Abstract
Viruses are a therapeutic challenge since their life cycles occur within the host cells and often utilize cellular proteins and hence it is harder to identify therapeutic targets compared to bacteria, which have their own cellular metabolism that is quite different from the host and often present unique targets such as enzymes, etc. Nevertheless, viral proteins may present useful targets for therapy, e.g., small molecule inhibitors of viral polymerases, or prevention, e.g., viral coat proteins for vaccination. However, some viruses may enter an inactive state of persistence or latency where no or very few viral proteins are produced. Thus, reagents that are specifically able to target nucleotide sequences within viral genomes would be a useful addition to the antiviral armamentarium. Such a reagent is the clustered regulatory interspaced short palindromic repeat (CRISPR)-associated 9 (Cas9) system, which is powerful, specific, and highly versatile and provides unprecedented control over genome editing. Here, we will discuss how CRISPR/Cas9 has been used against human viruses and future prospects for novel therapeutic approaches.
INTRODUCTION
Recent years have seen the rapid development of novel genetic-engineering technologies with a number of important clinical applications including the treatment of genetic diseases, cancers and viral infections. New classes of reagents have been developed that are specifically able to target nucleotide sequences within viral genomes and represent important additions to available antiviral strategies. These fall into three main categories. Zinc-finger nucleases (ZFN) are fusion proteins of the nonspecific cleavage domain of the FokI restriction endonuclease with custom-designed Cys2-His2 zinc-finger proteins (Gaj et a, 2012; Kim and Chandrasegaran, 1994), which cause sequence-specific DNA double-strand breaks (DSBs). DSBs are repaired by error-prone process nonhomologous end joining (NHEJ) to yield small alterations at targeted genomic loci. If two DSBs are generated, NHEJ will yield large fragment deletion at the targeted genome. Another class of reagents is known as the transcription activator-like effector nuclease (TALEN) system and are also FokI fusion proteins but the targeting domain is derived from the TAL effector proteins secreted by Xanthomonas bacteria (Wright et al., 2014). The third and most powerful class is the clustered regulatory interspaced short palindromic repeat (CRISPR)-associated 9 (Cas9), which provides unprecedented control over genome editing (Gaj et al., 2013; Mali et al., 2013b; Doudna and Charpentier, 2014; Hsu et al., 2014) and this review will focus on the use of the CRISPR/Cas9 system against human viruses.
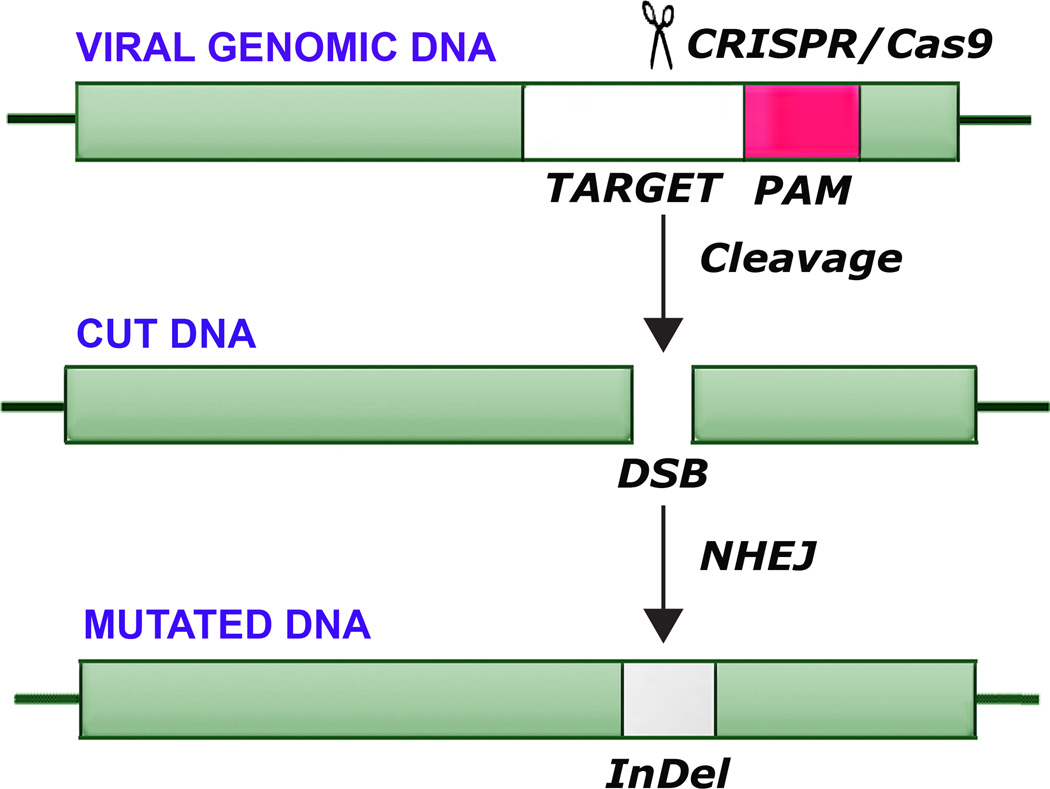
The CRISPR/Cas9 system is simple and easy to use, adaptable, flexible to different targets and continues to improve rapidly (Ran et al., 2013b). The CRISPR/Cas system evolved in bacteria and archaea as a defense mechanism to cope with virus attack (Bhaya et al., 2011). CRISPR loci and Cas proteins identified on sequenced genomes indicate that they are present in ~90% of archaeal and ~50% of bacterial genomes or their resident plasmids. Exciting breakthroughs have allowed this adaptive immune system to be developed into a very adaptable and precise genetic engineering tool, which uses a short guide RNA (gRNA) to direct degradation of specific nucleic acids. There are two distinct components to the CRISPR/Cas9 system: a guide RNA (gRNA) and an endonuclease (Cas9). When the gRNA and Cas9 are co-expressed in cells, the DNA target sequence can be modified or disrupted. The gRNA is designed to contain a 20 base-pair guide sequence, which recruits the gRNA/Cas9 complex to its target by Watson-Crick base-pairing. In addition to the complementary guide sequence, successful binding of Cas9 to the target and subsequent endonuclease disruption require the correct Protospacer Adjacent Motif (PAM) trinucleotide sequence immediately following the target sequence. Cas9 cuts both strands of DNA causing a DSB, which lies 3–4 nucleotides upstream of the PAM sequence. DSBs may be repaired by NHEJ DNA repair pathway, which is error-prone and often results in the generation of inserts/deletions (InDels) at the DSB site that can lead to frameshifts and/or premature stop codons. This can effectively disrupt the open reading frame (ORF) of the target gene. Since Cas9 functions as a general endonuclease, all that is needed is a small gRNA that can be chemically synthesized, in vitro transcripted or cellularly expressed to provide a highly specific gene-targeting tool. This ease of use has allowed it to be used in thousands of applications in the last two years (Doudna and Charpentier, 2014; Hsu et al., 2014; Kennedy and Cullen, 2015; Khalili et al., 2015). The operation of the CRISPR/Cas9 is shown diagrammatically in Figure 1.
Figure 1. Diagrammatic representation of mutagenesis of viral DNA by CRISPR/Cas9.
Viral DNA is shown indicating endonucleolytic cleavage by CRISPR/Cas9 at a target sequence adjacent to a protospacer adjacent motif (PAM) sequence. This is followed by nonhomologous end-joining (NHEJ), which introduces insertion/deletion mutations (InDel).
In addition to its versatility and simplicity, the Cas9 system is remarkable that it achieves a high degree of specificity and provides almost exclusive on-target cleavage given the large size of the human genome. Off-target cleavage can be assayed by either the SURVEYOR assay, which detects mismatched nucleotide pairs resulting from NHEJ, or whole genome sequencing. Thus, whole-genome sequencing at high coverage to assess the degree of off-target cleavage has revealed this to be a very rare event (Hu et al., 2014a; Veres et al., 2014; Yang et al., 2014) and the Cas9 system has proved to be remarkably specific. However, the Cas9 system continues to be modified and improved with respect to its genome editing efficiency and reduction of potential off-target effects. These include developing a “paired Cas9 nickase” strategy, which increases the site specificity DSB induction and can reduce off-target activity by 50- to 1,500-fold in cell lines (Mali et al., 2013a; Ran et la, 2013a; Cho et al., 2014; Shen et al., 2014). Another approach is to fuse a catalytically dead Cas9 to the FokI restriction endonuclease to create an RNA-guided nuclease with enhanced specificity (Guilinger et al., 2014; Tsai et al., 2014).
In principal, eradication of viruses from cells by CRISPR/Cas9 should be applicable to any DNA virus or RNA virus that has a DNA intermediate in its life cycle (Doudna and Charpentier, 2014; Hsu et al., 2014; Kennedy and Cullen, 2015; Khalili et al., 2015). This system has been used recently as a weapon against viruses that cause human diseases and future possibilities.
APPLICATION OF CRISPR/CAS9 TO SPECIFIC HUMAN VIRUSES
Human Papillomaviruses HPV16 and HPV18
Human papillomaviruses (HPVs) are an established etiological agent of human cancer and the high risk or oncogenic human papillomaviruses, HPV16 and HPV18, are responsible for the majority of HPV-associated cancers (White et al., 2014). Worldwide, HPV infections account for more than half of all infection-linked cancers in females, but less than 5% in males. The HPV genome is a linear double-stranded DNA of 7–8 Kbp (Danos et al., 1982) with about ten 10 ORFs and a long control region (LCR), which regulates epithelial-specific transcription. HPV transmission occurs by mucosal contact and by skin to skin contact and is facilitated by microabrasions. Anogenital HPV infections are transmitted by sexual contact. HPV is the major cause of cervical carcinoma and is also involved in other anogenital cancers (White et al., 2014). The oncogenic properties of high-risk HPVs involve the viral proteins E6 and E7 (Munger et al., 2004; Hamid et al., 2009). Cellular p53, the major cellular tumor-suppressor protein, is a target of E6, which binds and degrades p53 (Beaudenon and Huibregtse, 2008) while E7 complexes with proteins in the retinoblastoma family of tumor suppressors that regulate the cell cycle, pRb, p107 and p130 resulting in phosphorylation and release of the E2F transcription factors promoting cell cycle progression (Felsani et al., 2006). Thus the E6 and E7 genes are prime targets for therapeutic intervention in HPV-associated malignant disease.
Kennedy et al (2014) introduced Cas9 and E6- or E7-specific gRNAs into HeLa and SiHa cervical carcinoma cell lines which contain integrated HPV18 or HPV16 respectively. Inactivating deletion and insertion mutations were induced in E6 or E7 resulting in activation of p53 or pRb, resulting in cell cycle arrest and subsequently cell death (Kennedy et al., 2014). Hu et al (2014b) reported that the CRISPR/Cas system using an HPV16-E7-specific gRNA could disrupt HPV16-E7 DNA at specific sites, resulting in the induction of apoptosis and growth inhibition in HPV-positive SiHa and CaSki cells, but not in HPV negative C33A and HEK293 cells. Zhen et al (2014) established CRISPR/Cas9 targeting promoter of HPV16 E6/E7 and targeting E6, E7 transcribed regions. Transduction into SiHa cells showed that CRISPR/Cas9 targeting the promoter, as well as targeting E6 and E7 genes resulted in accumulation of p53 and p21 protein (cyclin-dependent kinase inhibitor 1, a cell cycle inhibitor protein which promotes growth arrest), and reduced cell proliferation in vitro and dramatic inhibition of tumorigenesis in nude mice (Zhen et al., 2014). Thus the CRISPR/Cas9 approach has proven effective when used on HPV-transformed cell lines in culture and has the potential to be developed as an effective therapy for HPV-associated tumors in the clinic.
Hepatitis B virus (HBV)
HBV was recognized in the 1940s as a bloodborne infectious disease causing jaundice and the HBV virus was identified in 1970 (Dane et al., 1970). HBV causes acute and chronic liver infections, which can lead to liver failure, cirrhosis and hepatocellular carcinoma (HCC). It is a hepadnavirus with a small, circular, partially double-stranded DNA genome (Robinson et al.,1974). Of people who are infected with HBV, ~1% develop fulminant hepatitis B, which is associated with liver failure requiring liver transplantation and ~5% develop chronic infections, which may lead to chronic hepatitis, cirrhosis and HCC (Zur Hausen, 2006). Chronic hepatitis B involves persistence of HBV DNA in an episomal form known as the covalently closed circular DNA (cccDNA), which is a hurdle for eradication of the virus. The DNA genome of HBV is small and encodes only a few proteins including HBV Protein X (HBx) and HBV surface antigen (HBsAg), which exert multiple pleiotropic effects that are important in transformation but are not well understood. The HBV genome may integrate into the host genome and this can occur at many different sites and may causes disruption of key genes that regulate proliferative signaling, which is thought to be an important mechanism in the development of HCC in HBV-infected individuals. HBx is oncogenic and can transform rodent hepatocytes and NIH 3T3 cells (Kew, 2011). HBx may act by activating signal transduction pathways involved in the regulation of cell proliferation such as ERKs, SAPKs and p38 protein kinase (Diao et al., 2001). It has also been reported to bind to p53 (Feitelson et al., 1993).
Curing chronic HBV infection will require the specific eradication of the persistent HBV cccDNA from infected cells. Lin et al (2014) designed eight gRNAs against HBV and showed that the CRISPR/Cas9 system significantly reduced the production of HBV core and HBsAg proteins in the Huh-7 hepatocyte-derived cellular carcinoma cell cells transfected with an HBV-expression vector. Further this system could cleave intrahepatic HBV genome-containing plasmid and facilitate its clearance in vivo in a mouse model resulting in a reduction in serum HBsAg level (Lin et al., 2014). Thus, CRISPR/Cas9 system could disrupt the HBV templates both in vitro and in vivo and may have a potential in eradicating persistent HBV infection. In another study, Seeger and Sohn (2014) tested in HepG2 hepatoma cells expressing the HBV receptor with different HBV-specific gRNAs and found inhibition of HBV infections up to eightfold, which was due to mutations and deletions in the cccDNA caused by Cas9 cleavage and repair by NHEJ. Kennedy et al. (2015) used lentiviral transduction of Cas9 and HBV-specific gRNAs and showed effective inhibition of HBV DNA production for in vitro models of both chronic and de novo HBV infection. Total HBV viral DNA levels were reduced by up to ~1000-fold while cccDNA levels were reduced by up to ~10-fold and the majority of the residual viral DNA was mutationally inactivated. In the most recent study of HBV and CRISPR, Zhen et al. (2015) targeted the HBsAg and HBx-encoding region of HBV, both in a cell culture system and in vivo. HBsAg levels in the cultures media of cells and in the sera of mice were reduced as shown by ELISA. The HBV DNA levels and HBsAg expression in mouse livers were reduced as shown by qPCR and immunohistochemistry respectively. In conclusion, studies from several labs have shown the potential of CRISPR/Cas9 to treat HBV-associated diseases.
Epstein-Barr virus (EBV)
EBV is a DNA virus of the herpesvirus family that causes Burkitt’s lymphoma and nasopharyngeal carcinoma (Pagano, 1999). EBV infections are very common with more than 90% of people becoming infected by their twenties (Henle et al., 1969). EBV has a large, 168–184 Kbp linear double-stranded DNA genome with 85 genes, terminal repeat regions and an internal repeat region (Baer et al., 1984). During latency the genome circularizes to form an episome that is maintained at constant copy number and (Adams and Lindahl, 1975). EBV has two subtypes, EBV-1 and EBV-2, which differ at the EBNA locus and infects lymphocytes and also epithelial cells, which are the primary site of initial replication (Lemon et al., 1977). EBV can enter a latent state in B cells, where the circular episomal EBV genome expresses only a few of the proteins that it encodes. The proteins expressed during latency may be involved in the events which lead to cellular transformation, especially EBV nuclear antigen 1 (EBNA-1, Rowe et al., 1992), which is a sequence-specific DNA-binding phosphoprotein that is involved in the maintenance of the latent EBV episome, viral DNA replication and cellular transformation (Rawlins et al., 1985). Another viral protein, EBNA-2 regulates expression of the other latency-associated EBV genes, LMP-1 and LMP-2 (Abbot et al., 1990). LMP-1 activates a number of pathways involved in cell transformation, induces invasion and metastasis factors and acts together with other EBV latency-associated proteins and RNAs (EBV-encoded small RNA-1 and -2 (EBER1 and EBER2), and the BamHI rightward transcripts (BARTs)) via multiple molecular mechanisms to effect cellular transformation by EBV in Burkitt’s lymphoma and nasopharyngeal carcinoma (Marquitz and Raab-Traub, 2102; Wakisaka and Pagano, 2003).
EBV has also been investigated using the CRISPR/Cas9 system (Wang and Quake, 2014). Cells derived from a patient with Burkitt’s lymphoma with latent EBV infection (Raji cells) showed a marked reduction in proliferation and decline in viral load as well as restoration of the apoptosis pathway in the cells after treatment with a CRISPR/Cas9 vector targeted to the viral genome using gRNAs specific for EBNA1, EBNA3C and LMP1 and others. SURVEYOR assays confirmed efficient editing of individual sites within the EBV genome. Yuen et al (2015) reported CRISPR/Cas9-mediated editing of EBV in human cells using two gRNAs to make a targeted deletion of 558 bp in the promoter region for BART in several human epithelial cell lines latently infected with EBV including nasopharyngeal carcinoma C666-1 cells. This resulted in the loss of BART miRNA expression and activity indicating the feasibility of CRISPR/Cas9-mediated editing of the EBV genome. No off-target cleavage was found by deep sequencing.
HIV-1
After 30 years since its discovery, HIV-1/AIDS remains a major public health problem worldwide. The development of highly effective Combination Antiretroviral Therapy that powerfully inhibits viral replication in cells that support HIV-1 infection and robustly reduces plasma viremia, has meant that HIV-1 infection is now survivable rather than always fatal. Despite this however, AIDS remains incurable since HIV-1 can undergo permanent integration into the host genome, thus providing a persistent reservoir of virus and the risk of viral reactivation even after effective antiretroviral therapy. As well as DNA viruses, retroviruses are amenable to CRISPR/Cas9 manipulation because they have a DNA intermediate in their life cycle. The CRISPR/Cas9 system has been used to target the HIV-1 LTR integrated in the genome of latently infected cells in culture (Ebina et al., 2013; Hu et al., 2014a). In this way, it was possible to inactivate viral gene expression and replication in latently infected microglial, promonocytic, and T cells (Hu et al., 2014a). This is a potential therapeutic advance in overcoming the stumbling block in eliminating all virus from HIV-1-infected individuals, i.e., permanently integrated DNA proviral copies of HIV-1 in the host genome, the reservoir of virus that is unreachable by current antiviral therapies. With CRISPR/Cas9, it was possible to identify specific targets in the HIV-1 proviral genome that completely excised integrated provirus (Hu et al., 2014a). Moreover, the presence of HIV-1-specific gRNAs and Cas9 in cells prevents HV-1 infection, i.e., CRISPR/Cas9 has both therapeutic and prophylactic potential (Hu et al., 2014a). Cas9/gRNAs caused neither genotoxicity nor off-target editing to the genome of host cells as revealed by whole-genome sequencing analysis and SURVEYOR assays. Recently, Zhu et al. (2015) tested 10 sites in HIV-1 DNA for CRISPR/Cas9 in JLat10.6 cells that are latently infected by HIV-1. Sequencing results showed that each target site in HIV-1 DNA was efficiently mutated by CRISPR/Cas9 with the target site in the second exon of Rev exhibiting the highest degree of mutation, with the result that HIV-1 gene expression and virus production were reduced 20-fold. In another recent study, Liao et al. (2015) also demonstrated that CRISPR/Cas9 system disrupted latently integrated HIV-1 genome and provided long-term adaptive defense against new viral infection, expression and replication in human cells. They showed that engineered human-induced pluripotent stem cells stably expressing HIV-targeted CRISPR/Cas9 could be efficiently differentiated into HIV reservoir cell types and maintain their resistance to HIV-1 challenge.
Another approach to employ CRISPR/Cas9 against HIV-1 would be to target cellular genes encoding proteins required for HIV-1 infection such as CCR5, a coreceptor for HIV-1 entry (Manjunath et al., 2013; Stone et al., 2013). Wang et al. (2014) showed that transduction of lentiviral vectors expressing Cas9 and CCR5-specific gRNAs into HIV-1 susceptible human CD4+ cells gave a high frequency of CCR5 gene disruption and became not only resistant to R5-tropic HIV-1, including transmitted/founder (T/F) HIV-1 isolates but also had a selective advantage over cells with undisrupted CCR5 during R5-tropic HIV-1 infection. No mutation was detected at potential off-target sites that were highly homologous, particularly CCR2.
Thus the CRISPR/Cas9 system has the potential to be developed to provide a specific and efficacious approach against HIV-1/AIDS that can be employed both prophylactically and therapeutically. The feasibility of HIV-1 eradication by CRISPR/Cas9 in animal models and in a clinical setting with individuals with HIV-1/AIDS remains under investigation.
CONCLUSION
The CRISPR/Cas9 system is a new and powerful tool for precise, simple and easy to use genetic engineering of different DNA targets. Although it has only been available to be used in eukaryotic cells for only about two years, it has already found many potential applications to human diseases including genetic disorders, cancer and viruses. So far, CRISPR/Cas9 has been applied to four human viruses as discussed above and summarized in Table 1.
TABLE 1.
Application of CRISPR/CAS9 to specific human viruses
| Virus | Target | Cells | Results | Reference |
|---|---|---|---|---|
| HPV18 | E6 or E7 | HeLa | Induction of p53 or Rb Cell cycle arrest | Kennedy et al., 2014 |
| HPV16 | E6 or E7 | SiHa | Induction of p53 or Rb Cell cycle arrest | Kennedy et al., 2014 |
| HPV16 | E7 | SiHa and CaSki | Apoptosis and growth inhibition | Hu et al., 2014b |
| HPV16 | E6, E7 or E6/E7 promoter | SiHa | p53 and p21 induction, Inhibition of cell proliferation and tumorigenesis | Zhen et al., 2014 |
| HBV | P1 (1,292–1,314) & XCp (1,742–1,764) | Huh-7 | Reduction in HBsAg level in medium | Lin et al., 2014 |
| P1 (1,292–1,314) & XCp (1,742–1,764) | Mouse liver in vivo | Reduction in HBsAg level in serum | Lin et al., 2014 | |
| ENII-CP/X & Pre-C | HepG2 with HBVR | 8-fold inhibition of HBV infection | Seeger and Sohn, 2014 | |
| HBsAg, core, and/orRT. | HepAD38 | Reduction in viral DNA and cccDNA levels | Kennedy et al., 2015 | |
| HBsAg, HBx | HepG2.2.15 | Reduction in HBsAg level In medium | Zhen et al., 2015 | |
| Mouse liver | Reduction in HBsAg level In serum | |||
| EBV | EBNA1, EBNA3C LMP1 and others | Raji | Reduction in proliferation and viral load, apoptosis | Wang and Quake, 2014 |
| BART promoter | C661-1 cells and others | Loss of miRNA expression | Yuen et al., 2015 | |
| HIV-1 | LTR | T cells | Loss of HIV-1 gene expression | Ebina et al., 2013 |
| LTR | Microglia, promonocytes, T cells | Excision of provirus Prevention of HIV-1 infection | Hu et al., 2014a | |
| CCR5 | CD4+ cells | Resistance to R5-tropic HIV-1 | Wang et al., 2014 | |
There are many other human disease-associated viruses that are likely susceptible to this approach. Examples include herpesviruses such as Kaposi's sarcoma-associated herpesvirus (KSHV, HHV8), which causes Kaposi’s sarcoma (Chang et al., 1994) and polyomaviruses such as Merkel cell carcinoma virus (MCV), which causes Merkel cell carcinoma (Feng et al., 2007), polyomavirus JC (JCV), which causes progressive multifocal leukoencephalopathy (Padgett et al., 1971) and polyomavirus BK (BKV), which causes polyomavirus-associated nephropathy (PVAN, Gardner et al., 1971).
While there are many examples of the efficacious use of CRISPR/Cas9 in cell culture as described in this review, application to the clinic will require a method of safe and efficient delivery. For basic research studies in cell culture, plasmid transfection or viral transduction with lentiviral or adenoviral vectors have been employed. Lentivirus transduction in particular has been used in many labs because of the high efficiency of delivery and stable gene expression. However, the safety of lentiviruses is compromised by its unfavorable insertion-mediated mutagenesis effect, whereby the silencing or activation of unexpected genes may occur. The generation of integrase-deficient lentivirus vectors (IDLV) has recently provided a means to reduce the risk of virus-mediated insertions and improve the safety of gene delivery (Liu et al., 2014). Immunogenicity of the viral vector is another problem both for lentiviruses (Rothe et al., 2013) and adenoviruses (Campos and Barry, 2007). Another possibility is to use adeno-associated virus (AAV), which is only mildly immunogenic, to deliver Cas9 (Howes and Schofield, 2015; Platt et al., 2014; Senis et al., 2014; Swiech et al., 2015). AAV is a small virus but the widely used Cas9 gene from Streptococcus pyogenes (spCas9) is 4.1 Kbp long so the constructs used in these studies necessarily had to use minimal promoter and polyadenylation signals. To improve delivery, it is possible to split spCas9 into functional N-terminal and C-terminal fragments and deliver and express them as separate polypeptides, which can be recruited by the gRNA into a ternary complex that recapitulates the activity of full-length spCas9 and catalyzes site-specific DNA cleavage (Wright et al., 2015; Zetsche et al., 2015). In addition, shorter size of functional Cas9 from Staphylococcus aureus (saCas9) has been identified and exhibits similar cleaving efficiency to spCas9. Thus the field of the CRISPR/Cas9 system is rapidly evolving and holds much promise for the future.
ACKNOWLEDGEMENTS
We wish to thank past and present members of the Department on Neuroscience and Center for Neurovirology for their continued support and insightful discussions. We also acknowledge the intellectual contributions of the Temple University School of Medicine Comprehensive Neuroaids Center (Basic Science Cores I and II). This work was supported by grants R01AI077460 (MKW), R01NS087971 (WH, KK) and P30MH092177 (KK) awarded by the NIH.
Footnotes
CONFLICTS OF INTEREST
None
REFERENCES
- Abbot SD, Rowe M, Cadwallader K, Ricksten A, Gordon J, Wang F, Rymo L, Rickinson AB. Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein. J Virol. 1990;64(5):2126–2134. doi: 10.1128/jvi.64.5.2126-2134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A, Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc Natl Acad Sci USA. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, Tuffnell PS, Barrell BG. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Beaudenon S, Huibregtse JM. HPV E6, E6AP and cervical cancer. BMC Biochem. 2008;9(Suppl 1):S4. doi: 10.1186/1471-2091-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–997. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Campos SK, Barry MA. Current advances and future challenges in Adenoviral vector biology and targeting. Curr Gene Ther. 2007;7(3):189–204. doi: 10.2174/156652307780859062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1(7649):695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- Danos O, Katinka M, Yaniv M. Human papillomavirus 1a complete DNA sequence: a novel type of genome organization among papovaviridae. EMBO J. 1982;1(2):231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Diao J, Garces R, Richardson CD. X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev. 2001;12(2–3):189–205. doi: 10.1016/s1359-6101(00)00034-4. [DOI] [PubMed] [Google Scholar]
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson MA, Zhu M, Duan LX, London WT. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8(5):1109–1117. [PubMed] [Google Scholar]
- Felsani A, Mileo AM, Paggi MG. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene. 2006;25(38):5277–5285. doi: 10.1038/sj.onc.1209621. [DOI] [PubMed] [Google Scholar]
- Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, Chang Y, Moore PS. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81:11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9(8):805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1(7712):1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid NA, Brown C, Gaston K. The regulation of cell proliferation by the papillomavirus early proteins. Cell Mol Life Sci. 2009;66(10):1700–1717. doi: 10.1007/s00018-009-8631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle G, Henle W, Clifford P, Diehl V, Kafuko GW, Kirya BG, Klein G, Morrow RH, Munube GM, Pike P, Tukei PM, Ziegler JL. Antibodies to Epstein-Barr virus in Burkitt's lymphoma and control groups. J Natl Cancer Inst. 1969;43(5):1147–1157. [PubMed] [Google Scholar]
- Howes R1, Schofield C. Genome engineering using Adeno-Associated Virus (AAV) Methods Mol Biol. 2015;1239:75–103. doi: 10.1007/978-1-4939-1862-1_5. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA. 2014a;111(31):11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang C, Wang L, Jiang X, Shen H, He D, Li K, Xi L, Ma D, Wang H. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int. 2014b;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88(20):11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Cullen BR. Bacterial CRISPR/Cas DNA endonucleases: A revolutionary technology that could dramatically impact viral research and treatment. Virology. 2015 doi: 10.1016/j.virol.2015.02.024. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2011;26(Suppl 1):144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- Khalili K, Gordon J, Kaminski R, Cosentino L, Hu W. Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J Neurovirol. 2015 doi: 10.1007/s13365-014-0308-9. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Li L, Chandrasegaran S. Insertion and deletion mutants of FokI restriction endonuclease. J Biol Chem. 1994;269(50):31978–31982. [PubMed] [Google Scholar]
- Lemon SM, Hutt LM, Shaw JE, Li JL, Pagano JS. Replication of EBV in epithelial cells during infectious mononucleosis. Nature. 1977;268(5617):268–270. doi: 10.1038/268268a0. [DOI] [PubMed] [Google Scholar]
- Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR, Young J, Izpisua Belmonte JC. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015 doi: 10.1038/ncomms7413. (In Press) [DOI] [PubMed] [Google Scholar]
- Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, Wu FY, Kao JH, Chen DS, Chen PJ. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KC, Lin BS, Gao AD, Ma HY, Zhao M, Zhang R, Yan HH, Yi XF, Lin SJ, Que JW, Lan XP. Integrase-deficient lentivirus: opportunities and challenges for human gene therapy. Curr Gene Ther. 2014;14(5):352–364. doi: 10.2174/1566523214666140825124311. [DOI] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013b;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N, Yi G, Dang Y, Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5(11):2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquitz AR, Raab-Traub N. The role of miRNAs and EBV BARTs in NPC. Semin Cancer Biol. 2012;22(2):166–172. doi: 10.1016/j.semcancer.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78(21):11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Zu Rhein GM, Walker DL, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet. 1971;1(7712):1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- Pagano JS. Epstein-Barr virus: the first human tumor virus and its role in cancer. Proc Assoc Am Physicians. 1999;111(6):573–580. doi: 10.1046/j.1525-1381.1999.t01-1-99220.x. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013a;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013b;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Robinson WS, Clayton DA, Greenman RL. DNA of a human hepatitis B virus candidate. J Virol. 1974;14(2):384–391. doi: 10.1128/jvi.14.2.384-391.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Modlich U, Schambach A. Biosafety challenges for use of lentiviral vectors in gene therapy. Curr Gene Ther. 2013;13(6):453–468. doi: 10.2174/15665232113136660006. [DOI] [PubMed] [Google Scholar]
- Rowe M, Lear AL, Croom-Carter D, Davies AH, Rickinson AB. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol. 1992;66(1):122–131. doi: 10.1128/jvi.66.1.122-131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Sohn JA. Targeting Hepatitis B Virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senís E, Fatouros C, Große S, Wiedtke E, Niopek D, Mueller AK, Börner K, Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J. 2014;9(11):1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS. 2013;8(3):217–223. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15(1):27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakisaka N, Pagano JS. Epstein-Barr virus induces invasion and metastasis factors. Anticancer Res. 2003;23(3A):2133–2138. [PubMed] [Google Scholar]
- Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci USA. 2014;111(36):13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9(12):e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Pagano JS, Khalili K. Viruses and human cancers: a long road of discovery of molecular paradigms. Clin Microbiol Rev. 2014;27(3):463–481. doi: 10.1128/CMR.00124-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Li T, Yang B, Spalding MH. TALEN-mediated genome editing: prospects and perspectives. Biochem J. 2014;462(1):15–24. doi: 10.1042/BJ20140295. [DOI] [PubMed] [Google Scholar]
- Yang L, Grishin D, Wang G, Aach J, Zhang CZ, Chari R, Homsy J, Cai X, Zhao Y, Fan JB, Seidman C, Seidman J, Pu W, Church G. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507. doi: 10.1038/ncomms6507. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KS, Chan CP, Wong NH, Ho CH, Ho TH, Lei T, Deng W, Tsao SW, Chen H, Kok KH, Jin DY. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J Gen Virol. 2015;96(3):626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015 doi: 10.1038/gt.2015.2. (In Press) [DOI] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33(2):139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Lei R, Le Duff Y, Li J, Guo F, Wainberg MA, Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 12(1):22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Hausen H. Infections causing human cancer. Wiley, Wilmington, DE: 2006. [Google Scholar]