junctate), a truncated isoform that binds calcium and promotes adhesion [19, 20], and a ~52–56 kD C-terminal fragment that has catalytic activity [21]. The C-terminal catalytic domain of AAH promotes cell motility by hydroxylating specific aspartate (Asp) and asparagine (Asn) residues contained within a consensus sequence of epidermal growth factor (EGF)-like domains, including those found in Notch and Jagged [16, 22, 23]. Correspondingly, AAH expression is functionally linked to Notch pathway activation in HCC [16].
Notch signaling starts by binding of Jagged (or Delta-like family of proteins) to Notch’s extracellular domain, triggering two rapid cleavage events. The Notch extracellular domain is first cleaved from the transmembrane domain in a metalloprotease-dependent manner. This rapidly promotes the second cleavage event, which is protease-dependent and releases the Notch intracellular domain (NID) [24]. The NID then translocates to the nucleus where it complexes with the transcriptional activator CBF1, suppressor of hairless or lag-1 (CSL), and transcriptional co-activators of the mastermind-like family of proteins. The complex binds to CSL consensus sequences on DNA, displaces co-repressors and recruits additional co-activators to promote transcription of Notch targets such as hairy and enhancer of split-1 (HES-1) and hairy/enhancer-of-split related with YRPW motif protein-1 (HEY-1) [25].
Insulin and IGF regulate AAH at both transcriptional and post-translational levels by signaling through PI3K [26]. With regard to post-translational processes, inhibition of glycogen synthase kinase-3β (GSK-3β) rather than activation of Akt is critical for increasing AAH protein, independent of its mRNA [26–28]. A potential role for direct phosphorylation of AAH as a means of regulating its protein expression and function was suggested by the findings that: 1) chemical or siRNA-targeted inhibition GSK-3β increases AAH protein expression and motility, while over-expression of constitutively active GSK-3β inhibits AAH protein expression and motility; 2) AAH protein has a number of consensus sequence sites for phosphorylation by GSK-3β, protein kinase A (PKA), protein kinase C (PKC), and casein kinase 2 (CK2); and 3) AAH migrates as a ~140 kD protein on SDS-PAGE, which is larger than its predicted ~86 kD mass.
Therapeutic targeting of AAH protein could provide a novel means of regulating HCC’s malignant infiltrative growth. Importantly, the potential exists to interfere with AAH’s function by altering its phosphorylation, which may be important for Notch pathway activation. The present study characterizes insulin/IGF-1 stimulation of AAH protein, AAH phosphorylation, and Notch activation, and examines the roles of specific kinases in relation to these responses in Huh7 human HCC cells. Given the relatively high levels of endogenous AAH in HCC cells and inability to distinguish the N-terminal domain of AAH from Humbug, we generated N-tagged and C-tagged AAH cDNA constructs to conduct these experiments.
Materials and Methods
Materials
Recombinant IGF-1 was purchased from Sigma-Aldrich (St. Louis, MO, USA). pcDNA 3 vector, Escherichia coli DH5α cells, Dulbecco’s Modified Eagle Medium, Lipofectamine 2000 Transfection Reagent, Hoechst 33342, and Amplex UltraRed were purchased from Invitrogen (Carlsbad, CA, USA). Non-essential amino acid mixture was purchased from Gibco-BRL (Grand Island, NY, USA). pcDNA 3 vector with a 6× Myc-tag was a gift from Dr. Y. Eugene Chin from Brown University (Providence, RI, USA) [29]. QIAquick Gel Extraction Kit and QIAprep Spin Miniprep Kit were purchased from Qiagen (Valencia, CA, USA). MaxiSorb plates, OptiPlates (96-well), BD Falcon culture inserts, and Nunc culture supplies were obtained from Thermo Scientific (Rochester, NY, USA). Polyvinylidene fluoride membranes were purchased from Perkin-Elmer (Waltham, MA, USA). Myc antibody purchased from Cell Signaling Technologies (Danvers, MA, USA) and HA antibody purchased from Santa Cruz Biotechnologies (Dallas, TX, USA). The A85G6 and FB50 mouse monoclonal antibodies to AAH were characterized as previously described [30, 31]. SuperBlock, bicinchoninic assay, enhanced chemiluminescence reagents, and Dylight 547 Conjugated to Streptavidin were purchased from Pierce (Rockford, IL, USA). Fisherbrand Superfrost Plus Stain Slides were purchased from Fisher Scientific (Pittsburgh, PA, USA) and SpectraMax M5 Microplate Reader from Molecular Dynamics (Sunnyvale, CA, USA). Histofix was purchased from Amresco (Solon, Ohio, USA) and Shandon Cytospin Centrifuge 3 from Thermo Shandon (Pittsburgh, PA, USA). Other fine chemicals were purchased from CalBiochem (Carlsbad, CA, USA) or Sigma-Aldrich (St. Louis, MO, USA).
Recombinant AAH plasmid constructs
The coding region of human AAH was amplified from a 293T cell cDNA library by the polymerase chain reaction (PCR) using the forward primer 5′-CGGAATTCATGGCCCAGCGTAAGAATGCCA-3′, reverse primer 5′-CCGCTCGAGCTAAATTGCTGGAAGGCTGC-3′ and Pfu DNA polymerase [13]. The AAH PCR product was digested with EcoRI and XhoI restriction enzymes and gel purified with the QIAquick Gel Extraction Kit. A pcDNA 3 vector with a 5′-end 6× Myc-tag insert was received as a gift and original pcDNA 3 vector was also engineered to contain a 3′-end 2× HA tag using the forward primer: 5′-GCAGGATCCTACCCATACGATGTTCCTGACTAT-3′ and reverse primer: 5′-TAACGGTACCAAGCTTGCATAGTC-3′. The AAH PCR product was cloned into the Myc-modified (pCMV-N-Myc-AAH) or the HA-modified pcDNA 3 vector (pCMV-C-HA-AAH). The recombinant plasmids were transformed into Escherichia coli DH5α competent cells and positive clones cultured in Luria Broth media. The plasmids were purified with the QIAprep Spin Miniprep Kit and correct insert sequence and orientation was verified by DNA sequencing. Lastly, protein expression was verified in 293T and Huh7 cells by Western blot.
Cell culture
Huh7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with heat-inactivated 5% fetal bovine serum (FBS), 10 mM non-essential amino acid mixture, and 2 mM L-glutamine in 5% CO2 at 37°C. Huh7 cells were transiently transfected with pCMV-N-Myc-AAH, pCMV-C-HA-AAH, or empty vector (EV) control at semi-confluency using Lipofectamine 2000 and co-transfected with a green fluorescent protein plasmid to monitor transfection efficiency. To determine the effects of growth-factor stimulation on recombinant AAH protein, 24-hr cultures expressing N-Myc-AAH or C-HA-AAH were placed in media with 1% FBS for 4 hrs and treated with vehicle or stimulated with IGF-1 (50 ng/mL) for up to 60 min. To assess the role of kinase inhibition on IGF-1-stimulated AAH, Huh7 were pre-treated with inhibitors of GSK-3β (20 mM LiCl), casein kinase 2 (CK2; 5 μM TBCA/TBB), protein kinase A (PKA; 20 μM H-89), protein kinase C (PKC; 1 μM Gö6983), or MAPK/ERK (20 μM PD98059) control for 4 hrs, followed by stimulation with IGF-1 (50 ng/ml). To assess the role of long-term of insulin stimulation and GSK-3β inhibition on N-Myc-AAH, cells were placed in 1% FBS media for 4 hrs, followed by insulin stimulation and 20 mM LiCl treatment for 16 hrs.
Protein studies
Huh7 protein homogenates were prepared in radio-immunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 2 mM EGTA) in the presence of protease (1mM PMSF, 0.1 mM TPCK, 1 mg/ml aprotinin, 1 mg/ml pepstatin A, 0.5 mg/ml leupeptin, 1 mM NaF, 1 mM Na4P2O7) and phosphatase (2 mM Na3VO4) inhibitors [32]. Protein homogenates were centrifuged at 14000×g for 10 min at 4°C and protein concentration in supernatant was measured by the bicinchoninic acid (BCA) assay. For Western blot analysis, 30 μg of protein were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes that were subsequently incubated with SuperBlock in TRIS buffered saline (TBS) to prevent non-specific binding. After washing, the membranes were incubated with primary antibodies (0.1 μg/ml) overnight at 4°C. Immunoreactivities were detected with horseradish peroxidase (HRP)-conjugated secondary antibody, SuperSignal enhanced chemiluminescence reagents, and film autoradiography.
Cellular ELISA
The cellular ELISA assay is an efficient method to rapidly and directly quantify protein under several treatment conditions in 96-well plate format [33]. Huh7 96-well cultures were fixed with Histofix, permeabilized with 0.05% saponin in TBS, and endogenous peroxidase activity blocked with 0.03% hydrogen peroxide in phosphate buffered saline (PBS). Non-specific protein binding was prevented by incubation with SuperBlock-TBS. After washing, the cells were incubated with primary antibodies (0.1 μg/ml) overnight at 4ºC and target protein immunoreactivity was detected with HRP-conjugated secondary antibodies and Amplex UltraRed fluorophore (Ex 530 nm/Em 590 nm). Ratios of target protein/cell density with Hoechst 33342 dye (Ex 360 nm/Em 460 nm) and fluorescence measured with SpectraMax M5 microplate reader. Ratios of target protein/cell density allowed for inter-well comparisons. 4–6 replicate cultures were analyzed for each experiment.
Statistical analysis
Data depicted in the graphs represent the means ± standard error of the mean (S.E.M.) for each group. Data (N=4 independent cultures/group) were analyzed using GraphPad Prism 5 software. Inter-group comparisons were made using repeated measures of one-way or two-way analyses of variance (ANOVA) with Fisher’s Least Significant Difference (LSD) post test. Computer software generated P-values are indicated within the graph panels (*P<0.05; **P<0.01; ***P<0.001; and **** P<0.0001).
Results
Effects of IGF-1 stimulation on AAH protein expression
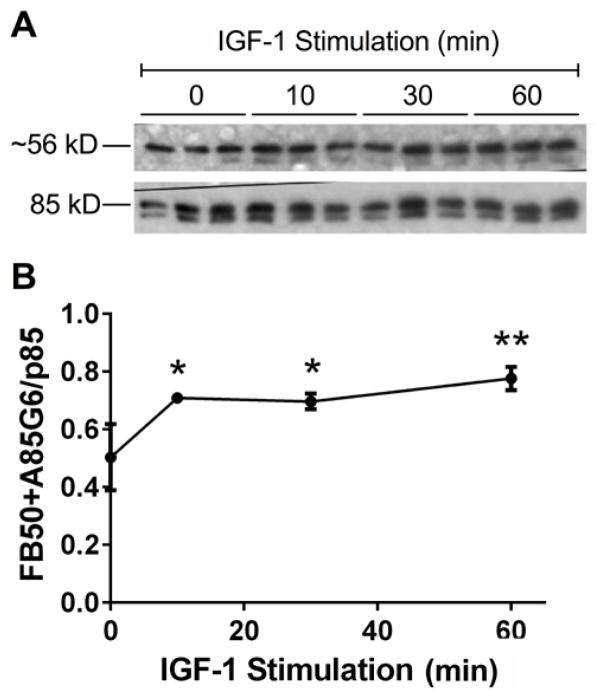
Western blot analysis was used to measure effects of IGF-1 stimulation on AAH protein expression (Figure 1). Short-term stimulation with IGF-1 significantly increased endogenous AAH (FB50+A85G6) expression relative to unstimulated cells. The ~56 kD band corresponds to the C-terminal catalytic domain of AAH, which is the main cleavage product detected in Huh7 cells. These results demonstrate that endogenous AAH accumulates in response to IGF-1 stimulation.
Figure 1. IGF-1 Stimulated AAH protein expression.
Sub-confluent Huh7 cultures were stimulated with IGF-1 (10 ng/ml) for 0–60 min and used for Western blot analysis. (A) FB50+A85G6 monoclonal antibodies were used to detect AAH (3 cultures/time point). The blots were stripped and re-probed with rabbit polyclonal anti-p85-PI3K as a loading control. (B) Western blot signals were quantified by digital imaging. The graph depicts the mean (± S.E.M.) relative levels of AAH protein (FB50+ A85G6/p85-PI3K pixel intensity ratios) per point. Data were analyzed by 1-way ANOVA with the Dunnett post-hoc test (*P<0.05 and **P<0.01 relative to control (0 time point).
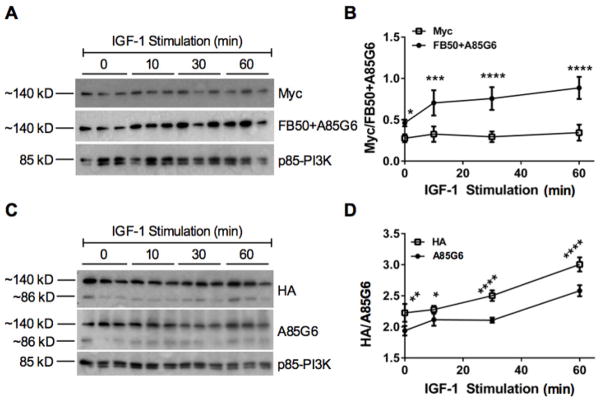
To increase the signal intensity of full-length AAH, N-terminally tagged (N-Myc-AAH) or C-terminally tagged (C-HA-AAH) AAH cDNAs (CMV promoter) were transiently expressed in Huh7 cells. Blots generated with N-Myc-AAH transfected cells were probed with antibodies to Myc, FB50+A85G6 (AAH), and p85 subunit of PI3K (p85-PI3K) (Figure 2A), and blots made with C-HA-AAH transfected cells were probed with antibodies to HA, A85G6, and p85-PI3K (Figure 2C). The p85-PI3K immunoreactivity was used as an internal “housekeeping” control as the levels are invariant with experimental treatments. Graphs (Figure 2B and 2D) depict changes (Mean ± S.D.; N=3) in FB50+A85G6/p85, Myc/p85, or HA/p85 signal intensities over time following IGF-1 stimulation. The FB50 antibody detects the N-terminus of AAH, while the A85G6 detects the C-terminal region of AAH [31, 34]. Short-term IGF-1 stimulated increases in Myc and HA immunoreactivity were more difficult to detect than the signals generated with the AAH-specific antibodies. Following both N-Myc-AAH and C-HA-AAH transfections, AAH immunoreactivity significantly increased over time relative to unstimulated cells (0 min), and relative to the signals detected with either the Myc or HA antibodies. One reason for the discrepancies is that the AAH-specific antibodies have very high affinities and specificities [31, 34]. As mentioned, although AAH has a calculated molecular weight of ~86 kD, it migrates at ~120–140 kD by Western blot analysis, possibly due to post-translational modifications.
Figure 2. AAH expression in N-Myc-AAH and C-HA-AAH transfected cells.
Huh7 cells were transfected with recombinant plasmid DNA carrying the full-length human AAH cDNA (CMV promoter) fused in-frame with either an N-terminal Myc tag (N-Myc-AAH) or C-terminal HA tag (C-HA-AAH). 24 hours after transfection, cultures stimulated with IGF-1 (10 ng/ml) for 0–60 min were subjected to Western blot analysis using antibodies to (A) Myc or FB50+A85G6, or (C) HA or A85G6. Blots were stripped and re-probed with antibodies to p85-PI3K (loading control). (B, D) Digital imaging was used to quantify the Western blot signals. Graphs depict calculated mean ± S.E.M. signal intensity ratios of AAH/p85-PI3K. Data were analyzed by 1-way ANOVA with the Dunnett post-hoc significance test (*P<0.05, **P<0.01, ***P<0,001; ****P<0.0001 relative to control (0 time point).
IGF-1 stimulated intra-cellular trafficking of AAH and Notch-1
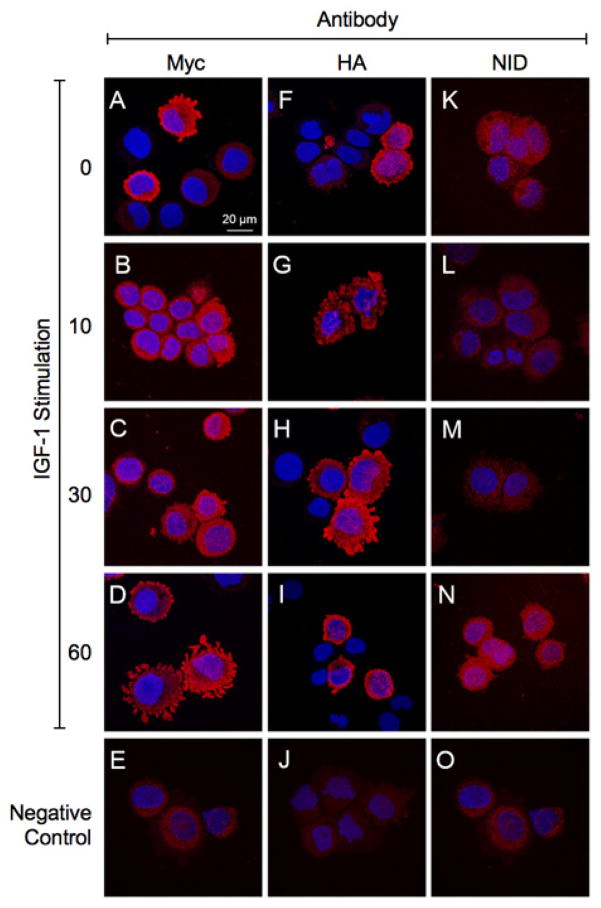
We next examined time course-dependent intra-cellular trafficking of N-Myc-AAH and C-HA-AAH in IGF-1 stimulated cells. In addition, we examined NID immunoreactivity in C-HA-AAH transfected cells. These studies required the use of Myc and HA antibodies to detect immunoreactivity and distinguish responses in recombinant from endogenous AAH protein. The IGF-1 stimulated intra-cellular trafficking patters of the N-terminus (Myc-tagged) and C-terminus (HA-tagged) fragments of AAH differed (Figure 3). In unstimulated cells (0 min), the N-terminus of AAH (Myc-tagged) was distributed at the periphery of cells (Figure 3A). After IGF-1 stimulation, Myc immunoreactivity shifted to the ER/perinuclear zone (10 and 30 min) (Figures 3B and 3C), followed by its return to the cell periphery, including within prominent cell processes (60 min) (Figure 3D). In contrast, C-HA-AAH immunoreactivity was mainly distributed in the perinuclear zone and nucleus (Figure 3F), as was previously described using the FB50 antibody to detect endogenously express AAH in HCC [34]. After IGF-1 stimulation, HA immunoreactivity appeared particulate or speckled, corresponding to microsomal or mitochondrial patterns (10 min) (Figure 3G). Subsequently, HA immunoreactivity increased in perinuclear, nuclear, and cell periphery, including processes/podocytes (30 min) (Figure 3H). HA immunoreactivity was tightly localized in the perinuclear zone and nucleus, 60 min after IGF-1 stimulation (Figure 3I). Note that C-HA-AAH contains the catalytic subunit which hydroxylates Notch. Notch-1 immunoreactivity increased and translocated from cytoplasm to the nucleus follows the trafficking of HA-AAH to the cell periphery (Figures 3K–3N). Negative control studies included parallel incubations in which the primary antibodies were omitted (Figures 3E, 3J, 3O).
Figure 3. Time-dependent shifts in the N- and C-terminal regions of AAH, and levels of Notch-1 immunoreactivity following IGF-1 stimulation.
Huh7 cells were transfected with (A–E, K–O) N-Myc-AAH or (F–J) C-HA-AAH and stimulated with IGF-1. Cells were cytospun onto glass slides and immunostained with antibodies to (A–D) Myc, (F–I) HA, or (K–N) Notch-1 intracellular domain (NID). Immunoreactivity was detected with biotinylated secondary antibody and Streptavidin-conjugated Dylight 547 (red). (E, J, O) Negative control studies primary antibodies omitted. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue) and imaged by confocal microscopy. Merged images are depicted. (600× original magnification, 2× digital zoom).
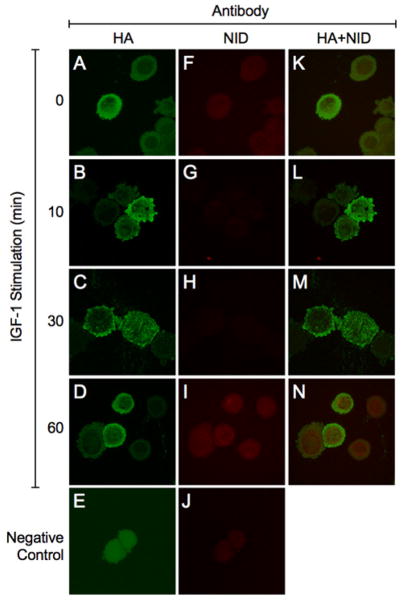
Further studies of Huh7 cells transfected with C-HA-AAH and stimulated with IGF-1 examined Notch-1’s intracellular domain (NID) expression and nuclear localization in relation to intracellular trafficking of C-HA-AAH. To accomplish this, transfected Huh7 cells were IGF-1 stimulated for 0–60 minutes and double-stained by immunofluorescence to detect HA and NID (Figure 4). Confocal microscopic imaging demonstrated that in un-stimulated cells (0 min), C-HA-AAH was localized in the perinuclear and ER zones (Figure 4A) while low levels of NID immunoreactivity were detected in the nuclear and peri-nuclear zones (Figure 4F). Within 10 minutes of IGF-1 stimulation, C-HA-AAH immunoreactivity shifted to the cell periphery and podotytes (Figure 4B), whereas NID immunoreactivity was reduced and scarcely detectable (Figure 4G). At the 30-minute time point, C-HA-AAH immunoreactivity was detected in both podocytes (although less than at 10 minutes) as well as in a particulate pattern corresponding to the ER and microsomal structures (Figure 4C), whereas NID immunoreactivity was still scarcely detectable. However, at the 60-minute time point, C-HA-AAH immunoreactivity returned to the nuclear and perinuclear zones (Figure 4D), and NID was abundantly present in the nucleus (Figure 4I). Merged images revealed low levels of nuclear NID from 0 to 30 minutes after IGF-1 stimulation, and abundant nuclear NID immunoreactivity in cells with high levels of C-HA-AAH localized to the nucleus and peri-nuclear zones (Figure 4N).
Figure 4. Co-localization of C-HA-AAH and Notch-1 intracellular domain.
Huh7 cells transfected with C-HA-AAH (A–J) were stimulated with IGF-1. Cytocentrifuge preparations of the cells were double-immunostained with antibodies to (A–D) HA and (F–I) Notch-1 intracellular domain (NID). Immunoreactivity was detected with biotinylated secondary antibody and Streptavidin-conjugated Dylight 547 for HA (green) or Dylight 647 for NID (red). Cells were imaged by confocal microscopy. (K–N) Merged images depict co-localization of HA and NID. (E, J) Negative control studies showing results with primary antibodies omitted. (600× original magnification, 2× digital zoom).
Kinase inhibitor effects on endogenous AAH protein expression
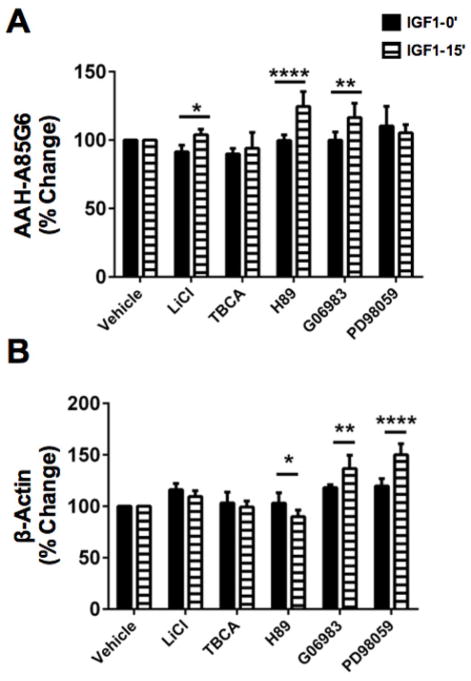
Given the finding that AAH protein has multiple phosphorylation motifs corresponding to mainly GSK-3β, but also PKA, PKC, and CK2, we conducted further studies to examine endogenous AAH protein expression following treatment with specific kinase inhibitors. HuH7 cells seeded in 96-well cultures were treated with inhibitors of GSK-3β (LiCl) [35,36], CK2 (TBCA/TBB), PKA (H89), PKC (Gö6983), MEK (PD98059), or CK1 (D4476), and then stimulated with IGF-1 or nothing (serum-free media treated) for 15 minutes. PD98059 and D4476 served as controls since there are no CK1 or MEK phosphorylation sites on AAH. Immunoreactivity to AAH was measured by cellular ELISA using the A85G6 AAH monoclonal antibody (catalytic domain). Parallel studies measured β-actin expression. Immunoreactivity was normalized to H33342 fluorescence, which correlates with cell density [30, 35].
ANOVA tests demonstrated significant differences in AAH (F=18.72; P=0.0042) (Figure 5A) and β-actin (F=18.44; P<0.0001) (Figure 5B) expression in IGF-1 stimulated, kinase inhibitor treated relative to control (vehicle-treated unstimulated or stimulated) (Table 1). Post hoc Fisher’s LSD test showed AAH was significantly increased by IGF-1 stimulation and pre-treatment with LiCl (P<0.05), H-89 (P<0.0001), and Gö6983 (P<0.01), but not TBCA or PD98059 (Figure 5). Note that AAH is predicted to have GSK-3β, PKA, and PKC phosphorylation sites [30]. Both basal and IGF-1 stimulated β-Actin levels increased in cultures treated with Gö6983 (P<0.01) or PD98059 (P<0.0001), and decreased in IGF-1 versus vehicle stimulated, H-89-treated cultures (P<0.05). Therefore, except for the Gö6983 effects, the responses with respect to AAH expression were dissimilar from with those of β-actin, possibly reflecting specificity (Figure 5).
Figure 5. Kinase inhibitor modulation of AAH expression.
Huh7 96-well cultures were pre-treated with vehicle, LiCl (GSK-3β inhibitor), TBCA (CK2 inhibitor), Gö6983 (pan-PKC inhibitor), H-89 (PKA inhibitor), or PD98059 (MEK-1 inhibitor) for 4 hrs and then stimulated with nothing or of IGF-1 for 15 min. Immunoreactivity to (A) AAH-A85G6 and (B) β-Actin was measured by cellular ELISA with results normalized to Hoechst H33342 fluorescence (cell density). Graphs depict the mean ± S.E.M of 4 replicate cultures per group. Inter-group comparisons were made by two-way ANOVA (Table 1) and post hoc Fisher’s Least Significance Difference tests (*P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001).
Table 1.
Differential effects of kinase inhibitors on short-term IGF-1 stimulated AAH immunoreactivity
| IGF1 Stimulation | Kinase Inhibitor | IGF-1 × Inhibitor | ||||
|---|---|---|---|---|---|---|
| Protein | F-Ratio | P-Value | F-Ratio | P-Value | F-Ratio | P-Value |
| A85G6 | 13.99 | 0.0003 | 35.18 | <0.0001 | 18.72 | 0.0042 |
| A85E6 | 12.41 | <0.0001 | 59.28 | <0.0001 | 14.88 | <0.0001 |
| FB50 | 3.57 | N.S. | 57.67 | <0.0001 | 3.18 | N.S. |
| β-Actin | 1.40 | N.S. | 66.43 | <0.0001 | 18.44 | <0.0001 |
Huh7 human hepatoma cells grown in 96-well micro-cultures were pre-treated with vehicle (control), LiCl, TBCA, Gö6983, H-89, or PD98059 to inhibit GSK-3β, CK2, pan-PKC, PKA, or MEK, and then stimulated for 15 minutes with 10 ng/ml IGF-1. Cellular ELISAs measured immunoreactivity to AAH (A85G6-C-terminus; A85E6 and FB50-N-terminus) and β-actin. Data were analyzed by Two-way ANOVA. Corresponding graphs with post-hoc Fisher LSD results are shown in Figure 5.
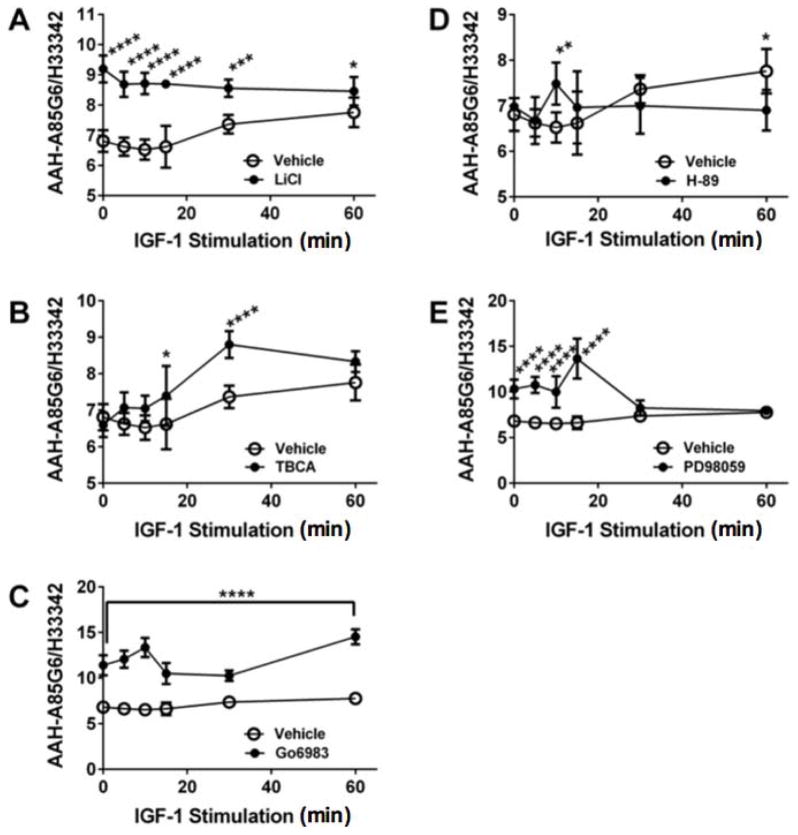
Further studies were performed using cellular ELISAs to characterize the time course of IGF-1 stimulated AAH protein (A85G6) in the presence or absence of LiCl to inhibit GSK-3β, TBCA to suppress CK2, Gö6983 to inhibit pan-PKC, H-89 to inhibit PKA, or PD98059 to inhibit MEK (Table 2; Figure 6). Four replicate cultures were assayed at each time point. Those studies demonstrated significant inter-group differences in AAH protein expression in cells stimulated with IGF-1 and treated with LiCl (F=8.92; 0.0008; Figure 6A), TBCA (F=9.18; P=0.032; Figure 6B), Gö6983 (F=6.42; P<0.0001; Figure 6C), H89 (F=25.81; P<0.016; Figure 6D), and PD98059 (F=24.42; P<0.0001; Figure 5E). LiCl (0.05<P<0.0001), Gö6983 (P<0.0001), and PD98059 (Figure 6E) pre-treatments significantly increased both basal and IGF-1 stimulated AAH. The responses to LiCl and Gö6983 were sustained throughout the time course (60 min), whereas for PD98059, the effect was only observed during the early time points. In contrast, the main effect of TBCA was to increase AAH expression at intermediate time points (15 and 30 min), and not the early or latest time points. Reponses to H-89 were significant at the 10- ad 60-minute time points, but overall, the effects of H-89+IGF-1 were modest. In essence, inhibition of GSK-3β or pan-PKC resulted in sustained increases in AAH protein; otherwise, the responses were transient (TBCA and PD98059) or limited (H-89) (Figure 6).
Table 2.
Differential effects of kinase inhibitors on long-term IGF-1 stimulated AAH immunoreactivity
| IGF-1 Stimulation | Kinase Inhibitor | IGF-1 × Inhibitor | ||||
|---|---|---|---|---|---|---|
| Inhibitor | F-Ratio | P-Value | F-Ratio | P-Value | F-Ratio | P-Value |
| H-89 | 16.95 | 0.083 | 0.21 | N.S. | 25.81 | 0.016 |
| PD98059 | 11.84 | 0.0003 | 49.81 | <0.0001 | 24.42 | <0.0001 |
| LiCl | 75.57 | <0.0001 | 3.69 | 0.071 | 8.92 | 0.0008 |
| Go6983 | 9.09 | <0.0001 | 79.37 | <0.0001 | 6.42 | <0.0001 |
| TBCA | 53.54 | <0.0001 | 13.55 | <0.0001 | 9.18 | 0.032 |
Huh7 human hepatoma cells grown in 96-well micro-cultures were pre-treated with vehicle (control), LiCl, TBCA, Gö6983, H-89, or PD98059 to inhibit GSK-3β, CK2, pan-PKC, PKA, or MEK, and then stimulated for 60 minutes with 10 ng/ml IGF-1. Cellular ELISAs measured immunoreactivity to AAH (A85G6-C-terminus; A85E6 and FB50-N-terminus) and β-actin. Data were analyzed by Two-way ANOVA. Corresponding graphs with post-hoc Fisher LSD results are shown in Figure 6.
Figure 6. Time course of kinase inhibitor modulation of AAH immunoreactivity in IGF-1 stimulated cells.
Huh7 96-well cultures were pre-treated with vehicle, (A) LiCl, (B) TBCA, (C) Gö6983, (D) H-89, or (E) PD98059 for 4 hrs and then stimulated with IGF-1 for 0–60 min. Immunoreactivity to (A) AAH-A85G6 and (B) β-Actin was measured by cellular ELISA with results normalized to Hoechst H33342 fluorescence (cell density). Graphs depict the mean ± S.E.M of 4 replicate cultures per group. Inter-group comparisons were made using two-way ANOVA (Table 2) and the post hoc Fisher’s Least Significance Difference tests (*p<0.05; **p<0.01; ***p<0.001; and ****P<0.0001).
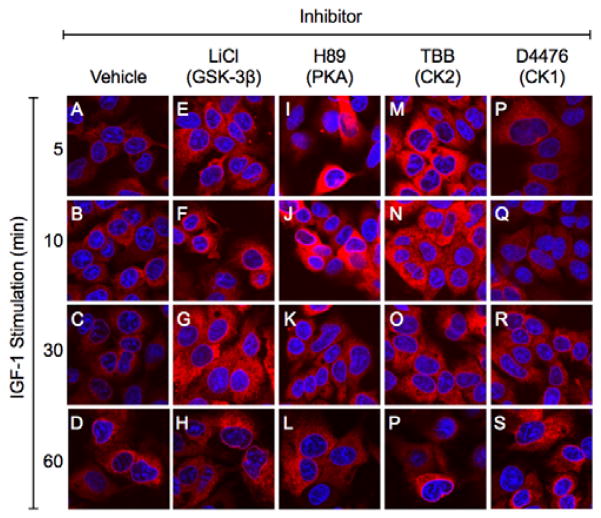
Immunofluorescence staining and confocal microscopy revealed low but progressive increases in IGF-1 stimulated AAH in cells treated with vehicle or D4476 (CK1 inhibitor) (Figures 7A–7D, 7P–7S). Corresponding with the cellular ELISA results, conspicuously higher levels of AAH immunoreactivity were observed in IGF-1 stimulated, LiCl-treated cultures throughout all time points (Figures 7E–7H). The effects of TBB (CK2 inhibitor were more robust (Figure 7M–7P) compared with TBCA (Figure 6B). The responses to H-89 were discordant with the findings by cellular ELISA, and instead suggest that inhibition of PKA prominently increases AAH protein expression in the earliest time periods following IGF-1 stimulation. In general, the increases in AAH immunoreactivity were distributed throughout the cytoplasm as well as the perinuclear zone. The pattern of diffuse cytoplasmic staining was reticular or particulate, consistent with the known microsomal and mitochondrial distributions of AAH.
Figure 7. Kinase inhibitor modulation of IGF-1 stimulated shifts in AAH intra-cellular immunoreactivity.
Huh7 chamber slide cultures were pre-treated for 4 h with vehicle, LiCl, H-89, TBB, or D4476, and then stimulated with IGF-1 for 5 to 60 min. Cells were stained to detect AAH using the FB50 antibody, biotinylated secondary antibody, and Cy-Dye fluorophore (red). Cells were counterstained with DAPI (blue) and imaged by confocal laser microscopy. Merged images are depicted.
Effects of LiCl-GSK-3β inhibition on insulin-stimulated AAH in HuH7 cells transfected with C-HA-AAH or N-Myc-AAH recombinant cDNA plasmids
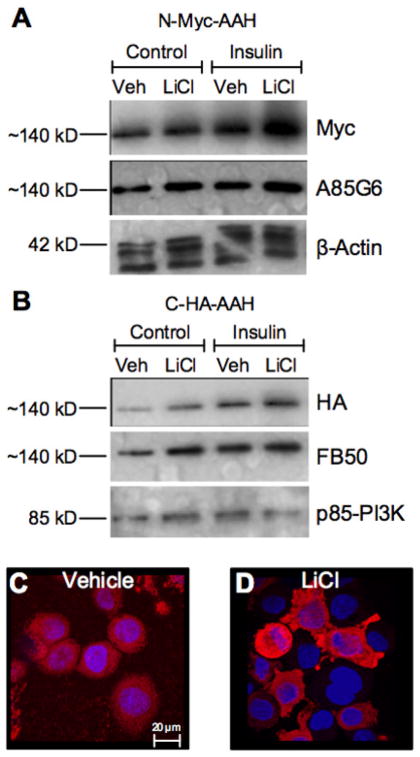
In the final experiment, we focused on the role of GSK-3β in relation to AAH expression since most of AAH’s predicted phosphorylation sites have GSK-3β consensus sequences, and previous studies showed that GSK-3β could directly phosphorylate AAH [30, 31]. We took advantage of our C-HA-AAH and N-Myc-AAH constructs for this experiment because most of the GSK-3β phosphorylation sites are located in the N-terminus of this Type 2 transmembrane protein. We used Western blot analysis to examine long-term effects of insulin stimulation on AAH protein, in the presence or absence of LiCl pre-treatment. In cells transfected with N-Myc-AAH or C-HA-AAH, both insulin and LiCl increased AAH protein expression relative to vehicle (Figures 8A–8B). Similar results were obtained using monoclonal antibodies to the C-terminus (A85G5) or N-terminus (FB-50) of AAH. Correspondingly, immunofluorescence staining and confocal imaging demonstrated higher levels of AAH immunoreactivity in insulin-stimulated, LiCl-treated versus vehicle treated cells (Figures 8C–8D), and a more prominent nuclear to perinuclear localization of AAH compared with short-term stimulation effects (Figures 8E–8H).
Figure 8. Effects of GSK-3β inhibition on AAH expression after long-term insulin stimulation.
Huh7 cells were transfected with N-Myc-AAH or C-HA-AAH cDNA, pre-treated with vehicle or LiCl, and insulin stimulated for 16 h. (A, B) Western blots were probed with antibodies to Myc or HA, then stripped and re-probed with the A85G6 or FB50 antibodies to AAH, followed by a third probing with anti-p85-PI3K. (C, D) N-Myc-AAH transfected cells cyto-centrifuged onto glass slides, were stained by immunofluorescence with anti-Myc (red) and counterstained with DAPI (blue). Images were acquired by confocal microscopy.
Discussion
Overarching concept
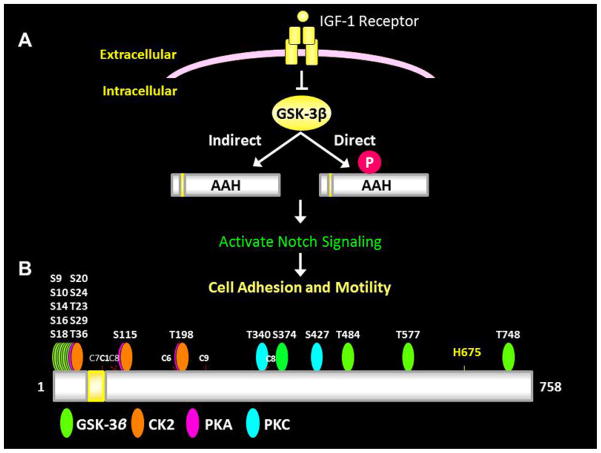
In HCCs, insulin and IGF signaling pathways are up-regulated [9, 36, 37]. AAH is over-expressed in HCC due to its regulation by insulin/IGF through both Erk-MAPK and PI3K-Akt pathways. Since AAH has functional roles in regulating cell adhesion, motility, and invasion [32], its over-expression HCCs could account for the relentless and aggressive infiltrative growth of these neoplasms. Correspondingly, AAH protein expression is abundantly expressed at infiltrating HCC tumor margin, which could be critical for mediating invasion of surrounding tissue and intrahepatic dissemination of HCC [38]. Although AAH stimulation occurs at the mRNA level, previous studies highlight the importance of distinct regulators of AAH’s protein that are independent of gene transcription. This phenomenon was made evident in studies showing that treatment of neuroblastoma cells with LiCl increased AAH’s protein but had no effect on its mRNA [26]. Further studies pointed to direct roles of GSK-3 regulation of AAH protein such that high levels of GSK-3β activity reduce AAH protein, while inhibition of GSK-3β increases AAH protein expression, again without altering the mRNA levels [30]. It was therefore logical that activation of insulin/IGF-1 signaling networks through PI3K-Akt could have long-term effects on AAH’s mRNA, and short-term effects on AAH’s protein due to PI3K-Akt inhibition of GSK-3β (Figure 9).
Figure 9.
(A) Diagram of insulin/IGF-1 regulation of AAH. Normally, insulin/IGF-1 stimulation inhibits GSK-3β via upstream PI3K-Akt activation. However, uncoupling or inhibition of signaling can lead to increased GSK-3β activity and phosphorylation of AAH (direct). In addition, oxidative stress levels increase with GSK-3β activation; this response can activate other kinases that phosphorylate AAH (indirect). Depending on the positions and nature of AAH phosphorylation, AAH’s protein levels can either increase (enhanced stability) or decreased (increased cleavage and degradation). With increased levels of AAH protein expression, correspondingly increased AAH catalytic activity hydroxylates Notch, causing it to cleave, i.e. it activates Notch signaling. The Notch intracellular domain then translocates to the nucleus where it promotes transcription of genes that increase cell adhesion, motility, and invasion. (B) Diagram of predicted predicted phosphorylation sites on AAH. Most of sites correspond to GSK-3β and are distributed in the N-terminus of the protein. CK2, PKA, and PKC also can phosphorylate AAH. The H675 residue is critical for catalytic (hydroxylase) activity.
The next consideration was to determine if short-term GSK-3β-mediated regulation of AAH protein could be due to direct effects of AAH protein phosphorylation by GSK-3β. Subsequence analysis of AAH protein indicated the presence of multiple potential phosphorylation sites, including those with consensus sequences corresponding to GSK-3β [30]. In addition, we determined that AAH has other potential phosphorylation sites with consensus sequences for PKA, PKC or CK2. Most of the predicted phosphorylation sites are located in the N-terminal region of the protein. In vitro kinase assays, metabolic labeling studies, and immunoprecipitation/Western blot analysis demonstrated that AAH could be phosphorylated by GSK-3β [30]. The present work was designed to characterize AAH protein expression in insulin/IGF-1 stimulated HCC cells to further examine the consequences of its phosphorylation, including the likely impact on Notch signaling.
Insulin/IGF-1 stimulation of AAH protein
Initial studies confirmed that IGF-1 stimulation increases endogenously expressed AAH protein within 5 minutes and sustained for at least 60 minutes. However, due to the relatively low signal, the studies were extended using Huh7 HCC cells transfected with C-HA-AAH or N-Myc-AAH in which gene expression was regulated by a CMV promoter. In a separate study, we showed that Huh7 cells transfected with C-HA-AAH or N-Myc-AAH had significantly increased AAH expression, catalytic activity, HES-1 expression, and directional motility relative to mock-transfected control cells [39]. As observed with respect to the endogenously expressed protein, AAH accumulated in response to short-term IGF-1 stimulation. In addition to increasing the signal for monitoring AAH protein, the approach enabled us to track cellular expression and distribution of AAH’s N- and C-terminal cleavage products with specific HA and Myc epitope antibodies. In Huh7 and other HCC lines, this strategy is important because, under physiological conditions, AAH is rapidly cleaved, resulting in the predominant ~56 kD C-terminal catalytic domain-containing fragment as observed by Western blot analysis of IGF-1 stimulated, non-transfected Huh7 cells. In contrast, in transfected cells expressed a ~140 kD AAH-immunoreactive protein (HA-tagged or Myc-tagged) that accumulated over time. The larger than predicted 86 kD protein corresponds with effects of post-translational modification, such as by phosphorylation.
Shifts in subcellular localizations of AAH and Notch-1 following IGF-1 stimulation
Previous studies linked AAH expression and hydroxylase activity to Notch-activated signaling [16]. In HCC, Notch-1 is the dominant isoform. Since hydroxylation of Notch by AAH requires the proteins to interact or be in close proximity, we anticipated that AAH protein would traffic from its perinuclear localization to the cell periphery or surface to interact with and hydroxylate Notch. Furthermore, we anticipated that the C-terminal fragment which contains the catalytic domain would likely move to the cell periphery in response to IGF-1 stimulation. Correspondingly, in cells transfected with recombinant AAH, IGF-1 stimulation resulted in the trafficking of C-HA-AAH from the perinuclear zone to the cell periphery, including podocytes, followed by nuclear accumulation of the Notch intracellular domain (NID). These results are consistent with our hypothesis that IGF-1 stimulation of AAH protein mediates Notch nuclear translocation and attendant regulation of gene expression.
IGF-1 plus kinase inhibitor modulation of AAH protein expression
Sub-sequence analysis of AAH protein predicts the presence of 17 sites for GSK-3β mediated phosphorylation, as well as several others corresponding to PKA, PKC, or CK2 phosphorylation sites [30]. Given the results obtained using LICl to inhibit GSK-3β, we next assessed whether other relevant kinase inhibitors would also increase AAH protein expression and alter its sub-cellular localization in response to IGF-1 stimulation. The combined results obtained by cellular ELISA and immunofluorescence/confocal microscopy demonstrated that, in addition to GSK-3b, PKA, PKC and CK2 inhibitors also increased AAH protein in IGF-1 stimulated Huh7 cells. However, the time course effects differed in that the PKA and CK2 inhibitor effects were more limited whereas the GSK-3b and PKC inhibitor effects were sustained. The early transient stimulatory effects of PD98059 on AAH protein are not fully understood. However, inhibition of MEK leads to oxidative stress and oxidative stress was previously shown to stimulate AAH expression via HIF-1α activated signaling [40].
Long-term effects of GSK-3β inhibition and insulin stimulation on AAH expression
To assess the differential roles of insulin or IGF-1 stimulation and GSK-3β inhibition on AAH protein expression, we examined AAH expression Huh7 cells transfected with C-HA-AAH or N-Myc-AAH and treated with LiCl, with or without insulin stimulation. Since this approach enabled abundant expression of full-length AAH, our ability to evaluate responses was not compromised by rapid cleavage and poor detection of relatively small amounts of protein. The studies demonstrated that LiCl inhibition of GSK-3β was sufficient to increase AAH protein levels in the absence of insulin (or IGF-1) stimulation. However, progressively higher levels of ~140 kD AAH were detected in insulin stimulated, followed by insulin+LiCl treated cultures, indicating additive effects of trophic factor stimulation and GSK-3β inhibition. Mechanistically, the additive effect of LiCl with insulin could have been due to reduced cleavage/degradation and increased stabilization of AAH protein. Consequences would include increased activation of Notch signaling and enhanced cell motility, as previously demonstrated in other cell types [22, 41].
Conclusions
The findings suggest that insulin/IGF signaling promotes the trafficking of AAH to the plasma where it may facilitate the activation of Notch signaling networks. The effects may occur through the post-translational regulation of AAH mainly by GSK-3β, CK2, and PKC. These proteins have the potential to serve as promising targets for controlling aggressive growth of HCC.
Acknowledgments
Supported by AA11431, AA12908 and 5T32DK060415-10 from the National Institutes of Health.
Footnotes
Conflict of interest: No conflicts declared.
References
- 1.Arkin DMP, Ray FB, Erlay JF, Isani PP. Mini review estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology (New York, NY 1943) 2007;132:2557. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: Consider the population. J Clin Gastroenterol. 2013;47(Suppl):S2–6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright WE. The two-stage mechanism controlling cellular senescence and immortalization. Experimental gerontology. 1992;27:383. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- 5.Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 6.Galle PR. CD95-induced apoptosis in human liver disease. Seminars in liver disease. 1998;18:141. doi: 10.1055/s-2007-1007150. [DOI] [PubMed] [Google Scholar]
- 7.Desbois-Mouthon C. Impact of IGF-1R/EGFR cross-talks on hepatoma cell sensitivity to gefitinib. International journal of cancer. 2006;119:2557. doi: 10.1002/ijc.22221. [DOI] [PubMed] [Google Scholar]
- 8.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 9.Tovar V. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. Journal of hepatology. 2010;52:550. doi: 10.1016/j.jhep.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvisi DF. Deregulation of signalling pathways in prognostic subtypes of hepatocellular carcinoma: novel insights from interspecies comparison. Biochimica et biophysica acta. 2012;1826:215. doi: 10.1016/j.bbcan.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Derianst CK, Vandusens W, Przysieckil CT, Walshu PN, Berknerll KL, Kaufman RJ, Friedman PA. Inhibitors of 2-ketoglutarate-dependent dioxygenases block aspartyl B-hydroxylation of recombinant human factor IX in several mammalian expression systems. J Biol Chem. 1989;264:6615–6618. [PubMed] [Google Scholar]
- 12.Hu T-H. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer. 2003;97:1929. doi: 10.1002/cncr.11266. [DOI] [PubMed] [Google Scholar]
- 13.Lavaissiere L, Jia S, Nishiyama M, de la Monte S, Stern aM, Wands JR, Friedman Pa. Overexpression of human aspartyl(asparaginyl) beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. J Clin Invest. 1996;98:1313–1323. doi: 10.1172/JCI118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sepe PS, Lahousse SA, Gemelli B, Chang H, Maeda T, Wands JR, Monte SMD. Role of the aspartyl-asparaginyl-B-hydroxylase gene in neuroblastoma cell motility. Lab Invest. 2002;82:881–891. doi: 10.1097/01.lab.0000020406.91689.7f. [DOI] [PubMed] [Google Scholar]
- 15.Palumbo KS, Wands JR, Safran H, King T, Carlson RI, Monte SMD. Human aspartyl (asparaginyl) B-hydroxylase monoclonal antibodies: potential biomarkers for pancreatic carcinoma. Pancreas. 2002;25:39–44. doi: 10.1097/00006676-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 16.Cantarini MC, De La Monte SM, Pang M, Tong M, D’Errico A, Trevisani F, Wands JR. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and Notch signaling mechanisms. Hepatology. 2006;44:446–457. doi: 10.1002/hep.21272. [DOI] [PubMed] [Google Scholar]
- 17.Jia S, VanDusen WJ, Diehl RE, Kohl NE, Dixon Ra, Elliston KO, Stern aM, Friedman Pa. cDNA cloning and expression of bovine aspartyl (asparaginyl) beta-hydroxylase. J Biol Chem. 1992;267:14322–14327. [PubMed] [Google Scholar]
- 18.Wang Q, Vandusen WJ, Petroski CJ, Garsky VM, Stern AM, Friedman PA. Bovine liver aspartyl B-hydroxylase purification and characterization. J Biol Chem. 1991;266:14004–14010. [PubMed] [Google Scholar]
- 19.Treves S, Feriotto G, Moccagatta L, Gambari R, Zorzato F. Molecular cloning, expression, functional characterization, chromosomal localization, and gene structure of junctate, a novel integral calcium binding protein of sarco(endo)plasmic reticulum membrane. J Biol Chem. 2000;275:39555–39568. doi: 10.1074/jbc.M005473200. [DOI] [PubMed] [Google Scholar]
- 20.Lee J-H. Overexpression of humbug promotes malignant progression in human gastric cancer cells. Oncol Rep. 2008;19:795–800. [PubMed] [Google Scholar]
- 21.Dinchuk JE, Henderson NL, Burn TC, Huber R, Ho SP, Link J, O’Neil KT, Focht RJ, Scully MS, Hollis JM, Hollis GF, Friedman Pa. Aspartyl beta -hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J Biol Chem. 2000;275:39543–39554. doi: 10.1074/jbc.M006753200. [DOI] [PubMed] [Google Scholar]
- 22.Silbermann E, Moskal P, Bowling N, Tong M, de la Monte SM. Role of aspartyl-(asparaginyl)-β-hydroxylase mediated notch signaling in cerebellar development and function. Behav Brain Funct. 2010;6:68. doi: 10.1186/1744-9081-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundogan F, Elwood G, Longato L, Tong M, Feijoo A, Carlson RI, Wands JR, de la Monte SM. Impaired placentation in fetal alcohol syndrome. Placenta. 2008;29:148–157. doi: 10.1016/j.placenta.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwald I, Kovall R. Notch signaling: genetics and structure. WormBook. 2013:1–28. doi: 10.1895/wormbook.1.10.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Lahousse Sa, Carter JJ, Xu XJ, Wands JR, de la Monte SM. Differential growth factor regulation of aspartyl-(asparaginyl)- beta-hydroxylase family genes in SH-Sy5y human neuroblastoma cells. BMC Cell Biol. 2006;7:41. doi: 10.1186/1471-2121-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen P. The role of protein phosphorylation in human health and disease. Eur J Biochem. 2001;268:5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 28.Taha C, Klip A. The insulin signaling pathway. J Membr Biol. 1999;169:1–12. doi: 10.1007/pl00005896. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z-L, Guan Y-J, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 30.Carter JJ, Tong M, Silbermann E, Lahousse Sa, Ding FF, Longato L, Roper N, Wands JR, de la Monte SM. Ethanol impaired neuronal migration is associated with reduced aspartyl-asparaginyl-beta-hydroxylase expression. Acta Neuropathol. 2008;116:303–315. doi: 10.1007/s00401-008-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Monte SM. Ethanol inhibition of aspartyl-asparaginyl-beta-hydroxylase in fetal alcohol spectrum disorder: potential link to the impairments in central nervous system neuronal migration. Alcohol (Fayetteville, NY) 2009;43:225. doi: 10.1016/j.alcohol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Monte SM, Tamaki S, Cantarini MC, Ince N, Wiedmann M, Carter JJ, Lahousse Sa, Califano S, Maeda T, Ueno T, D’Errico A, Trevisani F, Wands JR. Aspartyl-(asparaginyl)- beta-hydroxylase regulates hepatocellular carcinoma invasiveness. J Hepatol. 2006;44:971–983. doi: 10.1016/j.jhep.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 33.de la Monte S, Ganju N, Wands JR. Microtiter immunocytochemical ELISA assay. BioTechniques. 1999;26:1073–1078. doi: 10.2144/99266bm15. [DOI] [PubMed] [Google Scholar]
- 34.Wands JR. Immunological approach to hepatocellular carcinoma. Journal of viral hepatitis. 1997;4(Suppl 2):60. doi: 10.1111/j.1365-2893.1997.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 35.Cox DP. High Throughput Method for Assessment of Cellular Reduced Glutathione in Mammalian Cells. Journal of environmental protection science. 2007;1:23. [PMC free article] [PubMed] [Google Scholar]
- 36.Dürr R, Caselmann WH. Carcinogenesis of primary liver malignancies. Langenbecks Arch Surg. 2000;385:154–161. doi: 10.1007/s004230050259. [DOI] [PubMed] [Google Scholar]
- 37.Boyault S. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology (Baltimore, Md) 2007;45:42. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 38.Guo Y, Klein R, Omary Ra, Yang G-Y, Larson AC. Highly malignant intra-hepatic metastatic hepatocellular carcinoma in rats. Am J Transl Res. 2010;3:114–120. [PMC free article] [PubMed] [Google Scholar]
- 39.Borgas DL, Gao J-S, Tong M, de la Monte SM. Potential Role of Phosphorylation as a Regulator of Aspartyl-(Asparaginyl)- β-Hydroxylase: Relevance to Infiltrative Spread of Human Hepatocellular Carcinoma. Liver Cancer. 2015 doi: 10.1159/000367731. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawton M, Tong M, Gundogan F, Wands JR, De La Monte SM. Aspartyl-(asparaginyl) β-hydroxylase, hypoxia-inducible factor-1α and notch cross-talk in regulating neuronal motility. Oxid Med Cell Longev. 2010;3:347–356. doi: 10.4161/oxim.3.5.13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gundogan F, Bedoya A, Gilligan J, Lau E, Mark P, De Paepe ME, de la Monte SM. siRNA inhibition of aspartyl-asparaginyl β-hydroxylase expression impairs cell motility, Notch signaling, and fetal growth. Pathol Res Pract. 2011;207:545–553. doi: 10.1016/j.prp.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]