Abstract
We present a novel low-cost biosensor for rapid, sensitive and selective detection of nucleic acids based on an ionic diode feature of an anion exchange nanoporous membrane under DC bias. The ionic diode feature is associated with external surface charge inversion on the positively charged anion exchange nanomembrane upon hybridization of negatively charged nucleic acid molecules to single-stranded oligoprobes functionalized on the membrane surface resulting in the formation of a cation selective monolayer. The resulting bipolar membrane causes a transition from electroconvection-controlled to water-splitting controlled ion conductance, with a large ion current signature that can be used to accurately quantify the hybridized nucleic acids. The platform is capable of distinguishing two base-pair mismatches in a 22-base pairing segment of microRNAs associated with oral cancer, as well as serotype-specific detection of dengue virus. We also show the sensor’s capability to selectively capture target nucleic acids from a heterogeneous mixture. The limit of detection is 1 pM for short 27 base target molecules in a 15-minute assay. Similar hybridization results are shown for short DNA molecules as well as RNAs from Brucella and E.coli. The versatility and simplicity of this low-cost biosensor should enable point-of-care diagnostics in food, medical and environmental safety markets.
Keywords: Biosensing, DNA/RNA, Electrokinetics, Charge Inversion, Nanoporous Membrane
1. Introduction
DNA and RNA-based pathogen diagnostics remain as one of the most active fields of nucleic acid research (Gingeras et al. 2005; Wei et al. 2010). PCR amplification remains a gold standard in nucleic acid-based diagnosis, but it requires expensive labels and trained personnel--- and is time consuming because of multiple steps involved. In recent years, there has been interest in developing probe-based non-optical sensors that can obviate the use of PCR and fluorophore labeling to improve the detection time and lower the cost of diagnostic tests. A large number of papers have been published on amplification-free nucleic acid biosensors employing different transduction sensing mechanisms (Drummond et al. 2003; O'Connor and Glynn 2010; Palchetti and Mascini 2008; Peng and Miller 2011) of which label-free technologies are of special interest (Kataoka-Hamai and Miyahara 2011; Ricci and Plaxco 2008).
Nucleotides in DNA/RNA molecules are linked together through a sugar-phosphate backbone. The presence of the phosphate groups in this backbone renders both DNA and RNA molecules negatively charged making them suitable for manipulation (e.g. gel electrophoresis) as well as detection under electrical field. This intrinsic negative charge of DNA/RNA molecules can have a profound effect on both electronic and ionic conductivity of a system and has been explored to develop label-free nucleic acid sensors by measuring the electrical signal of the system. One such example is the field-effect-transistor (FET) that uses the electronic conductivity of the system for the detection of DNA molecules. In this system, the current passing between a source and a drain is controlled by the potential connected to a gate. This potential is sensitive to the negative charge of nucleic acid molecules present within the Debye length of the gate (Fritz et al. 2002). This changes the capacitance of the gate-electrolyte interface resulting in a change in the total current passing through the transistor (Kim et al. 2004; Pandana et al. 2008). Further, Li et al. demonstrated the detection of DNA by monitoring the change in conductivity of silicon nanowires (Li et al. 2004).
Researchers have also demonstrated nucleic acid detection by directly monitoring the influence of DNA/RNA molecules on the ionic conductivity of nanochannels. These nanochannels are often single nanopores or an array of nanopores, where the charge and conductance of the nucleic acids within the nanochannels affects the intra-channel ion conductance. The immobilized DNA can render the nanochannel, or part of it, ion selective (Hou et al. 2013). Jagerski et al. (Jagerszki et al. 2007) measured flux of easily traceable anionic dye in a nanochannel functionalized with neutral PNA (peptide nucleic acid) probe and its hybridization to target DNA. While the PNA probe did not hinder the flux of anionic dye, the hybridized target DNA molecules efficiently decreased the flux through the channel. Further, Ali et al.(Ali et al. 2010) observed a significant change in current voltage characteristics (CVC) upon hybridization of target DNA to uncharged peptide nucleic acid probes functionalized to a conical nanopore. Wang et al. employed an ion selective alumina nanochannel for the detection of DNA molecules(Wang and Smirnov 2009) and used the change in conductivity of nanochannel upon DNA hybridization as a detection signal. Other groups have demonstrated sensors involving biological nanopores where DNA hybridization causes opening or closing of the nanopore which can be tracked by measuring the ionic current going through the nanopore (Krishnamurthy et al. 2010a, b; Steller et al. 2012). This switch mechanism can essentially be used to detect any biomolecules including proteins and nucleic acids (Lucas and Harding 2000). A similar but simpler approach was presented by Weiss Wichert et al. where the binding of the target molecules blocked the entrance to the gramidicin pore resulting in diminished flux of ions (WeissWichert et al. 1997).
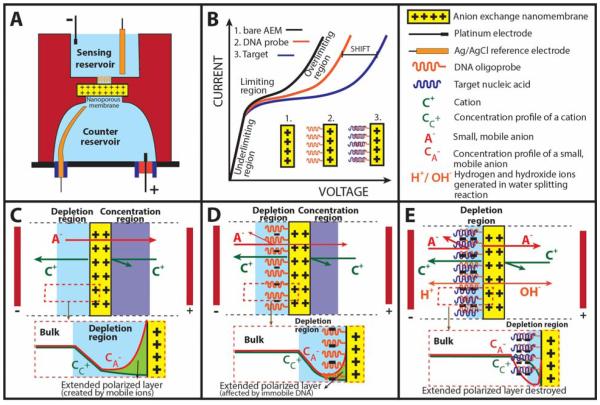
Both FET and nanochannel sensors remain expensive to fabricate—and hence have not been commercialized for field-use diagnostics. Their small current (nA) and voltage (mV) signals also render both platforms sensitive to noise and contamination. While translocation time for single molecule through a nanochannel is short (ms), the total time to interrogate every molecule in the sample is still prohibitively long. Herein we report a novel, low-cost and label-free biosensing platform for the detection of negatively charged nucleic acids using a positively-charged, heterogeneous anion exchange nanoporous membrane (Figure 1A). The sensor is based on a charge inversion phenomenon (Slouka et al. 2013) that occurs on the surface of a positively charged nanomembrane (not within the nanopores of the membrane) when negatively charged nucleic acid molecules bind to its surface. It operates at a much higher voltage than the nanochannel sensors because ion conductance is controlled by the surface charge of the membrane surface and two unique non-equilibrium ion transport phenomena described later. Anion exchange membranes are known to exhibit interesting non-linear current-voltage characteristics (Figure 1B – black curve) that arise due to the differences in fluxes of ions in the solution and the membrane. Small counterions (anions) are the main carriers of ionic current in the positively charged membrane (Figure 1C), while large anions and cations do not contribute to this ionic current due to size exclusion and electrostatic repulsion effects respectively. At low voltages, the current increases linearly with voltage showing Ohmic behavior which is also known as the under-limiting region on the CVC (Figure 1B black curve). Nanochannel sensors operate in this low-voltage Ohmic region. With increasing voltage, the concentration of the electrolyte on one side of the nanomembrane decreases (depletion side) while the concentration of the electrolyte on the other side increases (concentration side). This establishes a concentration gradient, known as concentration polarization, across the membrane (Krol et al. 1999; Tanaka 2004). The depletion side of the nanomembrane starts controlling the behavior of the system. At a certain current, the electrolyte concentration within the depletion region approaches zero, resulting in an increase in current resistance, which is reflected by a sudden decrease in the slope of the CVC. This region is denoted as the limiting region on the CVC (Figure 1B – black curve). The system gradually develops a very large differential resistance causing the electrical current to almost saturate out. In an ideal case, further increase in the voltage would result in broadening of the depletion region far into the electrolyte. However, experimental results show that the current does not saturate completely but instead starts to increase abruptly giving rise to a region known as overlimiting region. The explanation for this behavior is quite complex (Belova et al. 2006) as it usually involves a mechanism that destroys the depletion region. Recently, we reviewed possible mechanisms involving the overlimiting current and experimentally demonstrated that electroconvection phenomenon (Chang et al. 2012; Slouka et al. 2013; Yossifon et al. 2009) developed at the depletion side was the major contributing factor for the appearance of sudden increase in current in the overlimiting region (Figure 1B – black curve). This electroconvection mechanism results from an action of a very strong electrical field developed in the depletion region on an extended polarized layer where an excess of anions creates a space charge (see insert in Fig 1C for concentration profiles of anions and cations) (Chang et al. 2012; Rubinstein and Zaltzman 2010). This extended polarized layer is only a few times larger than the Debye layer and is no more than 100 nm in thickness, even though the electroconvection microvortices can reach 100 microns in dimension. This suggests that the overlimiting current and to a lesser extent, the limiting current are both sensitive to surface charge changes on the membrane surface making it ideal for rapid surface assays, as the molecules do not need to enter the nanopores of the membrane. Its non-equilibrium features also endow them with large voltage/current signatures with correspondingly high dynamic range and noise insensitivity.
Figure 1. Schematics and working principle of the nanomembrane sensor.
(A) Diagrammatic representation of a nanomembrane electrokinetic sensor consisting of the top sensing reservoir and bottom counter reservoir bridged together by a positively charged nanomembrane. (B) Current-voltage characteristics (CVC) showing changes in ohmic, limiting and overlimiting regions for bare anion-exchange membrane (black), membrane functionalized with oligoprobe (red) and hybridization of DNA/RNA with oligoprobe (blue). We utilize these changes in CVC for detection of nucleic acids. (C) Mechanism for the ohmic relationship at low voltages known as the underlimiting regions. (D) Changes in CVC as a result of DNA adsorption on the membrane surface leading to changes in the limiting regions. (E) Mechanism for the overlimiting region at high DNA concentrations causing electroconvection and water splitting phenomenon.
We recently demonstrated significant CVC changes when large, negatively charged molecules (e. g. DNA) are added to the surface of a positively charged membrane(Slouka et al. 2013). The size of these molecules prevents them from passing through the membrane (size exclusion), however they can easily adsorb to the membrane surface. The adsorption is facilitated by strong electrostatic interactions between positively charged membrane and negatively charged molecules. By running real time fluorescence imaging on membrane surface, we observed a gradual suppression of electroconvection of the system for DNA concentrations lower than 1 μM (for 29 nucleotide long DNA). This behavior is related to the effect of DNA on extended polarized layer, where the adsorbed, immobile DNA replaces small, mobile anions (Figure 1D). The corresponding CVC of such a system is plotted in Figure 1B (red curve) and is characterized with a shift of the over-limiting region to the right. At high DNA concentrations (over 1 μM) the layer of DNA becomes ion selective and creates a bipolar junction which at sufficiently large voltages splits water (Figure 1E), a phenomenon observed in bipolar membranes (Cheng and Chang 2011; Desharnais and Lewis 2002). We note that the transition from electroconvection-controlled to water splitting-controlled regime is gradual and water splitting reaction was detected even for concentrations below 1 μM (Slouka et al. 2013).
Figure 1B represents our strategy of using the behavior described above for detection of nucleic acids. Our strategy involves covalent immobilization of negatively charged DNA oligoprobes onto the membrane surface which results in a shift in the overlimiting region to the right (black to red curve in Figure 1B). Upon hybridization with specific DNA/RNA target molecules, the amount of negative charge increases corresponding to a further shift in the overlimiting region as depicted by the blue curve in Figure 1B. Such a change in the over-limiting region of CVC was used to record the signature of the hybridization event for the detection of short DNA molecules, microRNA molecules associated with oral cancer as well as RNA from dengue virus, Brucella bacteria and Escherichia coli (E.coli). The proposed sensor is novel because, for the first time, the signal from the surface charge change is significantly amplified with charge inversion and with the large “overlimiting currents” that arise from microvortices and water splitting phenomena that accompany the surface charge inversion. In terms of voltage signals, typical Field-Effect-Transistor sensors record up to 100 mV signals with surface charge changes compared to ~2 V shifts with our unique nanoporous membrane sensor. Additionally, novel mechanisms like water splitting reactions at the junction of a bipolar membrane (Cheng et al. 2011) and also vortices at the external boundary of a bipolar membrane (Senapati et al. 2011) are designed to enhance the signal. We demonstrate the unique performance of the sensor with high stability (8 days) and reproducibility, current limit of detection of 1 pM, selectively of 2 base pair mismatch as well as capability of identifying targets from a heterogeneous sample and a current time of detection of 20 minutes per sample. As the sensing principle is based on hybridization of negatively-charged nucleic acid molecules, the versatility of this platform makes it amenable for detection of other pathogenic targets in food, medical and environmental safety.
2. Experimental Procedure
2.1. Reagents and Chemicals
Commercially-available heterogeneous anion exchange nanomembrane composed of polystyrene-divinylbenzene fine particles with strong basic quaternary ammonium groups (R-(CH3)3N+) supported by polyethylene as a binder and polyamide/polyester textile fiber was obtained from Mega a.s., Czech Republic (http://www.mega.cz/heterogenous-ion-exchange-membranes-ralex.html). The thickness of the membrane was ~450 μm when dry and ~700 μm after swelling in water with an exchange capacity of 1.8 mval/g and a pore size of around 1 nm. Two component silicon RTV system and Quick cast polyurethane resin were purchased from TAP Plastic Inc. (San Leandro CA, USA). Phosphate Buffer Saline (PBS) 10X solution containing 1.37 M sodium chloride, 27 mM potassium chloride, 100 mM disodium hydrogen phosphate, and 18 mM potassium dihydrogen phosphate was purchased from Fisher Scientific (Waltham, MA, USA). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), Benzophenone-3,3′,4,4′-tetracarboxylic acid, sodium hydroxide were purchased from Sigma Aldrich (St. Louis MO, USA). Maintenance media including supplements, fetal bovine serum, streptomycin, penicillin, TRIzol®, amine-coupled oligoprobes and complementary single stranded DNA targets were obtained from Life Technologies (Grand Island NY, USA). RNeasy Mini Kit was obtained from Qiagen (Valencia CA, USA).
2.2. Fabrication of biochip
A polyurethane-based microfluidic device with top and bottom reservoirs bridged together by an anion exchange nanomembrane was used as the sensing biochip. To fabricate the biochip, two separate silicone molds representing the two reservoirs were cast from a primary mold created using CAD designs and 3D printing. A circular anion exchange membrane was sandwiched between the two silicone molds creating the reservoirs. A two-component polyurethane resin (1:1 ratio) was poured inside the mold and allowed to cure for 30 minutes. Release of the silicone mold yielded the polyurethane-based microfluidic biochip with an exposed membrane sensing area of either 3.5 or 1 mm2 depending on the initial CAD designs. The biochip was then mounted onto a glass slide with inlet and outlet. The schematic of the biochip is depicted in Figure 1A.
2.3. Attachment of oligoprobe on the sensing surface
The nanomembranes are made of divinylbenzene/polystyrene particles functionalized with quaternary ammonium groups that are embedded inside a polyethylene-polyamide/polyester matrix, where polyethylene acts as a binder and polyester/polyamide fibers provide mechanical stability. We used the inherent C-H bonds present on the membrane surface to covalently attach target specific oligoprobes using EDC chemistry(Sigal et al. 1996) and a carboxyl-terminated photo crosslinker(Lin et al. 1988). First, COOH groups were created on the surface of the anion exchange nanomembrane by adsorbing 10 μl of photo-reactive Benzophenone-3,3′,4,4′-tetracarboxylic acid (1 mg in 10 μl water at pH 6-7) and exposing it to UV light for 20 minutes. This allowed selective functionalization of carboxyl-benzophenone through radical polymerization to the C-H bonds on the membrane surface forming a carboxyl-terminated membrane surface. ElectroCure 500 UV Flood Curing Chamber with UV output of 30 mW/cm2 at 356 nm wavelength was used to perform the photocuring reaction (Electro-Lite Corp., Bethel CT, USA). Next, the membrane was incubated with 0.4 M EDC in MES buffer for 30 minutes followed by an overnight incubation with 10 μM of the amine-coupled oligoprobe in 0.1X PBS solution to covalently attach the probe to the sensing surface by amide linkage. The successful functionalization of the probe on the membrane surface was characterized using IR spectroscopy (Bruker FTIR) and by measuring the CVC of nanomembrane before and after functionalization.
2.4. Isolation of target RNA
Aedes albopictus C6/36 were grown in 75 cm3 tissue culture flasks at 28°C to ~80% confluency and infected with dengue serotype 2 (DENV-2 strain JAM1409) at a multiplicity of infection of 0.1. Maintenance media consisting of L-15 (Leibovitz) media supplemented with 10% tryptose phosphate broth and 2% FBS was added to the flasks after virus inoculation. Media was renewed 7 days post-infection. Cells were harvested day 14 post infection by scraping them from the flask. To isolate the DENV-2 RNA, TRIzol® lysing solution was added to the infected cells, followed by spin column purification using Qiagen RNeasy kit. For isolation of microRNA associated with oral cancer, oral squamous cell carcinoma cell line USCC-1 was engineered to overexpress miR-146a with viral vectors. The cells were cultured with MEM medium containing 10% fetal bovine serum, penicillin (100U/ml) and streptomycin (100μg/ml). Total RNA extraction was performed with TRIzol® reagent per instructions from the manufacturer. Purified Brucella RNA was obtained from Purdue University and E. coli RNA was obtained from the Construction Engineering Research Laboratory at the US Army Engineer Research and Development Center.
2.5. Measurement of current-voltage characteristics
All electrical measurements were made using the Gamry 500 potentiostat in a four-electrode setup (Gamry Instruments, Warminster PA, USA). Two reference Ag-AgCl electrodes were used for the measurement of the voltage across the membrane whereas two platinum electrodes were used to apply the current load. The reference electrodes were positioned close to the membrane surface in both reservoirs of the biochip, whereas the source electrodes were placed farther away to prevent any interference. Both reservoirs of the biochip were filled with 0.1X PBS solution prior to the measurement of the CVC. Two sensing areas of 3.5 mm2 and 1 mm2 were used to study the characteristics of the sensor. The CVC was obtained by applying current load from 0 to 75 μA for 3.5 mm2 sensing area and 0 to 40 μA for sensing area of 1 mm2 at the step rate of 1 μA/s and measuring voltage drop across the membrane. The sensing reservoir (side of the anion exchange membrane functionalized with the oligoprobe) was connected to the ground whereas a positive potential was applied in the other reservoir. This connection ensured the formation of the depletion region in the sensing reservoir, making it sensitive to functionalization of the probe as well as its hybridization with the target of interest. In a typical experiment, CVC was measured in 0.1X PBS solution prior to functionalization of the oligoprobes on the sensing region. To confirm the functionalization, a second CVC was measured to observe changes in the signal. Finally, the DNA/RNA sample of interest was incubated for 15 minutes in the sensing reservoir to allow hybridization. Non-specifically bound DNA/RNA molecules on membrane surface were washed three times using 4X PBS solution. A final CVC was measured after equilibrating the reservoirs in 0.1X PBS solution. Any shift in CVCs in the overlimiting region before and after hybridization indicated the successful detection of the target nucleic acids. To confirm the observed CVC shift was indeed due to hybridization of target nucleic acid molecules, a basic pH 13 solution was used to dehybridize and re-generate the probe resulting in the shift of CVCs back to the probe level.
3. Results and Discussions
Our sensor works on the principle that target nucleic acid molecules upon hybridization to molecular probes functionalized to the positively charged nanomembrane alters the ion conductance across the membrane-solution interface resulting in a significant shift in CVC in the overlimiting region (Figure 1). As stated earlier, an increase in DNA concentration on the anion exchange nanomembrane has two major effects on the system in the over-limiting region: (i) suppression of electroconvection and (ii) enhancement of water splitting reaction (Cheng and Chang 2011; Slouka et al. 2013). The extent to which electroconvection is suppressed and water splitting enhanced is given by the total amount of DNA present on the membrane surface and the combined effect reflects the amount of shift in the overlimiting region of the CVC. Here, we demonstrate our ability to use this highly sensitive overlimiting region of the CVC for the specific detection of complementary DNA as well as RNA from dengue virus, Brucella bacteria and E. coli O157:H7, microRNA associated with oral cancer and heterogeneous nucleic acid samples. We also studied stability, reproducibility, selectivity, limit of detection and dynamic range of the nanomembrane sensor.
3.1. Sensor stability and reproducibility
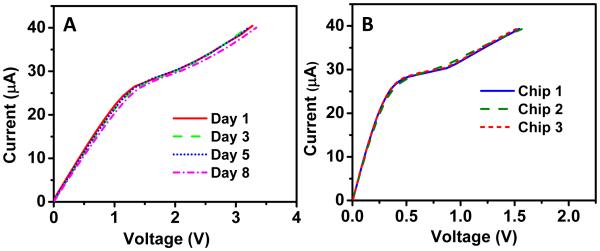
To validate the sensor stability, a bare sensor with 1 mm2 sensing area (data not shown for sensing area of 3.5 mm2) was fabricated and CVC was measured on the same sensor for 8 days at room temperature. For each measurement, the sensor was rinsed and equilibrated with a 0.1x PBS analyte solution and several CVC measurements were obtained on the same device to measure reproducibility (n=5). Figure 2A shows no significant change in CVC after 8 days of measurement suggesting the sensor was very stable over time. We observed an average voltage of 3.25V at 40 μA with a standard deviation of 0.06 V for the 20 measurements (five measurements on each of the four day points) thus demonstrating the sensors stability. Additionally, to study the sensor reproducibility, we again fabricated three different bare chips (without any analyte) with similar sensor area (1 mm2). As seen from Figure 2B, all three biochips showed similar CVCs (average of 1.54 V at 40 μA with a standard deviation of 0.03V), suggesting the nanomembrane sensor is reproducible for biochips with similar sensor area. This is a very unique characteristic of our sensor over electrochemical sensors. Electrochemical sensors have the advantage of signal amplification using redox reporters which is useful for enhancing the detection sensitivity but are often instable in detection signal(Bogomolova et al. 2009). In our case, sensing is based on transport of ions through the membrane and by keeping the source electrodes far away from the membrane surface we do not observe any electrochemical reactions on the sensor surface making the sensor highly stable and reproducible.
Figure 2. Stability and reproducibility of the sensor.
(A) A bare nanomembrane biosensing chip (1mm2) stable for 8 days at room temperature. (B) Three bare biochips with identical surface area (1mm2) showing reproducible curves.
3.2. Characterization of probe functionalization on membrane surface
The functionalization of oligoprobe was first evaluated by measuring the CVC before and after probe attachment and determining the shift in voltage at 50 μA. We observed a significant shift (0.5V) in the CVC after performing the functionalization process as compared to the CVC of the bare membrane confirming successful functionalization of probe onto the membrane surface (Figure 3A). It should be noted that the CVC was measured after washing the membrane with high ionic strength solution (4X PBS) to remove any non-covalently bound DNA oligos. The high salt buffer (4X) nullifies the electrostatic interaction between positively charge nanomembrane surface and negatively charge DNA molecules resulting in the removal of non-specifically adsorbed DNA molecules from the surface. We generally observed a shift of ~0.5-0.6 V after probe attachment on a sensing area of 1 mm2 and a smaller shift (~0.2 V, data not shown) for larger sensing area (3.5 mm2) since more oligoprobes are required to register the shift due to the charge inversion phenomenon described previously in Figure 1. The functionalization of oligoprobe on membrane surface was also characterized by IR-spectroscopy. Figure 3B shows the IR spectra of the oligoprobe in DI water and on a dry nanomembrane surface after functionalization. Upon hydration, DNA undergoes a structural transition (Pevsner and Diem 2001) resulting in symmetric and antisymmetric stretching vibrations of the phosphate linkage shifts progressively towards lower wavenumber. As seen in Figure 3B, the dry DNA probe-functionalized nanomembrane shows PO2− symmetric and antisymmetric stretching at 1092 cm−1 and 1236 cm−1 respectively while both the peaks shift for aqueous DNA sample and reaches at 1088 cm−1 and 1223 cm−1 respectively. Similarly, C-O stretching vibration of the phosphodiester observed at 1062 cm−1 is also affected by the presence of water molecules and shifts at 1054 cm−1 for the aqueous DNA solution. Further, the presence of the characteristic in-plane vibrations of C=O, C=C and C=N groups of the heterocyclic bases in the 1550-1750 cm−1 region clearly confirms the successful attachment of the oligoprobe onto the nanomembrane surface (Figure 3B).
Figure 3. Characterization of probe on membrane surface.
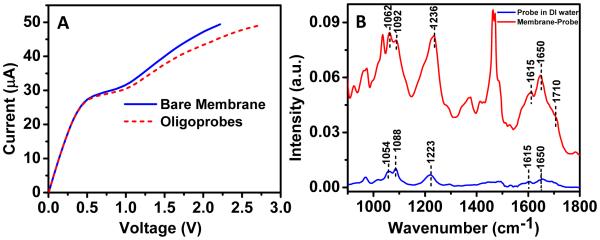
(A) CVC depicting a significant shift in voltage upon functionalization of probe indicating covalent attachment of probe onto the nanomembrane surface. (B) IR spectra confirming the probe immobilization by comparing probe-specific peaks on the nanomembrane surface compared to a probe sample in DI water.
3.3. Detection of pathogenic nucleic acids using nanomembrane technology
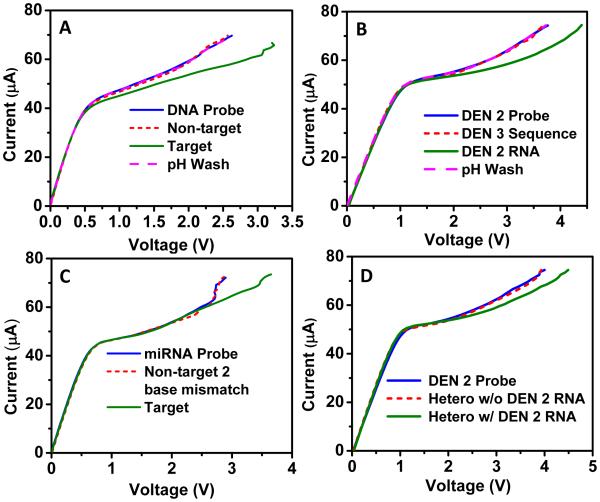
To demonstrate proof-of-concept, the nanomembrane sensor was first validated using a 27-base DNA sequence complementary to the probe functionalized on the membrane surface (Figure 4A). First, an oligoprobe was attached to the nanomembrane surface and CVC was measured and used as the baseline. The sensor was then exposed to 1μM non-complementary probe sequence for 15 min; no significant shift from probe signal was observed. However, upon incubation of the same biochip with 1μM complementary probe sequence for 15 min, the CVC showed a large shift of ~0.65 V, suggesting successful hybridization with the complementary probe sequence. To further confirm that the shift was due to hybridization, the biochip was treated with pH 13 solution to dehybridize and re-generate the probe. This resulted in the shifting of CVC back to the probe level. This clearly demonstrated the capability of the sensor to specifically detect short nucleic acid sequences. We would like to point out that since a larger sensing area (3.5 mm2) was used for all the experiments in Figure 4, a higher current load (0 to 75 μA) was applied for all biochip measurements. It should also be noted that changes in CVC for all experiments in Figure 4 were conducted at 70 μA.
Figure 4. Specificity of the nanomembrane sensor.
(A) A proof-of-concept demonstration of nanomembrane sensor using 27 base DNA sequence complementary to the probe sequence attached on the nanomembrane surface. (B) Serotype-specific detection of dengue virus indicating the sensor’s capability to specifically detect RNA from dengue serotype 2 when incubated with a sensor pre-functionalized with a DENV-2 oligoprobe. No shift was observed when the sensor was incubated with a sequence corresponding to DENV-3. In addition, high pH wash resulted in dehybridization of target and probe resulting in a regeneration of CVC corresponding to the probe level. (C) The specificity of the sensor was challenged by using a non-target sequence differing by only two base pairs compared to the target sequence. Change in CVC was only observed for the target microRNA sequence indicating the sensor’s capability to distinguish two base pair mismatches. (D) The specificity of the sensor when exposed to a heterogeneous mixture of target sequence along with three non-target sequences. Again, a change in CVC was only observed when the target DENV-2 RNA was present in the heterogeneous sample but no shift was observed for heterogeneous sample without the target RNA.
3.4. Specificity of the nanomembrane sensor
Another important criterion for the success of any sensor is the detection specificity and its capability to discriminate closely related sequences. We first demonstrated the capability of the platform to specifically distinguish between two of the four serotypes of RNA-based Dengue virus (DENV-2 and DENV-3). Dengue virus is a rapidly emerging global public health threat with an estimated 390 million people infected annually - three times more than the current estimate by WHO (Bhatt et al. 2013; Mangold and Reynolds 2013). Early diagnosis of dengue therefore has significant clinical importance, and can help facilitate appropriate supportive therapies to reduce morbidity and mortality. Diagnostic sequences for serotype-specific oligonucleotides have been previously published (Seah et al. 1995a; Seah et al. 1995b; Shu and Huang 2004) and were used for the experiments described here. First, the specific oligoprobe for DENV-2 was functionalized on membrane surface as described earlier. A CVC was recorded after functionalization and used as the baseline. A non-target complementary sequence corresponding to DENV-3 was incubated for 15 minutes, followed by a 4X PBS solution wash. A subsequent CVC was measured to observe any changes. Similar protocol was then used with the target RNA obtained from dengue serotype 2. As shown in Figure 4B, a shift of ~0.6 V was observed in CVC for target RNA sample (DENV-2) incubated at a concentration of 50 pg/μl while no significant shift was observed for non-target sequence (DENV-3), suggesting the capability of the sensor for serotype-specific detection of dengue virus. To further confirm that the shift with DENV-2 RNA was indeed due to hybridization to the functionalized probe, a high pH wash was performed resulting in the regeneration of CVC back to the probe level confirming the specific detection of DENV-2 RNA (Figure 4B).
We further tested the specificity of the nanomembrane-based sensor by using closely related microRNA sequences that differ by only 2 bases (miR146a target sequence: UGAGAACUGAAUUCCAUGGGUU and miR146b non-target sequence: UGAGAACUGAAUUCCAUAGGCU). Changes in expression level of miR146a plays a significant role in oral cancer detection (Shi et al. 2012). As seen in Figure 4C, no significant shift was observed for the two-base mismatch non-target sequence whereas a shift of ~0.7 V was observed when the target sequence (50 ng/μl) was incubated in the biochip. This clearly demonstrated our sensor’s capability to successfully discriminate microRNA sequences with two base mismatch.
To further test the efficacy of the sensor, we designed an experiment mimicking a practical sample consisting of several nucleic acids. We wanted to test the ability of the nanomembrane sensor to capture and record the hybridization of target nucleic acids from a heterogeneous mixture. To test the selectivity of the sensor in a heterogeneous RNA mixture, we first modified a nanomembrane surface with DENV-2 probe. A lab-constructed heterogeneous sample consisting of equal proportions (10 pg/μl) of DENV-2 target RNA, oligo sequence corresponding to DENV-3, Brucella RNA and E. coli RNA was prepared. After incubating this heterogeneous sample in the biochip functionalized with DENV-2 probe, a shift in the CVC (0.45 V) was observed confirming the ability of the sensor to pick up the specific target RNA sequence from a heterogeneous mixture (Figure 4D). Importantly, the same heterogeneous mixture without the DENV-2 RNA produced no significant shift in voltage showcasing the ability of our sensor to be specific even in the presence of three non-target sequences.
3.5. Limit of detection and dynamic range
In the experiments described earlier, a biochip with sensing area of 3.5 mm2 was used and we observed a noticeable shift for rather large concentrations of the target molecules on the order of 10-100 nM for a 27 base DNA sequence. Since the sensing principle is dependent on the amount of negatively charged molecules on the membrane surface, we expect to see enhanced sensitivity simply by reducing the sensing area of the nanomembrane sensor. We fabricated biochips with a smaller sensing area of 1 mm2 and performed a concentration study to see the effect of the sensing area on the limit of detection. We observed an improvement in the limit of detection of the nanomembrane biochip down to 1 pM (107 copies per 100 μl) after reducing the sensing area from 3.5 mm2 to 1 mm2 (inset of Figure 5A). This limit of detection is comparable to traditional electrochemical nucleic-acid based biosensors (Cagnin et al. 2009). However, it is not clearly understood why 3.5 times reduction in sensing area improves the limit of detection by 4 to 5 folds and is the subject of current investigation in our laboratory. We also observed a dependence of the voltage shift on the concentration of target molecules, which allows calibration of the nanomembrane sensor. 27 base target ssDNA solutions with concentrations of 1 pM, 100 pM, 10 nM and 1 μM were allowed to hybridize sequentially with the specific probe functionalized on membrane surface for 15 minutes and the appropriate CVCs were measured. Figure 5A shows clear evidence that higher concentrations produce larger voltage shifts in the CVC evaluated at 40 μA. It should be noted that the same biochip was used to perform the limit of detection study after de-hybridizing and regenerating the probe using pH 13 solution. The experiment was repeated in triplicates which show the reproducibility of the membrane to consistently detect low concentrations of DNA. Thus we expect a further enhancement in the sensitivity (<pM) for even smaller sensing area as well as for longer RNA sequences (>1000 bases long) as the detection is based on the amount of negative charge attaching to the membrane surface. Figure 5B depicts the bar diagram showing changes in CVC from probe level with increasing DNA concentration clearly demonstrating the sensor’s dynamic range of six orders of magnitude.
Figure 5. Sensitivity of the nanomembrane sensor.
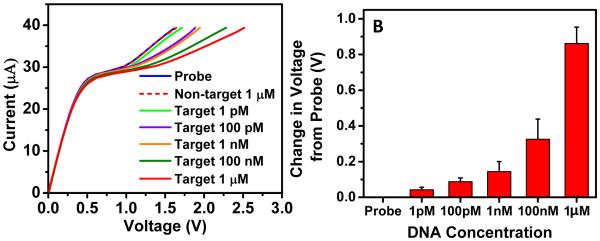
(A) Changes observed in CVC with gradual increase in the voltage shift upon incubating the same probe-functionalized biochip with increasing DNA concentration (27-base long target). As the sensing principle is based on charge inversion phenomenon, we observed that by decreasing the exposed sensing area, we were able to achieve sensitivity down to 1 pM concentration. Again, no change in CVC was observed for non-target sequence. (B) Bar diagram showing voltage shift from probe level with increasing DNA concentration (N=3 experiments).
3.6. Versatility of the nanomembrane technology
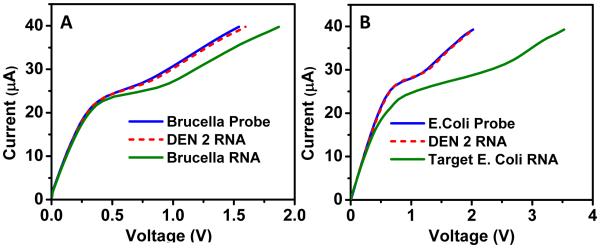
The versatility of the nanomembrane-based sensor was then demonstrated to detect target nucleic acids of various pathogens provided a target-specific oligoprobe sequence was known. To further extend the scope of our sensor, we tested the sensor against RNA samples of Brucella and E. coli O157:H7. Foodborne diseases are one of the most common causes of mortality worldwide, and have significant economic consequences with financial losses accounting to billions of dollars to food industry (Myint et al. 2006; Scharff 2012). Over the past decade, the noticeable increase in foodborne illnesses due to the bacterial contamination of foods have been reported with increasing illnesses resulting from consumption of fresh fruits and vegetables (Erickson et al. 2010; Heaton and Jones 2008) and furthermore, there is continued threat of transmission of emerging pathogens including Brucella sp., and E. coli O157:H7 through food (Altekruse et al. 1998; Skovgaard 2007). Similar to the experiment for dengue detection, previously-studied oligoprobes for Brucella were functionalized onto the membrane surface (Nelson et al. 2002; Wellinghausen et al. 2006). After confirming an improvement in sensitivity with smaller sensing area, experiments in Figure 6 were carried out with a smaller surface area (1 mm2) and hence the current load applied ranged from 0 to 40 μA. As expected, target Brucella RNA (2 pg/μl) caused a significant shift in voltage (0.3 V) at 40 μA while non-target RNA (DENV-2 RNA in this case) resulted in no shift from the probe level confirming the sensor’s ability to detect Brucella RNA (Figure 6A). Figure 6B shows similar results for detection of E.coli O157:H7. After functionalizing the probe (Sharma 2006), a significant shift in CVC (~1.5 V) was only observed for 40 pg/μl target RNA whereas no shift was observed when the biochip was incubated with non-specific DENV-2 RNA. It should be noted that for all the detection experiments, identical incubation (15 minutes) and washing conditions (high ionic strength 4X PBS solution) were used.
Figure 6. Versatility of the nanomembrane sensor for detection of pathogenic nucleic acids.
Specific shift in voltage observed in CVC upon incubation of a probe with specific target in case of (A) Brucella RNA and (B) E.coli RNA whereas no shift was observed for non-specific target (DENV-2 RNA in both cases).
4. Conclusions
We have successfully demonstrated a novel biosensor capable of rapid detection of nucleic acids (DNA/RNA) from any pathogen of interest, using the charge inversion phenomenon when negatively charged nucleic acids assemble on the surface of the positively charged membrane. Changes in current-voltage characteristics were used to identify and quantify targets that hybridize with specific complementary probes covalently functionalized on the membrane surface. In our platform, we observed large voltage and current signal changes suggesting better signal-to-noise ratio with more accurate quantification. We have shown that the nanomembrane sensor is stable, reproducible, and can specifically detect RNA of dengue virus, Brucella and E. coli as well as microRNA associated with oral cancer. The nanomembrane sensor is specific and able to distinguish two base mismatches in the target sequence as well as capable of capturing and recording the target sequence from a heterogeneous mixture. By reducing the sensing area, we demonstrated a limit of detection of 1 pM and a 6-order dynamic range. The versatility, specificity, sensitivity and its capability of rapid detection without prior amplification provide several future applications of this nanomembrane sensor in environmental, food and medical safety.
Acknowledgement
Authors would like to thank Ramesh Vemulapalli of Purdue University for providing Brucella RNA. Authors would also like to thank Travis King and Stephen Grimme of US Army Engineer Research and Development Center for providing E. coli RNA samples. This work has been supported by Army Corps of Engineers W9132T, USDA 2012-67005-19589, NSF-CBET 1065652 and Walter Cancer Foundation Grant 0120.
References
- Ali M, Neumann R, Ensinger W. Acs Nano. 2010;4(12):7267–7274. doi: 10.1021/nn102119q. [DOI] [PubMed] [Google Scholar]
- Altekruse SF, Swerdlow DL, Wells SJ. Food animal practice. 1998;14(1):1–15. doi: 10.1016/s0749-0720(15)30275-9. [DOI] [PubMed] [Google Scholar]
- Belova EI, Lopatkova GY, Pismenskaya ND, Nikonenko VV, Larchet C, Pourcelly G. J Phys Chem B. 2006;110(27):13458–13469. doi: 10.1021/jp062433f. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolova A, Komarova E, Reber K, Gerasimov T, Yavuz O, Bhatt S, Aldissi M. Anal Chem. 2009;81(10):3944–3949. doi: 10.1021/ac9002358. [DOI] [PubMed] [Google Scholar]
- Cagnin S, Caraballo M, Guiducci C, Martini P, Ross M, SantaAna M, Danley D, West T, Lanfranchi G. Sensors. 2009;9:3122–3148. doi: 10.3390/s90403122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Yossifon G, Demekhin EA. Annu Rev Fluid Mech. 2012;44:401–426. [Google Scholar]
- Cheng LJ, Chang HC. Biomicrofluidics. 2011;5(4):046502–8. doi: 10.1063/1.3657928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desharnais BM, Lewis BAG. Soil Sci Soc Am J. 2002;66(5):1518–1525. [Google Scholar]
- Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21(10):1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- Erickson MC, Webb CC, Diaz-Perez JC, Phatak SC, Silvoy JJ, Davey L, Payton AS, Liao J, Ma L, Doyle MP. Journal of food protection. 2010;73(6):1023–1029. doi: 10.4315/0362-028x-73.6.1023. [DOI] [PubMed] [Google Scholar]
- Fritz J, Cooper EB, Gaudet S, Sorger PK, Manalis SR. P Natl Acad Sci USA. 2002;99(22):14142–14146. doi: 10.1073/pnas.232276699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR, Higuchi R, Kricka LJ, Lo YMD, Wittwer CT. Clin Chem. 2005;51(3):661–671. doi: 10.1373/clinchem.2004.045336. [DOI] [PubMed] [Google Scholar]
- Heaton JC, Jones K. Journal of applied microbiology. 2008;104(3):613–626. doi: 10.1111/j.1365-2672.2007.03587.x. [DOI] [PubMed] [Google Scholar]
- Hou G, Peng Z, Tian Y, Zhang H, Jiang L. Chin. Sci. Bull. 2013;58(13):1473–1482. [Google Scholar]
- Jagerszki G, Gyurcsanyi RE, Hofler L, Pretsch E. Nano Lett. 2007;7(6):1609–1612. doi: 10.1021/nl0705438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka-Hamai C, Miyahara Y. Ieee Sens J. 2011;11(12):3153–3160. [Google Scholar]
- Kim DS, Jeong YT, Park HJ, Shin JK, Choi P, Lee JH, Lim G. Biosens Bioelectron. 2004;20(1):69–74. doi: 10.1016/j.bios.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy V, Monfared SM, Cornell B. Nanotechnology, IEEE Transactions on. 2010a;9(3):303–312. [Google Scholar]
- Krishnamurthy V, Monfared SM, Cornell B. Ieee T Nanotechnol. 2010b;9(3):313–321. [Google Scholar]
- Krol JJ, Wessling M, Strathmann H. Journal of Membrane Science. 1999;162(1-2):145–154. [Google Scholar]
- Li Z, Chen Y, Li X, Kamins TI, Nauka K, Williams RS. Nano Lett. 2004;4(2):245–247. [Google Scholar]
- Lin AA, Sastri VR, Tesoro G, Reiser A, Eachus R. Macromolecules. 1988;21(4):1165–1169. [Google Scholar]
- Lucas SW, Harding MM. Anal Biochem. 2000;282(1):70–79. doi: 10.1006/abio.2000.4568. [DOI] [PubMed] [Google Scholar]
- Mangold KA, Reynolds SL. Pediatr Emerg Care. 2013;29(5):665–669. doi: 10.1097/PEC.0b013e31828ed30e. [DOI] [PubMed] [Google Scholar]
- Myint MS, Johnson YJ, Tablante NL, Heckert RA. Food microbiology. 2006;23(6):599–604. doi: 10.1016/j.fm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Nelson BP, Liles MR, Frederick KB, Corn RM, Goodman RM. Environmental microbiology. 2002;4(11):735–743. doi: 10.1046/j.1462-2920.2002.00350.x. [DOI] [PubMed] [Google Scholar]
- O'Connor L, Glynn B. Expert Rev Med Devic. 2010;7(4):529–539. doi: 10.1586/erd.10.22. [DOI] [PubMed] [Google Scholar]
- Palchetti I, Mascini M. Analyst. 2008;133(7):846–854. doi: 10.1039/b802920m. [DOI] [PubMed] [Google Scholar]
- Pandana H, Aschenbach KH, Lenski DR, Fuhrer MS, Khan J, Gomez RD. Ieee Sens J. 2008;8(5-6):655–660. [Google Scholar]
- Peng HI, Miller BL. Analyst. 2011;136(3):436–447. doi: 10.1039/c0an00636j. [DOI] [PubMed] [Google Scholar]
- Pevsner A, Diem M. Appl Spectrosc. 2001;55(11):1502–1505. [Google Scholar]
- Ricci F, Plaxco KW. Microchim Acta. 2008;163(3-4):149–155. [Google Scholar]
- Rubinstein I, Zaltzman B. Adv Colloid Interfac. 2010;159(2):117–129. doi: 10.1016/j.cis.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Scharff RL. Journal of food protection. 2012;75(1):123–131. doi: 10.4315/0362-028X.JFP-11-058. [DOI] [PubMed] [Google Scholar]
- Seah CL, Chow VT, Chan YC. Clinical and diagnostic virology. 1995a;4(2):113–120. doi: 10.1016/0928-0197(94)00063-z. [DOI] [PubMed] [Google Scholar]
- Seah CL, Chow VT, Tan HC, Can YC. Journal of virological methods. 1995b;51(2-3):193–200. doi: 10.1016/0166-0934(94)00104-o. [DOI] [PubMed] [Google Scholar]
- Sharma VK. Molecular and cellular probes. 2006;20(5):298–306. doi: 10.1016/j.mcp.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Shi Z, Johnson JJ, Stack MS. Journal of oncology. 20122012 doi: 10.1155/2012/903581. 903581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu PY, Huang JH. Clinical and diagnostic laboratory immunology. 2004;11(4):642–650. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal GB, Bamdad C, Barberis A, Strominger J, Whitesides GM. Anal Chem. 1996;68(3):490–497. doi: 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]
- Skovgaard N. International journal of food microbiology. 2007;120(3):217–224. doi: 10.1016/j.ijfoodmicro.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Slouka Z, Senapati S, Yan Y, Chang HC. Langmuir. 2013;29(26):8275–8283. doi: 10.1021/la4007179. [DOI] [PubMed] [Google Scholar]
- Steller L, Kreir M, Salzer R. Analytical and bioanalytical chemistry. 2012;402(1):209–230. doi: 10.1007/s00216-011-5517-y. [DOI] [PubMed] [Google Scholar]
- Tanaka Y. Journal of Membrane Science. 2004;244(1-2):1–16. [Google Scholar]
- Wang X, Smirnov S. Acs Nano. 2009;3(4):1004–1010. doi: 10.1021/nn900113x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Lillehoj PB, Ho CM. Pediatr Res. 2010;67(5):458–468. doi: 10.1203/PDR.0b013e3181d361c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WeissWichert C, Smetazko M, ValinaSaba M, Schalkhammer T. J Biomol Screen. 1997;2(1):11–18. [Google Scholar]
- Wellinghausen N, Nockler K, Sigge A, Bartel M, Essig A, Poppert S. Journal of clinical microbiology. 2006;44(5):1828–1830. doi: 10.1128/JCM.44.5.1828-1830.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yossifon G, Mushenheim P, Chang YC, Chang HC. Phys Rev E. 2009;79(4):046305–9. doi: 10.1103/PhysRevE.79.046305. [DOI] [PubMed] [Google Scholar]