Abstract
R-type pyocins are representatives of contractile ejection systems, a class of biological nanomachines that includes, among others, the bacterial type VI secretion system (T6SS) and contractile bacteriophage tails. We report atomic models of the Pseudomonas aeruginosa precontraction pyocin sheath and tube, and the postcontraction sheath, obtained by cryo-EM at 3.5-Å and 3.9-Å resolutions, respectively. The central channel of the tube is negatively charged, in contrast to the neutral and positive counterparts in T6SSs and phage tails. The sheath is interwoven by long N- and C-terminal extension arms emanating from each subunit, which create an extensive two-dimensional mesh that has the same connectivity in the extended and contracted state of the sheath. We propose that the contraction process draws energy from electrostatic and shape complementarities to insert the inner tube through bacterial cell membranes to eventually kill the bacteria.
Contractile ejection systems are ubiquitous in bacteria1. Pathogens such as Vibrio cholerae and Burkholderia pseudomallei use contractile T6SSs to translocate protein virulence factors into target eukaryotic cells2,3, and T6SSs are also used for interbacterial competition4. Myovirus bacteriophages, exemplified by phage T4, use a similar contractile machine to translocate DNA and proteins into bacterial cells5–7. Some bacteria secrete insecticidal protein complexes that deliver toxins by contraction8,9, and others induce metamorphosis in marine animals by using morphologically similar structures10. These nanomachines use a sheath-tube assembly to create an opening in the envelopes of target eukaryotic or bacterial cells to translocate molecules or ions across lipid membranes. These events are accompanied by a massive structural transformation that involves contraction of the sheath and linear motion of the tube. In the absence of atomic-resolution information, how these machines work has remained poorly understood.
R-type pyocins produced by P. aeruginosa use the same contractility to kill competing bacteria11. However, these pyocins are unique because, unlike other contractile systems, they are not known to be delivery vehicles for DNA or toxins but appear to function by creating a channel in the envelope of their target bacterial cell that dissipates the cell's proton potential. Five R-type pyocins (R1–R5) have been identified, and they differ primarily in the C terminus of the tail fiber that confers target-strain specificity12,13. Owing to their high killing capacity14, R-type pyocins have attracted attention for antimicrobial and bioengineering applications12,15–17.
All known contractile machines have a similar architectural organization18. Most details regarding assembly and contraction pathways have been derived from extensive studies of the phage T4 tail. Contraction has been hypothesized to be driven by energy stored in the extended state of the particle during assembly19. The sheath has been proposed to assemble into its initial, extended, high-energy metastable state by using the central tube as a scaffold because both sheath and tube appear to have the same symmetry, at least in phage T4 (ref. 20). In the contracted state, the sheath is an extremely stable oligomeric structure that is resistant to chemical dissociation21.
Structural studies of several bacteriophage tails have shown that contraction is accompanied by large changes in the orientation of sheath subunits. However, none of these studies were based on atomic descriptions of either the tube or the sheath, and the details of how energy is stored in the precontraction particle and how sheath structure is maintained during the massive conformational changes have remained unclear2,5–7,22. Finally, how the tubes of these seemingly similar contractile machines can be used for translocating such different cargos—protons and other cations for pyocins, proteins for T6SSs and nucleic acids for phages—has remained a mystery.
We set out to understand the mechanism of contraction for these nanomachine assemblies. Here we report the atomic structures of the pyocin R2 sheath and tube in its extended precontraction form, at 3.5-Å resolution, and the sheath in its postcontraction form, at 3.9-Å resolution, both obtained by cryo-EM. Our atomic model of the precontraction state describes sheath-sheath, sheath-tube and tube-tube interactions, and the model for the postcontraction state describes alternative sheath-sheath interactions. These structural data suggest how energy is stored in the extended state, how it is released during contraction and how the pyocin tube is optimized for dissipating proton motive force to kill bacteria, a task different from those of other contractile machines.
RESULTS
Overall structure
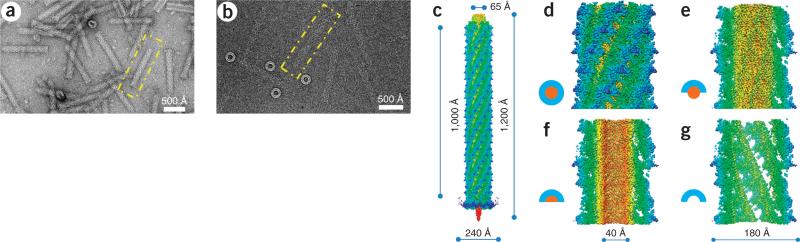
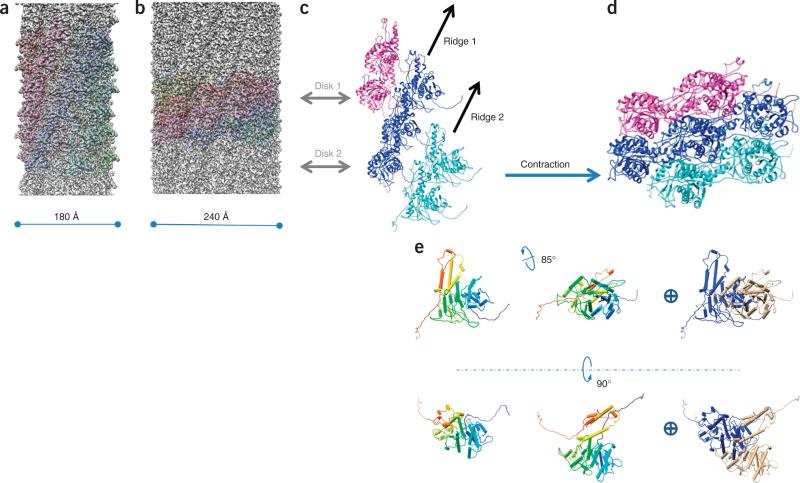
As seen in cryo-EM images and the montage three-dimensional (3D) model, the R2 pyocin can be divided into three major parts: baseplate, trunk and collar (Fig. 1a–c and Supplementary Video 1). The baseplate is a ring-like structure of 240 Å in diameter. Six tail fibers extend from the outer side of the baseplate. Their proximal parts are well resolved in the cryo-EM map. The inner side of the baseplate ring is connected to the central spike protein23 via spokes. Although many of the structural features of the central spike are lost, owing to averaging, the central metal ion at the tip of the spike complex23 is resolved when the cryo-EM map is viewed at a high density threshold. At the other end of the pyocin is its collar, where the trunk diameter becomes 65 Å. The precontraction trunk is formed by the sheath and tube and has a helical structure. We reconstructed the precontraction trunk to 3.5-Å resolution by averaging about 372,000 asymmetric units (Supplementary Videos 2–4 and Online Methods). Similarly to all known contractile ejection systems and to bacteriophage tails in general, the structure has an axial six-fold rotational symmetry and can be considered to be made of discs, each containing six sheath subunits and six tube subunits. The discs are related to one another by an 18.3° right-handed rotation and a 38.4-Å translation up the rotational axis. The subunits of the sheath protein display large globular protrusions, which create six prominent ridges with grooves in between (Fig. 1d,g). The internal tube is smooth and devoid of any prominent surface features (Fig. 1e,f). Each asymmetric unit of the helical structure consists of one copy of the sheath protein (41.2 kDa) and one copy of the tube protein (18.1 kDa) (further described below).
Figure 1.
Overall structure of precontraction pyocin R2. (a,b) Electron microscopy images of the pyocin R2 embedded in uranyl acetate stain (a) or vitreous ice (b). One precontraction pyocin particle is boxed. (c) Surface view of a 3D montage reconstruction of the entire precontraction pyocin. (d–g) Surface views of the 3D reconstruction of pyocin trunk. Each view corresponds to the segmentation patterns illustrated at left. 3D models and density maps of the attachment helix of the sheath and β-sheet region of the tube are shown in Supplementary Videos 1–4.
The tube as a proton-conducting channel
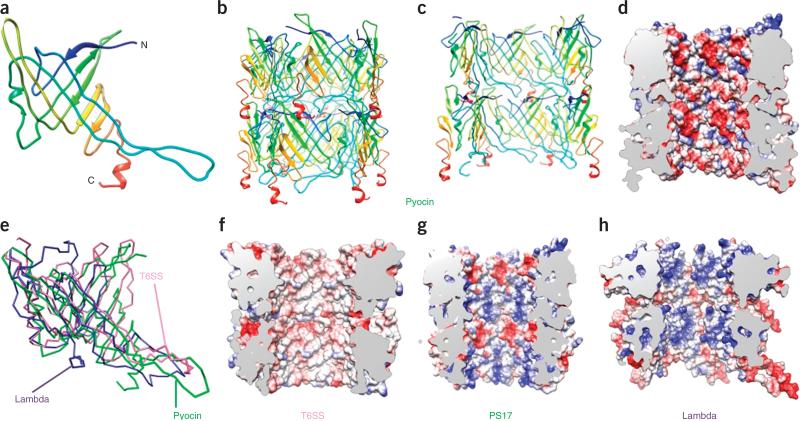
The tube protein has two major secondary-structure elements, which consist of two antiparallel β-sheets with an angle of ~90° between the directions of their strands (Fig. 2a and Supplementary Video 5). Two β-hairpins that originate from the first β-sheet join to form a second β-sheet, and the entire structure forms a roll. This β-sheet roll is distinct from the highly conserved jelly-roll fold24,25 found in many viral and cellular proteins26. The fold of the pyocin tube protein is very similar to that of the T6SS tube protein Hcp27, the bacteriophage λ tube protein28 and the T4 gp27 hub-protein tube domain29.
Figure 2.
Structure of the pyocin tube compared to other tubes. (a) Ribbon diagram of a pyocin tube-protein monomer. Additional ribbon diagram is in Supplementary Video 5. (b) Ribbon diagram of two hexamers (discs). (c,d) Cut-away views as ribbon (c) and electrostatic (d) diagrams showing the inner surface of the structure in b. Charge distribution (red, negative; blue, positive; white, neutral) is shown in d. (e) Superposition of tube-protein structures from pyocin, λ phage (PDB 2K4Q28, 29% residues superimposed, r.m.s. deviation 4.96 Å) and T6SS (Hcp, PDB 3EAA31, 39% residues superimposed, r.m.s. deviation 3.32 Å). (f–h) As in d, but for T6SS (f), PS17 phage (g) and λ phage (h).
Six tube-protein subunits create a ring-like structure that probably represents an assembly unit of the pyocin tube (Fig. 2b,c). The ring structures display complementary surface charges on their contacting interfaces: one is predominantly negatively charged, and the other is positively charged. This creates an electrostatic dipole, which is probably required for directional self-assembly (Supplementary Fig. 1).
24 β-strands line up along the inner surface of the tube to form a barrel, one of the largest β-structural pores ever resolved in a protein structure. An equally large pore is found in a bacterial pilus usher (PDB 2VQI30). This surface displays a prominent negative charge (Fig. 2c,d). We compared this property of the pyocin tube with that of other tail-like systems, such as the T6SS Hcp protein (PDB 3EAA31), bacteriophage λ (PDB 2K4Q28) and bacteriophage PS17 (with SWISS-MODEL homology modeling based on our pyocin tube structure) (Fig. 2e), all of which have high sequence homology. We found that none of the other structures are negatively charged. Consistently with its function, the inner surface of the T6SS tube is largely neutral (Fig. 2f), and, unlike those of pyocins and phages, T6SS tubes do not appear to involve active transport. Instead, the central channel and tip of the T6SS contain protein effectors, or effector domains, that are delivered into target cells in their loaded conformation32. By contrast, the two bacteriophage tubes are positively charged and function to translocate negatively charged DNA (Fig. 2g,h). Thus, internal charge differences appear to reflect adaptation to different functions.
The contractile sheath is a highly interwoven mesh
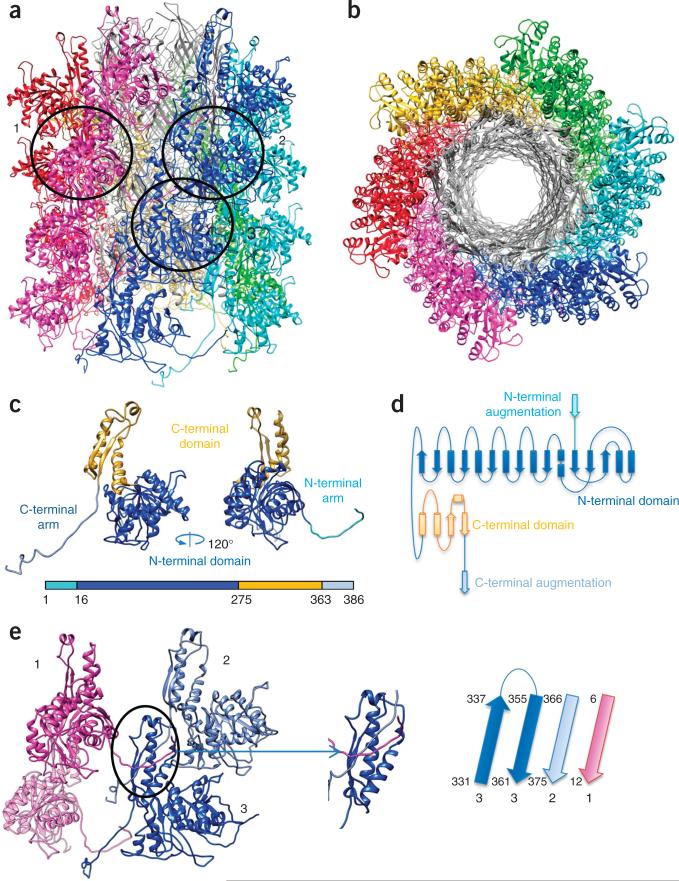
The sheath protein consists of a large N-terminal domain (N domain, residues 21–280), a smaller C-terminal domain (C domain, 281–361) (Fig. 3a–d and Supplementary Video 6) and long extension arms at both termini of the polypeptide chain (residues 2–20 and 362–386). The N domain contains a central six-stranded β-sheet sandwiched between six α-helices (three on each side of the sheet, Fig. 3c). A similar fold was reported for the sheath proteins of several bacteriophages18,22 and a T6SS33, although the latter contains two proteins instead of one. This domain forms the prominent ridge on the surface of the pyocin sheath. The inner C domain consists of a four-stranded β-sheet with two α-helices running along its length. Two strands in this sheet are formed by the extension arms that originate from two neighboring sheath subunits belonging to a disc above (Figs. 3c–e and 4). Residues 375–386 of the C-terminal arm extend even further to a disc below, and this arm thus connects three consecutive sheath subunits within the same ridge. Hydrogen bonds between the extension arms and the rest of the C domain are reinforced by extensive hydrophobic interactions. The C domains of the pyocin, several phages and T6SS VipB sheath proteins have a fold similar to that of the T4 gp25 baseplate protein (PDB 4HRZ), thus suggesting that all of these domains have a common function.
Figure 3.
Structure of the pyocin sheath. (a,b) Side (a) and top (b) views of the atomic model of the pyocin trunk. Sheath subunits are shown in colors, and the tube is shown in gray. (c) Ribbon structure of the sheath monomer. Additional ribbon diagram is in Supplementary Video 6. (d) Topological diagram of the sheath monomer. (e) Joining of three (with numbers matching those in a) of the four adjacent monomers of the sheath protein, via β-sheet augmentation in their C domains (oval and inset). The polarities and identities of the β-strands in this augmented sheet are illustrated on the right.
Figure 4.
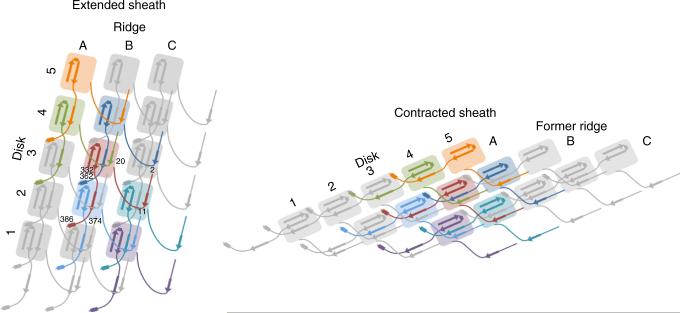
Schematic diagram for the pyocin sheath topology of the extended mesh created by the N- and C-terminal extension arms within the sheath in the pre- and postcontraction states. β-strands participating in the sheet augmentation of the C domain are shown. α-helices involved in intersubunit interactions are shown as rectangles. Residue numbers for the subunit in red are given for strategic locations.
The β-sheet augmentation is the most prominent interaction within the pyocin sheath. The free energy of this interaction in the extended state (–20 kcal/mol as calculated by PISA34) is twice that of all other sheath-sheath and sheath-tube interactions combined (a total of –10 kcal/mol). Apart from the extended arms, the sheath subunits do not interact with each other within a disk—all interactions are confined to a single ridge (Fig. 3a). Interestingly, oligomerization of bacterial pili involves a similar β-sheet–augmentation mechanism35. The β-sheet in each pilus subunit is augmented by a single strand to give rise to a linear structure that is strong enough to attach a bacterial cell to a surface. In contrast, the C domain of the pyocin sheath contains two strands that belong to two different subunits and create a highly interwoven two-dimensional mesh (Figs. 3e and 4). Indeed, this mesh- like organization by β-sheet augmentation has also been observed in the structures of postcontraction T6SSs from both Francisella novicida36 and V. cholerae37, and the N- and C-terminal extended arms have been shown by mutagenesis to be critical to contractile function but not assembly.
Sheath-tube interaction
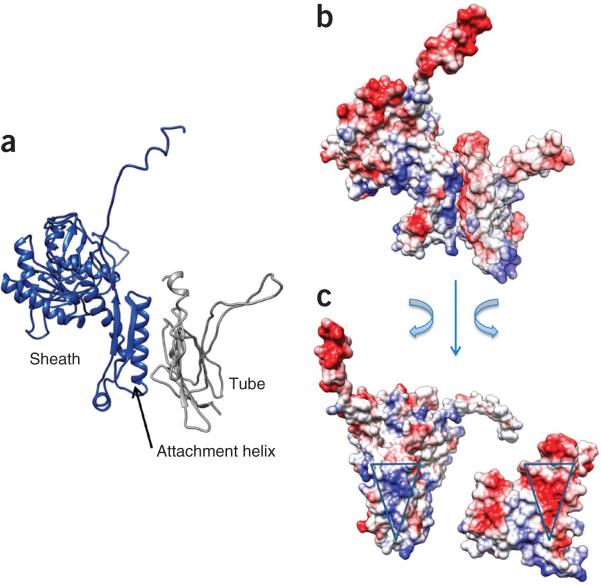
The sheath-tube interaction is largely electrostatic and has a free energy of –5 kcal/mol per pair of sheath-tube subunits. The outer surface of the tube subunit displays a triangular negatively charged patch (Fig. 5). The C domain of the sheath subunit binds to this patch via a complementary, triangular-shaped patch of positive charges on one of its two α-helices (‘attachment helix’) (Fig. 5).
Figure 5.
Sheath-tube interactions. (a,b) Side views of the interface between a sheath and a tube-protein subunit in ribbon diagram (a) and charged surface (b). (c) An open-book view of b. The complementary patches of interacting charges on both sheath and tube are marked with triangles.
Structure of the contracted sheath
To determine the mechanism of pyocin contraction, we also solved the structure of the contracted trunk to 3.9-Å resolution (Fig. 6a–d, Supplementary Fig. 2 and Supplementary Video 7). Upon contraction, the length of the sheath decreases by a factor of 2.4, its diameter increases from 180 to 240 Å, and it detaches from the tube, to allow the latter to extend beyond the plane of the baseplate (Supplementary Video 8). The relationship between the adjacent discs changed to a right-handed 33.1° rotation and a 16.2-Å translation up the rotational axis. The structure of the tube upon contraction is unknown, but the helical symmetry of the contracted sheath does not match that of the tube. In the figures presented here, the tube density is smeared out and is masked off for clarity. This quaternary sheath arrangement is similar but of opposite handedness to the contracted T6SS sheaths36,37.
Figure 6.
Contraction of pyocin. (a,b) Comparison between the precontraction (a) and postcontraction (b) sheath. The cryo-EM density map in a is filtered to a resolution comparable to that in b (3.9 Å). Animation of contracted sheath is in Supplementary Video 7. (c,d) Ribbon diagrams of seven adjacent sheath subunits in their precontraction (c) and postcontraction (d) states. Morphing between c and d is shown in Supplementary Videos 8 and 9. (e) Overall rotational and translational movement of a single sheath monomer during contraction, illustrated in side and top views. The precontraction (left) and postcontraction (middle) states of a monomer are superimposed at right (blue, precontraction state; beige, postcontraction state).
Except for the β-sheet–augmentation regions at the N and C termini, the structure of the sheath subunit in the pre- and post-contraction conformations is essentially unchanged. The subunits belonging to the two conformations can be superimposed with an r.m.s. deviation of 1.5 Å with all residues participating in the alignment (Supplementary Fig. 3a,b). Remarkably, the β-sheet augmentation of the C domain and the intersubunit mesh that binds the subunits together are both preserved in the contracted structure. Considering that the free energy of β-sheet augmentation is lower than that for any other intersubunit interaction within the sheath, and β-sheet augmentation is unchanged in the initial (extended) and final (contracted) state of the sheath, we propose that the mesh is preserved through the contraction event. This mesh conceivably gives the sheath stability and maintains the integrity of the entire structure during contraction (Fig. 4). Furthermore, because the contraction is known to propagate from the baseplate in a disc-by-disc manner38, the mesh is probably responsible for the pulling force that acts between discs during contraction.
DISCUSSION
A model for assembly
The tube is known to have a critical role in the assembly of the sheath in the extended state of the phage T4 tail18,39. However, the free energy of sheath-tube interactions alone is insufficient to direct the assembly of the extended conformation because the contracted state is energetically much more favorable (as described below). A possible solution to this free-energy puzzle can be inferred by considering that the assembly of the pyocin sheath probably starts from the baseplate, as is the case for the T4 tail39. Together, the baseplate and the tube create a platform that binds the first disc of sheath subunits in the extended conformation. The tube and the existing disc of sheath subunits then serve as a scaffold for assembly of the rest of the sheath. Thus, the assembly of the contracted state is avoided by the creation of a template in which sheath subunits interact along a ridge without any lateral contacts (which are more energetically favorable and are eventually realized in the contracted state). This template-driven assembly probably results in a metastable oligomeric structure, which could possibly be unlocked from this state by the baseplate upon interaction with the target-cell surface.
A model for contraction
In contrast to minimal changes in sheath subunit structure, the gross subunit movement of the sheath protein upon contraction is profound. In addition to translational movement along and perpendicular to the helical axis, both domains, in concert, turn 85° about an axis roughly perpendicular to the helical axis (Fig. 6e). The extended arms connecting the subunits function as hinges in this transformation (Fig. 4). The rest of the protein probably maintains its structure through the contraction event and moves as a rigid body (Fig. 6c,d and Supplementary Videos 8–10). This rigid-body movement was hypothesized earlier for the phage T4 sheath, on the basis of cryo-EM structures at 15- to 17-Å resolution40. The current atomic structures suggest how the mesh created by the extended arms makes this massive structural transformation possible.
The rearrangement of sheath subunits upon contraction leads to a much more densely packed structure in which the ridges seen in the extended conformation are brought closer together to become partly interdigitated (Fig. 6c,d). Each subunit in the contracted sheath interacts with ten other subunits. Despite losing contact with the tube, each sheath subunit in the contracted state buries about 1,145 Å2 more of its surface than in the extended state. Contraction leads to the formation of many new charged pairs of complementary residues. Unlike those in the sheath-tube interaction, these residues do not form large matching patches. Instead, the charged interactions are rather complex and are spread out over the entire interface between sheath proteins (Supplementary Figs. 3c–g, 4 and 5).
The free energy of all interfaces for each subunit in the contracted sheath (–42 kcal/mol) is much lower than that of the extended state (–30 kcal/mol, including the sheath-tube interface). The difference in the free energy characterizing the two states is –12 kcal/mol, which is comparable to a value of –25 kcal/mol obtained in earlier microcalori-metric measurements of phage T4 tail contraction19. The T4 sheath protein is about twice the size of the pyocin sheath, and its subunits form additional interactions that are likely to result in a larger difference in free energy between the two states. There are 162 sheath subunits in the pyocin particle, giving a value of ~2,000 kcal/mol of energy released during the contraction event. Most of this energy is probably converted into a linear power stroke that drives the tube out of the sheath and into the target-cell membrane. Because T6SSs can be five times longer than pyocins2, a similar contractile mechanism may allow them to develop the tremendous force required for translocation of large protein molecules across target-cell membranes.
Any molecular machine, such as a pyocin or viral fusion protein41, that uses stored (potential) energy for transforming its structure from one state to another needs to overcome the challenge of assembly (and arrest) in a high-energy state that is stable enough to maintain its conformation until a switch signal is received. Pyocins probably respond to this challenge by using several strategies. First, the sheath protein avoids self-assembly into the energetically favored lower-energy contracted state by dispersing complementary charges over the surface of the subunit. This strategy ensures that freshly expressed sheath-protein subunits do not electrostatically attract and bind each other to form contracted sheath-like structures. Second, the assembly of the sheath is directed by a scaffold created by the preassembled tube. Third, the baseplate may provide a template that positions the initial disk of sheath subunits in a high-energy conformation that attracts subsequent disks in a directional manner.
The R2 pyocin is an extraordinary molecular machine that performs work by using energy stored within its own biological ‘battery’. Its receptor specificity makes it an ideal system for engineering narrow-spectrum antibiotics for targeted killing, thus minimizing concerns regarding transmissible resistance or disruption of protective microbiota. Given its simplicity, it also serves as an excellent model for understanding the energetics and mechanisms of assembly of other more complex contractile nanomachines.
ONLINE METHODS
Purification of pyocin
R2 pyocin was produced from P. aeruginosa strain PAO1 and purified as described previously12. Briefly, the cells were grown in G medium to an OD600 of 0.2 at 37 °C with 250 r.p.m. shaking, at which time mitomycin C was added to a final concentration of 3 μg/ml. After 3 h, cell lysis was noted, DNase 1 (Invitrogen) was added to a concentration of 2 U/ml, and the culture was incubated for an additional 30 min. Cell debris was removed by centrifugation at 17,000g for 1 h, after which pyocin was precipitated from the supernatant by addition of saturated ammonium sulfate to a final concentration of 1.6 M. The precipitate was pelleted by centrifugation at 17,000g for 1 h. This pellet was resuspended in 1/10 volume of TN50 (Tris-Cl, pH 7.5, and 50 mM NaCl) and centrifuged at 60,000g for 1 h to pellet the pyocins. The pyocins were then resuspended in 1/20 of the original culture volume in TN50.
This crude pyocin sample was further purified by sucrose gradient with 10% to 50% sucrose (w/v) concentration for 1 h at 100,000g and 4 °C. The pyocin-containing band was carefully collected with a 100-μl pipette tip, then diluted with 4 ml PBS buffer, pH 7.4, and concentrated with a 100-kDa molecular filter (Amicon). The concentrated sample was dialyzed (diluted with 4 ml buffer again, then concentrated) with (4 ml) PBS buffer, pH 7.4, twice in the same filter and reduced to a final volume of 30 μl. Possibly owing to the mechanical stress during purification, the resulting sample contained both pre- and postcontraction pyocins.
Cryo-electron microscopy and reconstruction
Each aliquot of 2.5-μl purified pyocin sample in pH 7.4 PBS was applied to a preirradiated (‘baked’) Quantifoil 1.2/1.3 grid that was never glow discharged after baking. The sample was blotted and flash frozen in a Vitrobot Mark IV (FEI) (blot time, 4 s; blot force, 1; humidity, 100%). Cryo-EM images were then taken in an FEI Titan Krios microscope operated at 300 kV, with a dosage of 25 e/Å2 and a nominal magnification of 59,000×. Images were recorded on Kodak SO-163 film and digitized in Nikon 9200 ED scanners. The final pixel size is 1.104 Å/pixel. The defocus range of these images is 0.6–2.5 μm underfocus.
Both pre- and postcontraction pyocin particles were selected manually with EMAN42 helixboxer from the same data set. Their structures were reconstructed with modified43,44 IHRSR45 with Relion46 as a refinement engine (in a procedure in which Relion is used to refine the reference without any helical symmetry for one iteration, modified IHRSR is used to apply the helical symmetry in real space to the resulting volume, and finally the helically symmetrized volume is passed back to Relion for the next iteration of refinement; described in ref. 36). Initial guesses of helical parameters were based on the parameters for the T4 bacteriophage tail, which were then refined to convergence by IHRSR45. An initial refinement was done for each state by modified IHRSR with EMAN42 as a refinement engine, and each resulting structure was used as the initial reference for its final refinement with modified IHRSR with Relion as the refinement engine. The latter refinements were governed by the ‘auto-refine’ function of Relion, which automatically determines working resolution for each iteration and final convergence. The refinements for pre- and postcontraction pyocin converged at 33 and 32 iterations, respectively. Pre- and postcontraction pyocin particles were extracted from the same total of 307 film images. The defocus range for these 307 images was 1.6–2.5 μm underfocus. 6,591 pre-contraction pyocin assemblies were included in the refinement process. They were segmented so that each box contained about two discs worth of independent data. A total of 30,771 such boxes, equivalent to about 372,000 asymmetric units, were included in the final reconstruction. The resolution of the final reconstruction was determined by gold-standard FSC 0.143 at 3.5 Å (Supplementary Fig. 6a). A total of 1,499 postcontraction pyocin assemblies were included in the refinement process for the postcontraction structure. The overlapping was set to 18 pixels. A total of 12,242 boxes, equivalent to about 90,346 asymmetric units, were included in the final reconstruction. The resolution was determined to be 3.9 Å (Supplementary Fig. 6b).
Atomic modeling
We built a model for the pyocin sheath and tube (precontraction) with Coot47 with no reference. This model was refined with CNS48 and then with Phenix49. Noncrystallographic symmetry, including the six-fold rotational symmetry and the helical symmetry, were applied as a restraint among identical chains. A total of four discs were included in the model. This model was refined to 3.5 Å eventually, with R factor, 27.8%; Rfree, 28.0%; and last-shell R factor, 48.9% (Ramachandran plot in Supplementary Fig. 6a). We also built a model for the contracted sheath by docking the precontraction sheath into its density and correcting/rebuilding mismatching regions. It was refined similarly to 3.9 Å (except that the model was refined with six discs), with R factor, 26.6%; Rfree, 27.5%; and last-shell R factor, 42.9% (Ramachandran plot in Supplementary Fig. 6b). Figures were prepared with the UCSF Chimera package50.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Martin of AvidBiotics for discussion and support throughout this project and UCLA undergraduate student J. Chiou for assistance in data processing. This research was supported in part by the US National Institutes of Health (NIH) (AI046420/AI094386 and GM071940 to Z.H.Z.). P.G. was supported in part by an American Heart Association Western States Affiliates Postdoctoral Fellowship (13POST17340020). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines, supported by UCLA and by instrumentation grants from the NIH (1S10RR23057). Recharge fees for access to this facility for imaging the pyocin samples were partially defrayed by an award to Z.H.Z. from the UCLA Clinical and Translational Science Institute core voucher program.
Footnotes
AUTHOR CONTRIBUTIONS
Z.H.Z., J.F.M., P.G., D.S. and P.G.L. designed the experiments. Z.H.Z. supervised the execution of the experiments. D.S. prepared the crude pyocin sample. X.Y. purified the same sample. P.G. performed cryo-EM, processed the images and built the atomic models. All authors interpreted the results. P.G. and P.G.L. drafted the manuscript. P.G., P.G.L., Z.H.Z. and J.F.M. edited the manuscript, and all authors reviewed the final manuscript.
Accession codes. Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank, under accession numbers EMD-6270 (precontraction) and EMD-6271 (postcontraction). Atomic models have been deposited in the Protein Data Bank under accession codes 3J9Q (precontraction) and 3J9R (postcontraction).
Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Bönemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol. Microbiol. 2010;76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 2.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.French CT, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc. Natl. Acad. Sci. USA. 2011;108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aksyuk AA, et al. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostyuchenko VA, et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- 7.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 8.Hurst MRH, Glare TR, Jackson TA. Cloning Serratia entomophila antifeeding genes: a putative defective prophage active against the grass grub Costelytra zealandica. J. Bacteriol. 2004;186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Dowling AJ, Gerike U, ffrench-Constant RH, Waterfield NR. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J. Bacteriol. 2006;188:2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shikuma NJ, et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail–like structures. Science. 2014;343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 12.Williams SR, Gebhart D, Martin DW, Scholl D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 2008;74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama K, et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 14.Kageyama M, Ikeda K, Egami F. Studies of a Pyocin. III. Biological properties of the pyocin. J. Biochem. 1964;55:59–64. doi: 10.1093/oxfordjournals.jbchem.a127841. [DOI] [PubMed] [Google Scholar]
- 15.Scholl D, et al. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob. Agents Chemother. 2009;53:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie JM, et al. An Escherichia coli O157-specific engineered pyocin prevents and ameliorates infection by E. coli O157:H7 in an animal model of diarrheal disease. Antimicrob. Agents Chemother. 2011;55:5469–5474. doi: 10.1128/AAC.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uratani Y, Hoshino T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 1984;157:632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 19.Arisaka F, Engel J, Klump H. Contraction and dissociation of the bacteriophage T4 tail sheath induced by heat and urea. Prog. Clin. Biol. Res. 1981;64:365–379. [PubMed] [Google Scholar]
- 20.Arisaka F, Tschopp J, Van Driel R, Engel J. Reassembly of the bacteriophage T4 tail from the core-baseplate and the monomeric sheath protein P18: a cooperative association process. J. Mol. Biol. 1979;132:369–386. doi: 10.1016/0022-2836(79)90266-3. [DOI] [PubMed] [Google Scholar]
- 21.To CM, Kellenberger E, Eisenstark A. Disassembly of T-even bacteriophage into structural parts and subunits. J. Mol. Biol. 1969;46:493–511. doi: 10.1016/0022-2836(69)90192-2. [DOI] [PubMed] [Google Scholar]
- 22.Aksyuk Anastasia A, et al. Structural conservation of the Myoviridae phage tail sheath protein fold. Structure. 2011;19:1885–1894. doi: 10.1016/j.str.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG. Phage pierces the host cell membrane with the iron-loaded spike. Structure. 2012;20:326–339. doi: 10.1016/j.str.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 25.Abad-Zapatero C, et al. Structure of southern bean mosaic virus at 2.8 Å resolution. Nature. 1980;286:33–39. doi: 10.1038/286033a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 Å resolution. eLife. 2013;2:e01299. doi: 10.7554/eLife.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 30.Remaut H, et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jobichen C, et al. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS ONE. 2010;5:e12910. doi: 10.1371/journal.pone.0012910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kube S, et al. Structure of the VipA/B type VI secretion complex suggests a contraction-state-specific recycling mechanism. Cell Rep. 2014;8:20–30. doi: 10.1016/j.celrep.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Remaut H, Waksman G. Protein–protein interaction through β-strand addition. Trends Biochem. Sci. 2006;31:436–444. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Clemens DL, Ge P, Lee B-Y, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudryashev M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moody MF. Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J. Mol. Biol. 1973;80:613–635. doi: 10.1016/0022-2836(73)90200-3. [DOI] [PubMed] [Google Scholar]
- 39.King J, Mykolajewyoz N. Bacteriophage T4 tail assembly: proteins of the sheath, core and baseplate. J. Mol. Biol. 1973;75:339–358. doi: 10.1016/0022-2836(73)90025-9. [DOI] [PubMed] [Google Scholar]
- 40.Aksyuk AA, et al. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge P, Zhou ZH. Chaperone fusion proteins aid entropy-driven maturation of class II viral fusion proteins. Trends Microbiol. 2014;22:100–106. doi: 10.1016/j.tim.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 43.Ge P, et al. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc. Natl. Acad. Sci. USA. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.