Abstract
Targeting of different cellular proteins for conjugation and subsequent degradation via the ubiquitin pathway involves diverse recognition signals and distinct enzymatic factors. A few proteins are recognized via their N-terminal amino acid residue and conjugated by a ubiquitin-protein ligase that recognizes this residue. Most substrates, including the N alpha-acetylated proteins that constitute the vast majority of cellular proteins, are targeted by different signals and are recognized by yet unknown ligases. We have previously shown that degradation of N-terminally blocked proteins requires a specific factor, designated FH, and that the factor acts along with the 26S protease complex to degrade ubiquitin-conjugated proteins. Here, we demonstrate that FH is the protein synthesis elongation factor EF-1 alpha. (a) Partial sequence analysis reveals 100% identity to EF-1 alpha. (b) Like EF-1 alpha, FH binds to immobilized GTP (or GDP) and can be purified in one step using the corresponding nucleotide for elution. (c) Guanine nucleotides that bind to EF-1 alpha protect the ubiquitin system-related activity of FH from heat inactivation, and nucleotides that do not bind do not exert this effect. (d) EF-Tu, the homologous bacterial elongation factor, can substitute for FH/EF-1 alpha in the proteolytic system. This last finding is of particular interest since the ubiquitin system has not been identified in prokaryotes. The activities of both EF-1 alpha and EF-Tu are strongly and specifically inhibited by ubiquitin-aldehyde, a specific inhibitor of ubiquitin isopeptidases. It appears, therefore, that EF-1 alpha may be involved in releasing ubiquitin from multiubiquitin chains, thus rendering the conjugates susceptible to the action of the 26S protease complex.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebersold R. H., Leavitt J., Saavedra R. A., Hood L. E., Kent S. B. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci U S A. 1987 Oct;84(20):6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel J. P., Kasper T. J., Bunn H. F. Cotranslational amino-terminal processing of cytosolic proteins. Cell-free expression of site-directed mutants of human hemoglobin. J Biol Chem. 1988 Jun 15;263(17):8443–8449. [PubMed] [Google Scholar]
- Brown J. L., Roberts W. K. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J Biol Chem. 1976 Feb 25;251(4):1009–1014. [PubMed] [Google Scholar]
- Dahlmann B., Kuehn L., Grziwa A., Zwickl P., Baumeister W. Biochemical properties of the proteasome from Thermoplasma acidophilum. Eur J Biochem. 1992 Sep 15;208(3):789–797. doi: 10.1111/j.1432-1033.1992.tb17249.x. [DOI] [PubMed] [Google Scholar]
- Dever T. E., Costello C. E., Owens C. L., Rosenberry T. L., Merrick W. C. Location of seven post-translational modifications in rabbit elongation factor 1 alpha including dimethyllysine, trimethyllysine, and glycerylphosphorylethanolamine. J Biol Chem. 1989 Dec 5;264(34):20518–20525. [PubMed] [Google Scholar]
- Ferber S., Ciechanover A. Role of arginine-tRNA in protein degradation by the ubiquitin pathway. Nature. 1987 Apr 23;326(6115):808–811. doi: 10.1038/326808a0. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989 Oct 5;264(28):16700–16712. [PubMed] [Google Scholar]
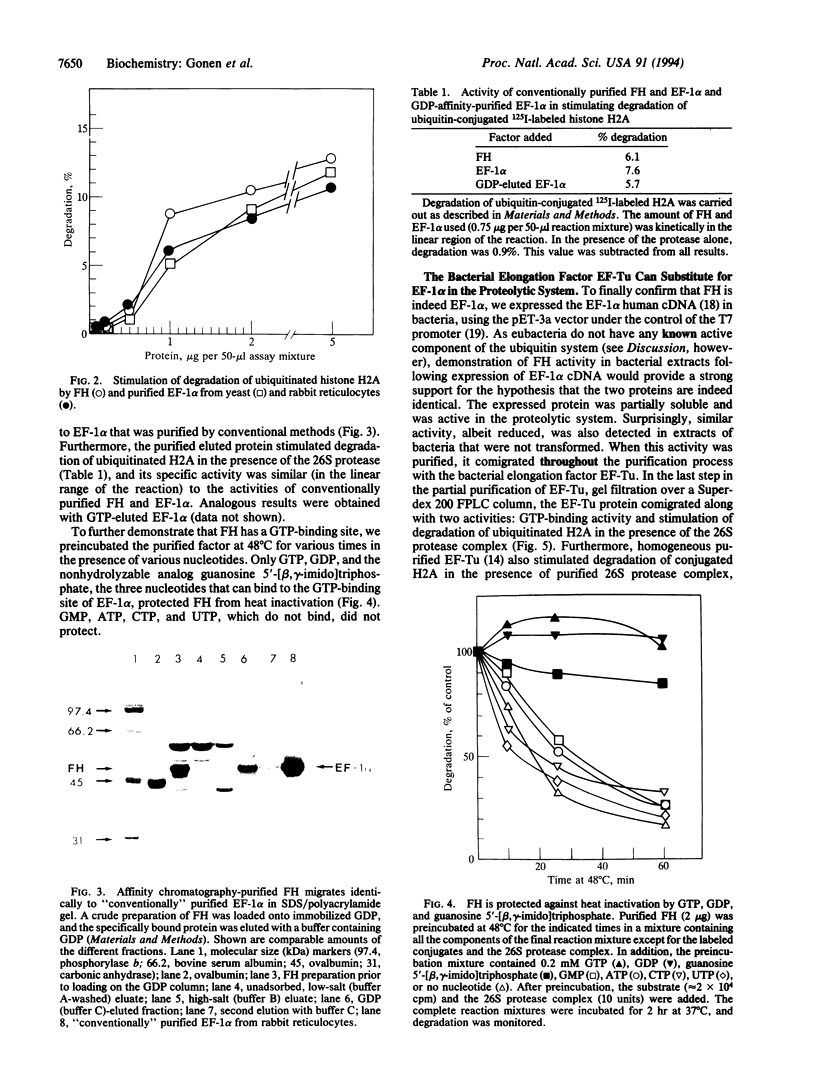
- Gonen H., Schwartz A. L., Ciechanover A. Purification and characterization of a novel protein that is required for degradation of N-alpha-acetylated proteins by the ubiquitin system. J Biol Chem. 1991 Oct 15;266(29):19221–19231. [PubMed] [Google Scholar]
- Heller H., Hershko A. A ubiquitin-protein ligase specific for type III protein substrates. J Biol Chem. 1990 Apr 25;265(12):6532–6535. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hershko A., Rose I. A. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
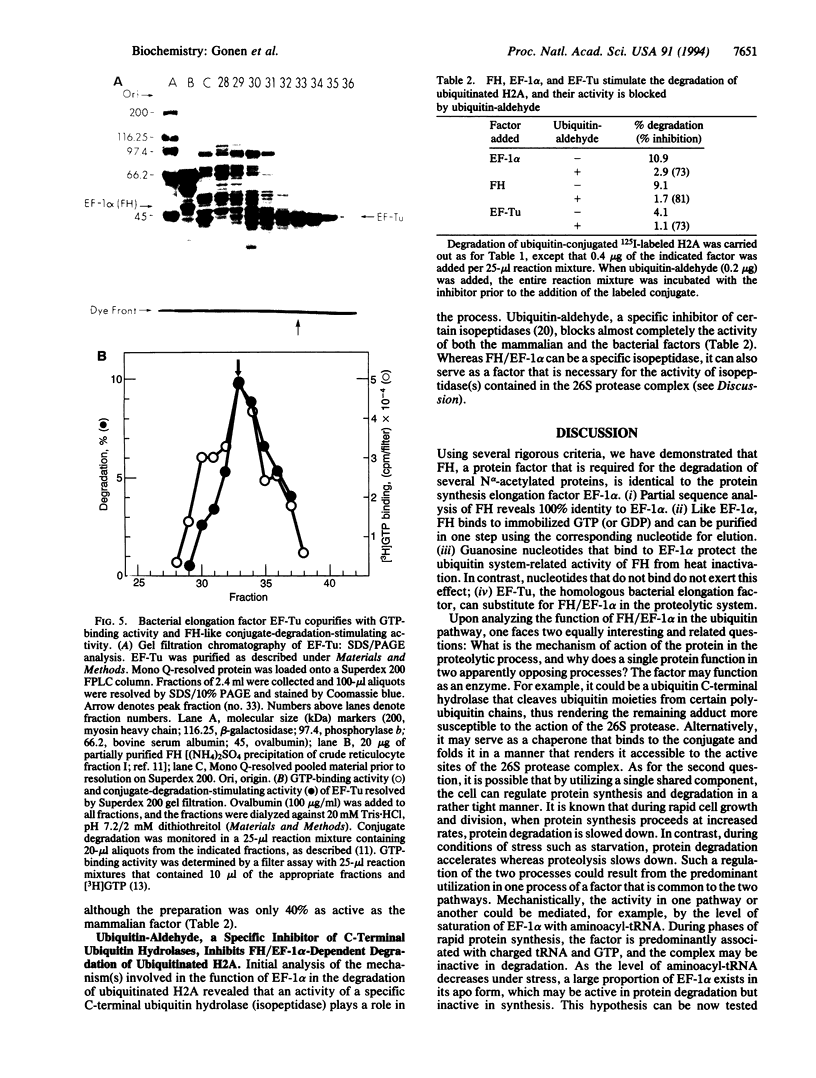
- Hunkapiller M. W., Hewick R. M., Dreyer W. J., Hood L. E. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- Louie A., Ribeiro N. S., Reid B. R., Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem. 1984 Apr 25;259(8):5010–5016. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Mayer A., Siegel N. R., Schwartz A. L., Ciechanover A. Degradation of proteins with acetylated amino termini by the ubiquitin system. Science. 1989 Jun 23;244(4911):1480–1483. doi: 10.1126/science.2544030. [DOI] [PubMed] [Google Scholar]
- Merrick W. C., Dever T. E., Kinzy T. G., Conroy S. C., Cavallius J., Owens C. L. Characterization of protein synthesis factors from rabbit reticulocytes. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):235–240. doi: 10.1016/0167-4781(90)90173-y. [DOI] [PubMed] [Google Scholar]
- Reiss Y., Kaim D., Hershko A. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988 Feb 25;263(6):2693–2698. [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Saha S. K., Chakraburtty K. Protein synthesis in yeast. Isolation of variant forms of elongation factor 1 from the yeast Saccharomyces cerevisiae. J Biol Chem. 1986 Sep 25;261(27):12599–12603. [PubMed] [Google Scholar]
- Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. The N-end rule in bacteria. Science. 1991 Nov 29;254(5036):1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992 May 29;69(5):725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Wolf S., Lottspeich F., Baumeister W. Ubiquitin found in the archaebacterium Thermoplasma acidophilum. FEBS Lett. 1993 Jul 12;326(1-3):42–44. doi: 10.1016/0014-5793(93)81757-q. [DOI] [PubMed] [Google Scholar]