Abstract
Background
Vocalization is a prominent social behavior among vertebrates, including in the midshipman fish, an established model for elucidating the neural basis of acoustic communication. Courtship vocalizations produced by territorial males are essential for reproductive success, vary over daily and seasonal cycles, and last up to hours per call. Vocalizations rely upon extreme synchrony and millisecond precision in the firing of a homogeneous population of motoneurons, the vocal motor nucleus (VMN). Although studies have identified neural mechanisms driving rapid, precise, and stable neuronal firing over long periods of calling, little is known about underlying genetic/molecular mechanisms.
Results
We used RNA sequencing-based transcriptome analyses to compare patterns of gene expression in VMN to the surrounding hindbrain across three daily and seasonal time points of high and low sound production to identify candidate genes that underlie VMN’s intrinsic and network neuronal properties. Results from gene ontology enrichment, enzyme pathway mapping, and gene category-wide expression levels highlighted the importance of cellular respiration in VMN function, consistent with the high energetic demands of sustained vocal behavior. Functionally important candidate genes upregulated in the VMN, including at time points corresponding to high natural vocal activity, encode ion channels and neurotransmitter receptors, hormone receptors and biosynthetic enzymes, neuromodulators, aerobic respiration enzymes, and antioxidants. Quantitative PCR and RNA-seq expression levels for 28 genes were significantly correlated. Many candidate gene products regulate mechanisms of neuronal excitability, including those previously identified in VMN motoneurons, as well as novel ones that remain to be investigated. Supporting evidence from previous studies in midshipman strongly validate the value of transcriptomic analyses for linking genes to neural characters that drive behavior.
Conclusions
Transcriptome analyses highlighted a suite of molecular mechanisms that regulate vocalization over behaviorally relevant timescales, spanning milliseconds to hours and seasons. To our knowledge, this is the first comprehensive characterization of gene expression in a dedicated vocal motor nucleus. Candidate genes identified here may belong to a conserved genetic toolkit for vocal motoneurons facing similar energetic and neurophysiological demands.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1577-2) contains supplementary material, which is available to authorized users.
Keywords: Vocalization, Motoneuron, Transcriptome, Gene expression, Seasonal reproduction
Background
Vocal-acoustic communication is a prominent feature of vertebrate social behavior. Despite having evolved diverse peripheral sonic organs, brainstem vocal networks that control vocalization are remarkably conserved across vertebrate taxa [1,2]. In tetrapods and several clades of fishes, sound production relies upon temporally stable modulations of acoustic waveforms over millisecond timescales [3-6]. Although vocal networks in several vertebrate lineages achieve high degrees of synchrony and precision (e.g., [7-10]), it is unknown whether these properties depend on a suite of conserved genetic/molecular mechanisms.
Recent microarray and genome analyses in a songbird, the zebra finch (Taeniopygia guttata), have advanced our understanding of the relationship between genes and vocal behavior by identifying transcription factors and gene networks activated in forebrain song nuclei [11-13], including during singing [13-15]. Similar large-scale gene expression studies in other species that aim to link gene expression to behavior typically use whole brains or regions containing multiple neuron types (e.g. [11-14,16-19]). While these studies provide significant insight into the genetic regulation of behavioral states, cellular and network level interpretations of results are confounded by the complexity of the underlying neural structure or behavior.
For the vast majority of vocal vertebrates that currently lack either genomic data or species-specific microarrays, no large-scale expression study has examined genes whose products directly support the function of a single neuronal population devoted to sound production. Here, we identify candidate molecular mechanisms contributing to the neural coding of vocal behavior in the plainfin midshipman fish (Porichthys notatus), an established model for the neural basis of acoustic communication. In order to link gene expression to neurophysiological events that directly translate to behavior, we focused RNA sequencing (RNA-seq) transcriptome analyses on a single hindbrain nucleus that is dedicated to the temporal patterning of a simple vocal behavior.
Midshipman belong to an order of highly vocal teleost fish known as toadfishes (Batrachoidiformes) [20] that produce different call types depending on social context [21,22]. Nest-guarding males, which are used in this study, produce several call types, including a long duration advertisement “hum”. The hum is a stable, high frequency (100 Hz at ~16°C) call that is energetically demanding and essential for mate attraction [23]. Hums are produced continuously for minutes to hours, including in captivity (e.g., see 1.85 h hum in Figure 1A), throughout the night during the summer breeding season [22,24-26].
Figure 1.
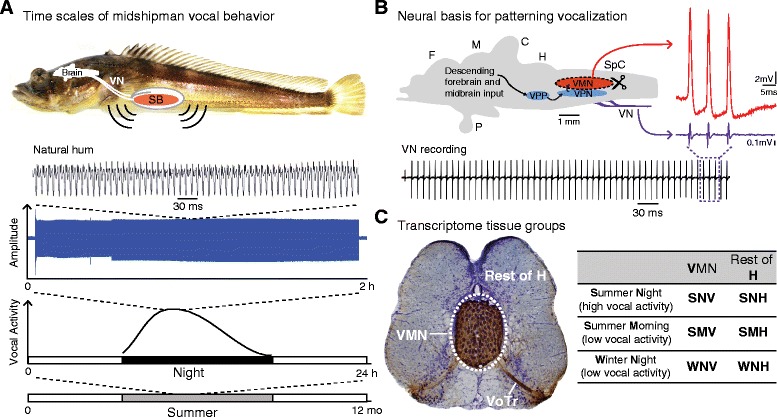
Midshipman is a neuroethological model for studying neural control of vocalization. A) Midshipman vocal behavior varies across a range of time scales. Picture of a midshipman fish (top) with schematic side view of the brain, vocal nerve (VN), and sonic swimbladder (SB), the sound-producing organ with vocal muscles (red) attached to the sidewalls. Continuous hums can last from mins to >1 h, as shown in the 1.85 h hum recorded from a captive male (blue trace), and are produced repetitively throughout the night during the summer breeding season (bottom). Close-up of natural hum is adapted from [25]. B) Schematic saggital view of the midshipman brain showing the hindbrain vocal pattern generator that consists of the vocal pre-pacemaker nucleus (VPP), vocal pacemaker nucleus (VPN), and vocal motor nucleus (VMN) (left). Forebrain and midbrain vocal centers drive the hindbrain pattern generator, which provides a precise and synchronous code that controls sonic muscle contraction in a one-to-one manner (compare the series of sound pulses in the natural hum in A to the vocal nerve potentials directly to the right in the bottom trace in B. Extreme temporal precision of motoneuron firing is shown by corresponding traces from an intracellular VMN recording (red) and VN recording (purple; adapted from [8]). Trace of a long duration VN recording (bottom) adapted from [39]. C) Tissue groups used for transcriptome analysis and their notations. Left: a transverse section at the level of VMN showing bilateral, transneuronal biocytin labeling in VMN and the vocal tract (VoTr) (see [27]); each VMN innervates the ipsilateral vocal SB muscle. In this study, we surgically separated the midline pair of VMN from the surrounding hindbrain tissue (Rest of H). Right: abbreviations of sample groups according to the tissue and time of collection, used throughout this paper.
In midshipman, well-delineated forebrain, midbrain, and hindbrain nuclei comprise a vocal network [27-29] (Figure 1B). The collective output of this network determines the natural vocalization’s fine (e.g. fundamental frequency) and gross (e.g. duration) temporal structure [30]. This motor command is relayed to a single pair of dedicated vocal muscles by way of the hindbrain vocal motor nucleus (VMN), which forms a hindbrain vocal central pattern generator (CPG) with separate premotor pacemaker and prepacemaker nuclei (Figure 1B) [8,30-36]. Although forebrain and midbrain vocal nuclei gate and modulate hindbrain vocal CPG output and are highly sensitive to hormone action [34,37-39], the surgically isolated hindbrain vocal CPG region is both necessary and sufficient for generating the vocal command signal, and remains sensitive to hormonal modulation [34].
VMN’s intrinsic and network neuronal properties drive synchronous and stable population-level firing that faithfully amplifies premotor temporal input [8]. Each VMN spike leads to a single sound pulse, resulting in a one-to-one translation between VMN output and the temporal properties of natural vocalization, such as call duration and fundamental frequency (compare Figure 1A and B). Terrestrial vertebrates such as songbirds, bats, and primates rely on vocal networks that integrate input from breathing circuits [40-44] to drive multiple vocal and respiratory muscles. In contrast, the simpler vocal systems of fishes do not rely on airflow and engage a single pair of sonic muscles, providing a straightforward link between gene expression, neural networks, and behavior [8,10].
In addition to the direct translation between neurophysiological activity and behavior, the midshipman VMN provides several advantages for revealing molecular mechanisms underlying vocal patterning. First, the paired midline VMN together (Figure 1C) form a highly interconnected, homogenous population of ~4000 vocal motoneurons [8,45], inclusive of surrounding glial cells and presynaptic inputs that support VMN network function, which can be excised in-toto for focal molecular analysis of a single brain nucleus [46]. Second, a set of intrinsic (low baseline excitability, rapid membrane repolarization) and network (dense excitatory and inhibitory inputs; electrotonic coupling) neuronal properties of the VMN are well characterized [8]. These properties guide the identification of candidate genes encoding specific molecular counterparts that likely contribute to VMN’s extreme population-level synchrony on a millisecond timescale (Figure 1B). Third, midshipman vocal behavior follows predictable daily and seasonal cycles [22,25,26,39]. This allows us to utilize temporal variation to identify genes driving changes in vocal network excitability at different daily and seasonal time points [25] (Figure 1C). Fourth, a large body of work that documents hormonal modulation of the vocal system at multiple levels of analyses (neuroanatomy, neurophysiology, qPCR quantification, behavior) informs the functional significance of gene expression patterns (see [47,48]).
We aimed to identify molecular pathways specific to VMN function by comparing patterns of gene expression in the VMN to the surrounding hindbrain (H) across daily and seasonal time points of high and low sound production (Figure 1A,C). As outlined in Figure 2, we focused first on between-tissue differences in gene expression to identify candidate gene pathways important for VMN function, then on within-tissue comparisons to identify candidate gene pathways with biologically relevant expression patterns across the day and season.
Figure 2.
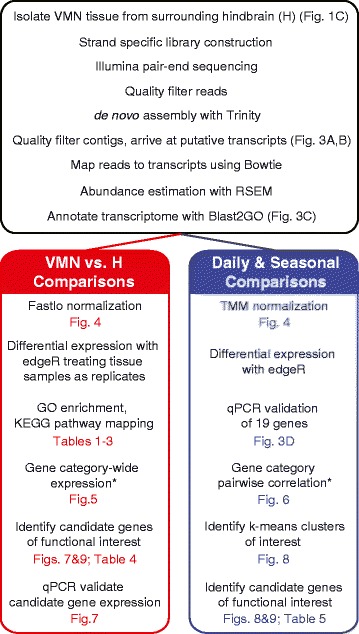
Schematic of analytic approaches with associated figure and table numbers. We took a two-pronged approach in analyzing the vast array of data generated from RNA-seq. First, for VMN vs. H comparisons, we used a cyclic loess method, fastlo, to eliminate a non-linear tissue-dependent skew between datasets (left, red). We then treated time point samples as replicates for each tissue for differential expression with edgeR. Second, we followed the Trinity-supported downstream analyses of clustering differentially expressed transcripts and focused these analyses on clusters with biologically relevant expression patterns across the day and season (right, blue). Additionally, we analyzed expression and correlation patterns of transcripts belonging to six broad gene functional categories. *Genes were grouped in broad categories regardless of whether they were differentially expressed.
By harnessing the extensive knowledge of the midshipman vocal network, the VMN transcriptome elucidated a set of molecular pathways underlying neuronal excitability in a motor nucleus that instructs vocal patterning across multiple timescales, from milliseconds to hours and seasons. Our results directly inform future studies using molecular, anatomical, and neurophysiological methods to validate the function and cellular localization of candidate genes in the VMN. We propose that the candidate genes and molecular pathways identified here may belong to a shared genetic toolkit for vocal motoneurons in many species that face similar energetic and neurophysiological demands. Thus, our results will inform future comparative studies to achieve a broader understanding of the molecular machinery required for vocalization.
Results and discussion
In order to globally characterize molecular pathways governing vocal motor patterning, we compared the transcriptome of surgically isolated VMN to H tissue at three time points corresponding to high (reproductive summer night) and low (reproductive summer morning and non-reproductive winter night) vocal activity (Figure 1C). In order to minimize activity-induced gene-expression, we used males who had not been humming prior to sacrifice. See Methods, and Figures 1 and 2 for explanations of sample groups and analysis pipelines.
Transcriptome assembly, annotation, and qPCR validation
Sequencing using the Illumina HiSeq2000 system yielded approximately 200 million 100 bp reads across all brain groups used here and ear sample groups used for a companion study [49]. Over 90% of the raw reads survived quality filtering and trimming, resulting in 21.4 ± 2.8 million (mean ± S.D.) paired-end reads per brain sample. Our initial assembly produced 293,702 contigs, with a mean length of 1006.86 ± 1259.72 bp. After discarding contigs with open reading frames (ORFs) of less than 50 amino acids and lowly supported transcripts (isoforms) with less than 1% mapped reads for the gene (component), the final transcriptome contained 83,967 assembled transcripts, with a mean length of 1713.57 ± 1585.21 bp (range 201–18,637; N50 = 2647), representing 40,656 gene components across brain and ear sample groups. The transcript length distribution and number of transcripts per gene are shown in Figure 3A and B. The mean GC content for this filtered transcriptome was 48.22%. Of the 83,967 transcripts, 46,629 (55.5%) contained complete ORFs, comparable to a recent zebra finch transcriptome [50]. Additionally, CEGMA (Core Eukaryotic Genes Mapping Approach) analysis of conserved eukaryotic genes (CEGs), determined that our filtered transcriptome contained 453 full-length CEGs of the complete set of 458 CEGs (99%), and 238 of the more-conserved set of 248 CEGs (96%) [51,52]. Blast2GO annotation resulted in 74,000 (88.1%) of our assembled transcripts with significant annotation hits, comparable to or above reported rates (see [18,53,54]). Seventeen of the top 26 blast hit species were teleost fish, with the top nine all being teleosts (Figure 3C). There were 77,285 transcripts, representing 35,983 gene components, expressed in brain samples (>0 counts in at least one brain sample).
Figure 3.
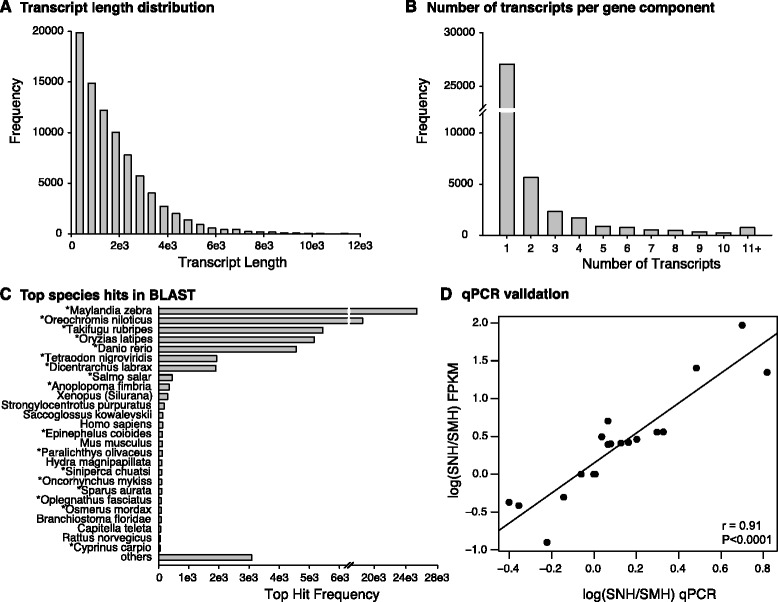
Transcriptome assembly quality assessment. A) Transcript length distribution. B) Number of transcripts per gene component as determined by Trinity. C) Top species hits from BLAST against NCBI’s nr database, first nine species are teleost fish sequences out of a total of 17 teleost species within the list, indicated by *. D) Quantitative PCR (qPCR) was used to verify transcriptome-derived FPKM values of 19 genes in SNH and SMH tissue groups. Pearson’s correlation analysis showed that qPCR and transcriptome measured ratios of SNH/SMH are significantly correlated with each other.
Using qPCR, we validated the relative abundances of 19 transcripts from two of the same, pooled H samples that were submitted for Illumina sequencing: reproductive summer morning (SMH) and reproductive summer night (SNH). Our results showed a tight and highly significant correlation between the log-normalized SNH/SMH ratios by qPCR and FPKM (fragments per kilobase of transcript per million reads mapped) (Pearson’s r = 0.91, P < 0.0001; Figure 3D). Additionally, expression levels of nine candidate genes also showed significant correlation between RNA-seq and qPCR values (see section below). Together, these results validated our transcriptome-predicted abundances and demonstrated the high quality of our de novo assembled transcriptome.
VMN versus H tissue comparisons
Overview
In order to identify molecular pathways and candidate genes important for VMN function, we first focused on between-tissue comparisons. The cyclic loess “fastlo” normalization method [55], as opposed to trimmed mean of M values (TMM) normalization [56], successfully removed the nonlinear tissue-dependent skew (Figure 4A-C), while preserving biologically relevant sample group similarities as seen in the clustering of samples by tissue and season in the multidimensional scaling plot (Figure 4D). By conservatively treating the three sample groups collected at different times as biological replicates from which to estimate transcript-wise dispersion values in edgeR’s differential expression analysis [57], we identified 1,717 transcripts significantly upregulated in H and 2,948 transcripts upregulated in VMN, which were the focus of downstream analyses.
Figure 4.
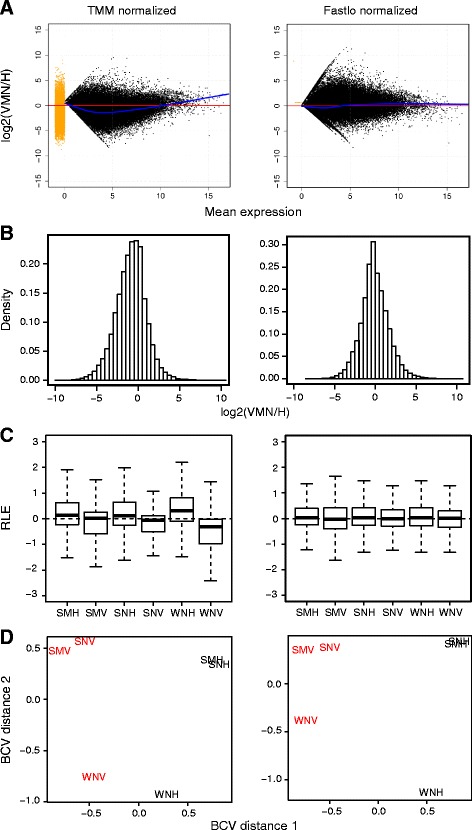
Evaluating TMM and fastlo normalization of tissue comparisons. A) Plots of log2(VMN/H) ratios (y axis) versus mean expression across all sample groups (x axis). N = 76,878 genes. Alignment along 0 horizontal is expected. Fitted loess lines are shown in blue. B) Histograms of VMN/H mean ratios. Even with peak at 0 indicates good normalization. C) Box plots of each sample group. Medians are expected to align along 0 with even distribution about the median across sample groups. RLE: Relative log ratio. D) Multidimensional scaling plots showing similarities between VMN (red) and H (black) sample groups. Samples show biologically relevant grouping by tissue and by season. BCV: biological coefficient of variation.
GO term enrichment
We performed Fisher’s exact test on GO terms to identify gene functions that are enriched in VMN or H. By comparing the subset of genes upregulated in VMN to the entire transcriptome, we found 279 over-represented GO terms (complete list in Additional file 1: Table S1). The enrichment results overwhelmingly highlighted the importance of ATP production in VMN, with 29% of the enriched GO terms relating to aerobic metabolism (Additional file 1: Table S1). This high metabolic activity in the VMN is exemplified by the 10 most significantly enriched GO terms (Table 1). Although few VMN enriched GO terms were related to neural transmission functions, the GO terms “neurotransmitter biosynthetic process” and “choline O-acetyltransferase activity” were enriched, consistent with VMN motoneurons being cholinergic [58] (Additional file 1: Table S1). Furthermore, we found several enriched GO terms related to steroid hormone signaling, including “gonadotropin secretion”, “endocrine hormone secretion”, and “sterol biosynthetic process”, indicating that the VMN is hormonally active (Additional file 1: Table S1). Finally, GO terms related to post-transcriptional processes were also enriched for VMN, including “translation” (Biological process, Table 1), “ribosome” (Cellular component, Table 1), “gene expression” (Additional file 1: Table S1), and “RNA processing” (Additional file 1: Table S1), indicating that VMN is translationally active.
Table 1.
Enriched GO terms of genes upregulated in VMN compared to H
| GO category | GO term | Name | FDR |
|---|---|---|---|
| Biological process | GO:0006412 | Translation | 2.90E-18 |
| GO:0006119 | Oxidative phosphorylation | 3.60E-18 | |
| GO:0022900 | Electron transport chain | 8.20E-18 | |
| GO:0042775 | Mitochondrial ATP synthesis coupled electron transport | 1.60E-17 | |
| GO:0042773 | ATP synthesis coupled electron transport | 1.60E-17 | |
| GO:0022904 | Respiratory electron transport chain | 2.30E-17 | |
| GO:0045333 | Cellular respiration | 2.30E-17 | |
| GO:0044283 | Small molecule biosynthetic process | 3.10E-15 | |
| GO:0055114 | Oxidation-reduction process | 5.10E-15 | |
| GO:0044711 | Single-organism biosynthetic process | 7.40E-15 | |
| Molecular function | GO:0003735 | Structural constituent of ribosome | 2.30E-17 |
| GO:0016491 | Oxidoreductase activity | 1.90E-15 | |
| GO:0003954 | NADH dehydrogenase activity | 6.20E-10 | |
| GO:0050136 | NADH dehydrogenase (quinone) activity | 6.20E-10 | |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | 6.20E-10 | |
| GO:0015078 | Hydrogen ion transmembrane transporter activity | 1.70E-07 | |
| GO:0016655 | Oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | 3.50E-07 | |
| GO:0015002 | Heme-copper terminal oxidase activity | 3.80E-07 | |
| GO:0016676 | Oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor | 3.80E-07 | |
| GO:0016675 | Oxidoreductase activity, acting on a heme group of donors | 3.80E-07 | |
| Cellular component | GO:0005739 | Mitochondrion | 8.80E-53 |
| GO:0044429 | Mitochondrial part | 1.20E-22 | |
| GO:0044444 | Cytoplasmic part | 1.70E-21 | |
| GO:0005840 | Ribosome | 2.10E-21 | |
| GO:0005743 | Mitochondrial inner membrane | 4.40E-20 | |
| GO:0019866 | Organelle inner membrane | 6.10E-19 | |
| GO:0070469 | Respiratory chain | 1.50E-16 | |
| GO:0005740 | Mitochondrial envelope | 1.50E-16 | |
| GO:0030529 | Ribonucleoprotein complex | 2.50E-16 | |
| GO:0005737 | Cytoplasm | 3.00E-16 |
Significantly over represented gene ontology (GO) terms found in transcripts upregulated in VMN compared to the surrounding hindbrain (H). Top 10 GO terms from each GO category (biological process, molecular function, and cellular component) are listed in order of decreasing significance by false discovery rate (FDR).
In contrast, 75% of the 130 GO terms enriched in transcripts upregulated in H were related to synaptic function and neurotransmission (see Additional file 1: Table S2 for complete list). The top 10 Biological Process GO terms were all related to cell signaling and synaptic transmission, the top 10 Molecular Function terms were related to membrane transporter activity, and the top 10 Cellular component GO terms were related to the synapse or membrane (Table 2). The observation that H contains a higher diversity of neurotransmission genes is consistent with H containing diverse motor and non-motor nuclei as well as abundant white matter (see [27]) compared to the high homogeneity of motoneurons in VMN. In contrast to VMN, we found no enriched terms related to steroid hormones in H, although “neuropeptide signaling pathway” was enriched (Additional file 1: Table S2).
Table 2.
Enriched GO terms of genes upregulated in H compared to VMN
| GO category | GO term | Name | FDR |
|---|---|---|---|
| Biological process | GO:0007154 | Cell communication | 7.30e-15 |
| GO:0044700 | Single organism signaling | 1.60e-14 | |
| GO:0023052 | Signaling | 1.60e-14 | |
| GO:0019226 | Transmission of nerve impulse | 6.50e-14 | |
| GO:0007267 | Cell-cell signaling | 3.50e-13 | |
| GO:0035637 | Multicellular organismal signaling | 6.90e-13 | |
| GO:0007268 | Synaptic transmission | 1.20e-12 | |
| GO:0044699 | Single-organism process | 1.20e-11 | |
| GO:0044763 | Single-organism cellular process | 7.10e-11 | |
| GO:0050877 | Neurological system process | 3.60e-10 | |
| Molecular function | GO:0005515 | Protein binding | 3.40e-17 |
| GO:0015291 | Secondary active transmembrane transporter activity | 1.80e-09 | |
| GO:0015293 | Symporter activity | 4.00e-08 | |
| GO:0022804 | Active transmembrane transporter activity | 6.90e-08 | |
| GO:0015075 | Ion transmembrane transporter activity | 2.70e-07 | |
| GO:0015294 | Solute:cation symporter activity | 4.00e-07 | |
| GO:0015081 | Sodium ion transmembrane transporter activity | 1.20e-06 | |
| GO:0022857 | Transmembrane transporter activity | 1.40e-06 | |
| GO:0022891 | Substrate-specific transmembrane transporter activity | 1.70e-06 | |
| GO:0008324 | Cation transmembrane transporter activity | 4.40e-06 | |
| Cellular component | GO:0031224 | Intrinsic to membrane | 8.00e-10 |
| GO:0016021 | Integral to membrane | 5.20e-09 | |
| GO:0044425 | Membrane part | 6.70e-09 | |
| GO:0016020 | Membrane | 2.40e-07 | |
| GO:0005886 | Plasma membrane | 1.70e-06 | |
| GO:0030054 | Cell junction | 8.30e-06 | |
| GO:0071944 | Cell periphery | 1.00e-05 | |
| GO:0045202 | Synapse | 1.60e-04 | |
| GO:0097060 | Synaptic membrane | 3.10e-04 | |
| GO:0044456 | Synapse part | 3.30e-04 |
Significantly over represented gene ontology (GO) terms found in sequences upregulated in the surrounding hindbrain (H) compared to the VMN. Top 10 GO terms from each GO category (biological process, molecular function, and cellular component) are listed in order of decreasing significance by false discovery rate (FDR).
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways
In order to compare the relative representation of biochemical signaling pathways in VMN vs. H, we mapped differentially expressed transcripts to KEGG pathways [59] (complete list in Additional file 1: Table S3). Transcripts upregulated in VMN mapped predominantly to pathways directly related to ATP production, including “oxidative phosphorylation”, “pyruvate metabolism”, “citrate cycle (TCA cycle)”, and “glycolysis/gluconeogenesis”, which is consistent with GO term enrichment results that also indicated high levels of metabolic activity in VMN. Pathways unique to transcripts upregulated in VMN also included those involved in antioxidant defense against oxidative stress, such as “glutathione metabolism” [60], indicating that VMN is capable of combating reactive oxygen species generated by aerobic metabolism. Additionally, we found steroid hormone related pathways (“steroid hormone biosynthesis”, “steroid biosynthesis”, “steroid degradation”; Table 3, Additional file 1: Table S3) that support GO term enrichment results highlighting VMN as a hormonally active nucleus. Transcripts in the “carbon fixation pathways in prokaryotes” found in VMN were confirmed by BLAST to have high identity to transcripts of other fish and likely represent metabolic genes conserved from bacteria through vertebrates. In contrast, transcripts upregulated in H mapped predominantly to phospholipid metabolism and signaling pathways (Table 3). Phospholipids are important components of cell membranes and precursors to important secondary messengers such as diacyl glycerol and inositol 1,4,5-triphosphate [61].
Table 3.
KEGG pathways mapped to transcripts differentially upregulated in VMN vs. H
| VMN | H | ||
|---|---|---|---|
| Pathway | # seqs | Pathway | # seqs |
| Purine metabolism | 60 | Phosphatidylinositol signaling system | 19 |
| Pyrimidine metabolism | 29 | Glycerophospholipid metabolism | 15 |
| Oxidative phosphorylation | 28 | Purine metabolism | 12 |
| Aminoacyl-tRNA biosynthesis | 25 | Lysine degradation | 12 |
| Carbon fixation pathways in prokaryotes | 16 | Glycerolipid metabolism | 11 |
| Pyruvate metabolism | 16 | T cell receptor signaling pathway | 9 |
| Valine, leucine and isoleucine degradation | 14 | Inositol phosphate metabolism | 7 |
| Citrate cycle (TCA cycle) | 13 | Alanine, aspartate and glutamate metabolism | 4 |
| Porphyrin and chlorophyll metabolism | 13 | Arginine and proline metabolism | 3 |
| Glycolysis/Gluconeogenesis | 13 | Fatty acid biosynthesis | 3 |
| Glutathione metabolism | 11 | Glutathione metabolism | 0 |
| Steroid hormone biosynthesis | 7 | Steroid hormone biosynthesis | 0 |
| Steroid biosynthesis | 5 | Steroid biosynthesis | 0 |
| Steroid degradation | 3 | Steroid degradation | 0 |
Top 10 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways with the highest number of mapped transcripts that were upregulated in VMN or H. Additionally, we show that more transcripts upregulated in VMN are mapped to glutathione methabolism, steroid and steroid hormone biosynthesis pathways.
Together, KEGG pathway and GO term enrichment results showed that VMN is metabolically, hormonally, and post-transcriptionally active. In contrast, transcription in the surrounding hindbrain is devoted to synaptic transmission and membrane processes. It has been estimated that acoustic courtship in ectotherms requires an eight-fold increase from resting metabolic rate [62,63]. Results highlighting the importance of cellular respiration in VMN are consistent with its ability to sustain high frequency firing that drives advertisement hums up to hours (Figure 1A) repetitively throughout a single night of courtship-related activity [22,24,26]. While studies have extensively characterized the high aerobic demands of “superfast” muscles in midshipman and closely related toadfish, among the fastest contracting skeletal muscles in vertebrates [64,65], our results indicate a concomitantly high metabolic demand in the motoneurons that drive these muscles.
Gene category analyses
Guided by the above results and previous studies of VMN characteristics, we wanted to compare expression levels of transcripts belonging to different functional categories with significant implications for cellular and network level excitability. Thus, regardless of differential expression, we selectively analyzed transcripts belonging to the following functional categories: neurotransmission (ion channels and neurotransmitter receptors), neuroendocrine (genes related to steroid and thyroid hormone function), neuropeptides, neuromodulators, cellular respiration, and antioxidants (Figures 5 and 6). Transcripts for each category are listed in Additional file 1: Table S4.
Figure 5.
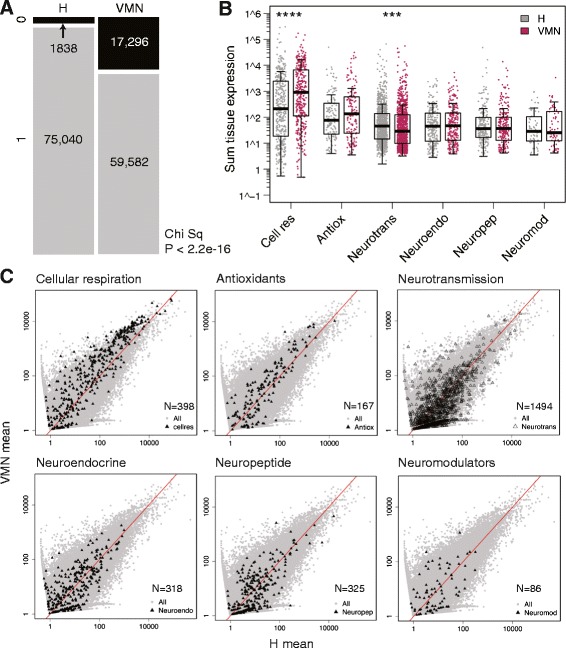
Tissue comparison of functional category expression. A) Raw counts from the brain transcriptome were dichotomized into expressed (1, gray) or not expressed (0, black) based on the summed tissue values. Compared to VMN, H has significantly more expressed genes. Chi sq: Chi squared test. B) Normalized abundances for each unique transcript expressed were summed from VMN or H sample groups, box-cox transformed, and subjected to one-way ANOVA for tissue comparison, with the Bonferroni corrected alpha level set at 0.008. Cellular respiration transcripts had significantly higher expression in the VMN while neurotransmission transcripts were higher in H. ****P < 0.0001, ***P = 0.0005. C) Mean VMN vs. H expression levels (fastlo-normalized) of transcripts within each of the six gene categories (black triangles) are plotted on top of all transcripts (gray dots). N: number of transcripts in each category. Cell res: cellular respiration; Antiox: antioxidants; Neurotrans: neurotransmission; Neuroendo: neuroendocrine; Neuropep: neuropeptide; Neuromod: neuromodulators.
Figure 6.
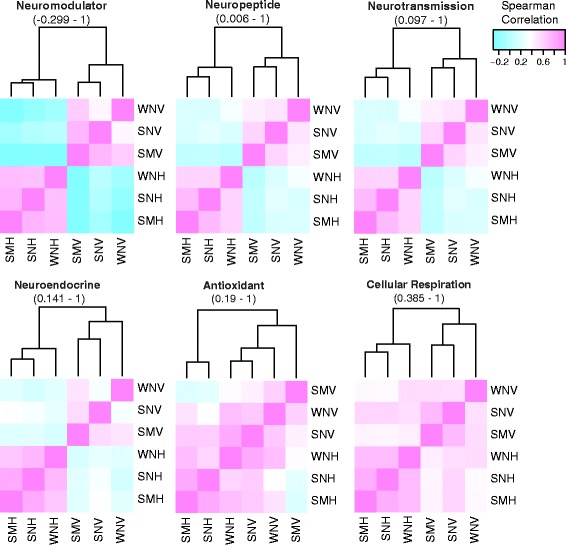
Spearman correlation heatmaps of functional gene categories. Heatmaps are generated from Spearman correlation coefficients (rho) for each pair of samples based on TMM-normalized FPKM values. Values in parenthesis are the ranges of rho.
While the overall pool of expressed transcripts was smaller in VMN than in H (Chi squared test, P < 0.0001) (Figure 5A), transcript abundances were comparable between tissues in most gene categories (Figure 5B). Two exceptions were cellular respiration, which had significantly higher expression in VMN (ANOVA, F = 30.84, P < 0.0001), and neurotransmission, which had significantly higher expression in H (ANOVA, F = 12.07, P = 0.0005) (Figure 5B,C), substantiating our interpretations of GO term enrichment results as discussed above (Tables 1–2).
Pairwise Spearman correlation analyses revealed that overall, cellular respiration had the highest correlation between sample groups (Figure 6). Because the Spearman coefficient is derived from calculating the Pearson correlation of transcript abundance ranks between two samples, these results demonstrate that although cellular respiration expression levels were higher in VMN (Figure 5), the relative abundances of transcripts to one another is conserved across sample groups (Figure 6). In contrast, the other functional categories, especially neuromodulators, showed lower correlation between VMN and H (Figure 6).
Candidate genes
Supporting the hypothesis that VMN expresses a unique molecular toolkit dedicated to vocal motor coding, the majority of functionally important transcripts upregulated in VMN (Table 4) were unique from those upregulated in H (Additional file 1: Table S5). Among the transcripts significantly upregulated in VMN relative to H (Figure 7A), we identified those with neurotransmission functions, including ligand- and voltage-gated ion channels as well as metabotropic neurotransmitter receptors (Table 4). We also identified transcripts with neuroendocrine functions, including steroid signaling and biosynthesis, neuropeptides and peptide hormones, and other neuromodulators (Table 4). Neurotransmission and neuroendocrine functions were the focus because of relevance to neural excitability and previous midshipman studies focusing on these mechanisms.
Table 4.
Functionally important candidate genes upregulated in VMN
| Category | Blast2GO description |
|---|---|
| Neurotransmission | Anoctamin-5-like isoform x4 |
| ATP-sensitive inward rectifier potassium channel 8-like | |
| Calcium-activated potassium channel subunit alpha-1-like (kcnma1) isoform 3 | |
| Gamma-aminobutyric acid receptor subunit alpha-3-like (gabra3) | |
| Gamma-aminobutyric acid receptor subunit alpha-5-like (gabra5) | |
| Gamma-aminobutyric acid receptor subunit pi-like (gabrp) | |
| Gamma-aminobutyric acid type b receptor subunit 1-like | |
| Gap junction beta-6 (cx30) | |
| Glutamate ionotropic kainate 3-like | |
| Glutamate ionotropic kainate 5-like | |
| Glutamate receptor delta-1 subunit-like | |
| Glycine receptor subunit beta | |
| Glycine receptor subunit beta-like | |
| Kv channel-interacting protein 4-like | |
| Muscarinic acetylcholine receptor m2 (chrm2) | |
| Neuronal acetylcholine receptor subunit alpha-2-like (chrna2) | |
| Neuronal acetylcholine receptor subunit alpha-3-like | |
| Neuronal acetylcholine receptor subunit alpha-9-ii-like | |
| Neuronal acetylcholine receptor subunit beta-2-like | |
| Neuronal acetylcholine receptor subunit non-alpha-3-like (chrnb3) | |
| Potassium sodium hyperpolarization-activated cyclic nucleotide-gated channel 2-like | |
| Potassium voltage-gated channel subfamily b member 2 (kcnb2/Kv2.2) | |
| Potassium voltage-gated channel subfamily c member 3-like (kcnc3/Kv3.3) isoform x1 | |
| Potassium voltage-gated channel subfamily e member 1-like | |
| Potassium voltage-gated channel subfamily h member 5 | |
| Potassium voltage-gated channel subfamily kqt member 2-like (kcnq2/Kv7.2) | |
| Potassium voltage-gated channel subfamily s member 1-like (kcns1/Kv9.1) | |
| Potassium voltage-gated channel subfamily s member 3-like | |
| Sodium channel protein type 5 subunit alpha | |
| Sodium channel subunit beta-4-like | |
| Transient receptor potential cation channel subfamily m member 2-like | |
| Two pore calcium channel protein 1-like | |
| Voltage-dependent l-type calcium channel subunit alpha-1f-like | |
| Voltage-dependent r-type calcium channel subunit alpha-1e | |
| Voltage-gated potassium channel subunit beta-1 isoform 1 (kcnab1) | |
| Voltage-gated potassium channel subunit beta-2-like isoform 1 | |
| Steroid pathway | 3-keto-steroid reductase-like (hsd17b7) |
| Androgen receptor alpha (ar-a) | |
| Androgen-induced gene 1 protein | |
| Dihydroxyvitamin d 24- mitochondrial-like isoform 1 | |
| Estrogen receptor beta 2 (esr2) | |
| Estrogen-related receptor alpha | |
| Hydroxysteroid 11-beta-dehydrogenase 1-like (hsd11b1l) | |
| Lanosterol 14-alpha demethylase-like | |
| Ovarian aromatase (cyp19a1a) | |
| Sterol regulatory element-binding protein 1 | |
| Sterol regulatory element-binding protein 2 | |
| Neuropeptides/peptide hormones | Atrial natriuretic peptide-converting enzyme-like |
| Calcitonin gene-related peptide precursor | |
| Cholecystokinin type a receptor | |
| Growth hormone receptor | |
| Inhibin beta b chain-like | |
| Insulin gene enhancer protein isl-1 | |
| Insulin-like growth factor binding protein 1 | |
| Insulin-like growth factor i | |
| Neuropeptide b precursor | |
| Peptide yy-like | |
| Pituitary adenylate cyclase-activating polypeptide type i receptor-like | |
| Vasoactive intestinal polypeptide receptor 1-like | |
| Thyroid hormone | Thyroid hormone receptor alpha |
| Thyroid hormone receptor-associated protein 3-like | |
| Thyroid receptor-interacting protein 6-like | |
| Other neuromodulators | Adenosine receptor a1-like |
| D-like dopamine receptor-like (drd2) | |
| Melatonin receptor type 1a-like | |
| Prostaglandin e synthase 2-like | |
| Prostaglandin f2-alpha receptor-like | |
| Antioxidants | Cu/Zn superoxide dismutase (sod1) |
| Glutaredoxin 3 | |
| Glutaredoxin-related protein 5 | |
| Glutathione peroxidase 4b (gpx4b) | |
| Glutathione s-transferase (gst) | |
| Peroxiredoxin 6 | |
| Thioredoxin domain-containing protein 17 | |
| Thioredoxin-dependent peroxide mitochondrial precursor | |
| Thioredoxin-like protein 1 | |
| Thioredoxin-like protein 4a |
Gene descriptions with neurotransmission, neuroendocrine, and antioxidant functions from transcripts upregulated in VMN compared to surrounding hindbrain. Genes that are mentioned in the text or shown in figures have gene symbols in parentheses.
Figure 7.
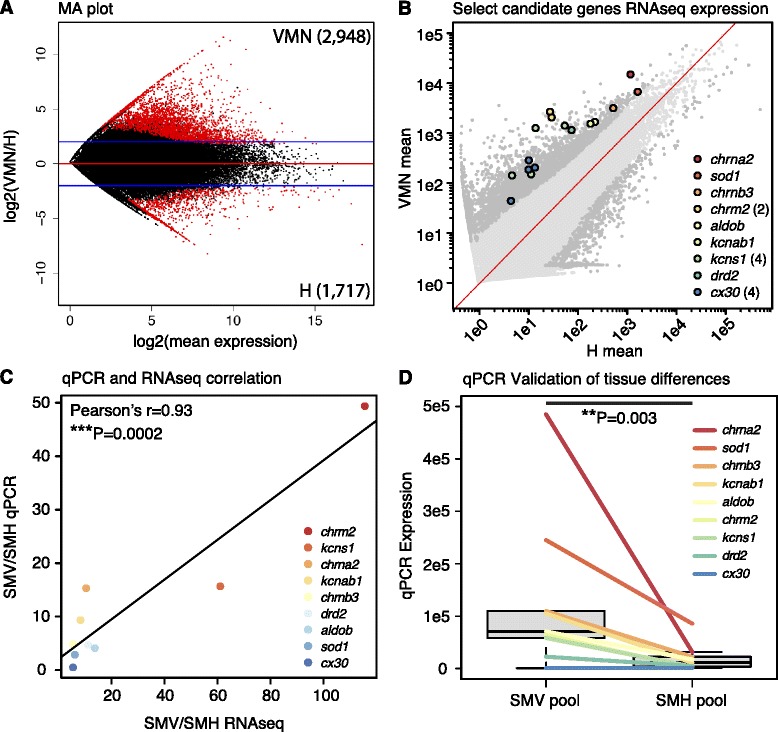
Candidate functional genes from tissue comparisons validated by qPCR. A) MA plot showing log2 VMN/H mean ratios by average expression level for each transcript. Mean values were calculated from the three sample groups for each tissue. Transcripts with positive log ratios are higher in VMN, while transcripts with negative log ratios are higher in H. The numbers of significantly upregulated transcripts for each tissue are shown in parentheses. Red dots indicate significantly differentially expressed transcripts in VMN or H. Blue lines indicate log2 values of +/− 2, representing fold-change of 4. B) Nine candidate genes that were chosen for qPCR verification are plotted on top of a scatter plot of VMN mean vs. H mean values for all transcripts, calculated from the three sample groups for each tissue. Number of differentially expressed isoforms per gene is indicated in parentheses. Dark gray dots are significantly differentially expressed transcripts in VMN or H. The line of unity is in red. C) Correlation of SMV/SMH ratios derived from qPCR data and fastlo-normalized RNAseq data. For RNAseq data, ratios were calculated from the average expression of all isoforms within a gene component. Linear regression line is in black. D) qPCR validation of SMV and SMH expression for the candidate genes showing significant upregulation of candidate genes in SMV.
Candidate neurotransmission genes
VMN’s synchronous and temporally precise activity is dependent on both synaptic interactions of the network as well as intrinsic properties of individual motoneurons [8]. There were 57 transcripts with 34 unique neurotransmission gene annotations upregulated in the VMN (Table 4). These candidate genes included those supporting two of VMN’s major network components: electrical synapses formed by gap junctions and GABAergic inhibition [8], both of which are also prominent mechanisms promoting network-level synchrony in other systems [66,67]. The presence of elevated gap junction subunit expression in VMN (Table 4) affirmed previous evidence of electrotonic coupling in VMN, demonstrated by electron microscopy [68], transneuronal dye diffusion [8,27], and intracellular neurophysiology [8]. The multiple GABA receptor subunits upregulated in VMN (Table 4) likely provide the prominent network-dependent afterhyperpolarization observed in VMN motoneurons, supported by the pharmacological blocking of GABAA receptors that severely reduced the duration and amplitude of VMN output [8].
VMN’s intrinsic neuronal properties are likely mediated by several voltage-gated potassium channel (KCN/Kv) subunits that have known functions in controlling neuronal excitability (Table 4; we report potassium channel gene names by both the KCN and Kv nomenclature systems [69,70]). First, VMN showed increased expression of potassium voltage-gated channel subfamily c member 3-like (kcnc3/Kv3.3) subunits (Table 4), which contributes significantly to action potential repolarization and recovery of sodium channels from inactivation (e.g. [71]), permitting high-frequency repetitive firing that is a key feature of VMN motoneurons [8]. Second, it has been shown that while nonfunctional alone, KCNS1/Kv9.1 subunits decrease KCNB1/Kv2.1 currents when coassembled in heteromeric channels, which was modeled to increase the firing fidelity of a simulated neuron to a 100 Hz stimulus [72]. The potential interaction of our candidate potassium voltage-gated channel subfamily s member 1-like (kcns1/Kv9.1) and potassium voltage-gated channel subfamily b member 2 (kcnb2/Kv2.2) subunits (Table 4) may therefore regulate VMN firing frequency that directly determines the pulse repetition rate and fundamental frequency of natural vocalizations (see [32]). KCNS1/Kv9.1 subunits also decrease the conductance of KCNC/Kv3 subunits, mentioned above, when they form heteromeric channels [73]. Third, channels composed of potassium voltage-gated channel subfamily kqt member 2-like (kcnq2/Kv7.2) subunits, also elevated in VMN (Table 4), dampen neural excitability by being partially activated at resting membrane potentials [74]. We propose kcnq2/Kv7.2 underlies the low baseline excitability of VMN neurons, which contributes to their ability to fire with high fidelity to presynaptic input [8] from the vocal pacemaker nucleus that sets call frequency [30]. In the fully aquatic frog Xenopus laevis, male laryngeal motoneurons that drive vocalizations also have a strong low-threshold potassium current [10], leading to the hypothesis that low intrinsic excitability conferred by high kcnq2/Kv7.2 expression may be a shared feature of vertebrate vocal motoneurons.
Although we identified transcripts of candidate neurotransmission genes predicted a priori to be important for VMN function, we also found candidates previously not well studied in this system. One example is evidence of cholinergic input to VMN, supported by choline acetyltransferase staining of somata in the vocal premotor pacemaker region of midshipman ([58], A. Bass, unpub observ), that has potentially significant contributions to VMN function. The alpha and beta subunits of neuronal nicotinic receptors found in our candidate list (Table 4) could mediate fast, postsynaptic action of acetylcholine [75], or modulation of presynaptic neurotransmitter release, including GABA [76], shown to play a prominent role in regulating VMN output (see above). Furthermore, evidence from other motor systems indicates cholinergic control of motoneuron excitability and firing frequency [77-79], including actions mediated via postsynaptic muscarinic receptors and KCNB1/Kv2.1 potassium channels [77,78,80,81], transcripts of which were elevated in VMN: muscarinic acetylcholine receptor m2 (chrm2) and kcnb2/Kv2.2 (Table 4). Another target of muscarinic modulation is the KCNQ/Kv7 family of voltage gated potassium channels that decrease baseline neural excitability, of which kcnq2/Kv7.2 is found among our candidates (discussed above; Table 4) [74].
Importantly, many of the candidate ion channels we note above are ideal targets for pharmacological validation using selective blockers (e.g. see [82] for kcnq2/Kv7.2 blockers, [71] for kcnc/Kv3 blockers, [69] for other potassium channel blockers; [83] for muscarinic and [84,85] for neuronal nicotinic acetylcholine receptor blockers; [86] for gabra5 blocker). It is also worth highlighting that both kcns1/Kv9.1 and kcnq2/Kv7.2 mRNA levels were elevated in four major telencephalic song control nuclei in the zebra finch [12], with kcns1/Kv9.1 showing additional label in the tracheosyringeal division of the hypoglossal motor nucleus (nXIIts) (see the ZEBrA database, Oregon Health & Science University, Portland, OR 97239; http://www.zebrafinchatlas.org), the brainstem region representing VMN’s analogue, if not homologue (see [3]).
Candidate neuroendocrine and neuromodulator genes
Both behavioral and neurophysiological studies have shown that steroids exert rapid and robust effects on vocal behavior and the output of the vocal CPG via classical steroid receptors in midshipman and a closely related toadfish species (e.g. [34,87]). Consistent with these studies, steroid receptors androgen receptor alpha (ar-a) and estrogen receptor beta 2 (esr2) were both upregulated in the VMN (Table 4). The estradiol synthetic enzyme ovarian aromatase (cyp19a1a) was upregulated in the VMN (Table 4), consistent with previous studies showing dense aromatase protein and mRNA expression in a dorsal layer of glial cells that surrounds and projects throughout the VMN [88]. The steroidogenic enzyme 3-keto-steroid reductase-like (hsd17b7), responsible for the interconversion between estrone and estradiol [89], was also elevated in the VMN (Table 4). Additionally, we found higher VMN expression of hydroxysteroid 11-beta-dehydrogenase 1-like (hsd11b1l), a crucial enzyme that converts 11-beta-hydroxytestosterone to 11-ketotestosterone, and cortisol to the inactive metabolite cortisone [90] (Table 4). 11-ketotestosterone and cortisol are the major circulating androgen and glucocorticoid, respectively, in type I male midshipman that are the source of tissues used here (see [91]), and cause a rapid increase in vocal CPG output duration within five minutes of systemic injection [33,34]. Thus, many of the candidate neuroendocrine transcripts upregulated in VMN have direct relevance to VMN function as corroborated by prior studies.
Additionally, we found previously under-studied neuroendocrine or neuromodulatory signaling pathways in VMN, including growth hormone, thyroid hormone, prostaglandin and dopamine, the latter of which is consistent with dense catecholaminergic input to VMN [92] (Table 4). One intriguing example is Prostaglandin f2-alpha receptor-like, because Prostaglandin f2-alpha is a potent sex pheromone that induces spawning behavior in fish [93], though little is known about its effects on vocal behavior.
Together, the steroid signaling candidate genes indicate that VMN exhibits high androgen and estrogen sensitivity by expressing steroid receptors and steroidogenic enzymes, consistent with prior studies showing their anatomical localization to VMN and influence on vocal CPG output (e.g. [46,94]). VMN function is also likely modulated by candidate gene products belonging to novel neuroendocrine and neuromodulatory pathways identified here.
Candidate antioxidant genes
Based on the enrichment of cellular respiration genes being upregulated in VMN, expected to generate harmful reactive oxygen species [95], we hypothesized that the VMN combats oxidative stress by expressing high levels of antioxidant enzymes and proteins. Supporting this hypothesis, we found the increased expression of several antioxidant genes in the VMN compared to H (Table 4). Notably, these included the well-studied Cu/Zn superoxide dismutase (sod1) enzyme (Figure 7B, C; Table 4), which catalyzes the conversion of two superoxide radicals into hydrogen peroxide [95]. The importance of sod1 in motoneuronal function is demonstrated by human sod1 mutations that lead to the motoneuron degenerative disease amyotrophic lateral sclerosis (ALS) [96,97]. The suite of antioxidants upregulated in VMN also included enzymes that produce or utilize glutathione for reducing reactive oxygen species, such as glutathione s-transferase (gst), which synthesizes glutathione [98], and glutathione peroxidase 4b (gpx4b), which detoxifies hydrogen peroxide [95] (Table 4). These results support our hypothesis that the VMN must combat oxidative stress incurred by high rates of cellular respiration resulting from prolonged activity [22,24,26]. Because the fish used in this study were not actively engaged in vocal activity at the time of sacrifice (see Methods), these results highlight a constitutive feature of the VMN. Based on the implication of antioxidant enzymes and proteins in motoneuron and neural degenerative diseases [96], we believe the VMN is a motor nucleus capable of withstanding extreme oxidative stress and therefore a useful model for studying the relationship between antioxidants and neural function.
Candidate gene qPCR validation
We chose nine candidate genes upregulated in VMN to validate expression levels with qPCR (Figure 7C). We used the same, pooled SMV and SMH RNA for generating our RNA-seq libraries to validate our findings of upregulated transcripts in the VMN. There was a high correlation of expression levels of all nine candidate genes measured by the two methods (SMV: Pearson’s r = 0.97, P < 0.0001; SMH: Pearson’s r = 0.89, P = 0.0002) (Additional file 2). Similarly, SMV/SMH fold changes calculated from qPCR correlated significantly with RNA-Seq derived fold changes (Pearson’s r = 0.93, P = 0.0002) (Figure 7C). Finally, in concordance with RNA-seq results, qPCR expression levels of candidate genes were significantly higher in SMV when compared to SMH (t = 4.22, P = 0.003) (Figure 7D). The only gene not showing the expected pattern, cx30, had 20 assembled isoforms and had the lowest expression among the tested candidates (Figure 7D). Altogether, qPCR results largely supported RNA-seq results.
Seasonal and daily variation in the VMN
Overview
We aimed to identify functionally important candidate genes whose expression levels change on a daily or seasonal basis. Hierarchical clustering of differentially expressed transcripts revealed that sample groups were most divergent across tissues (H vs. VMN samples) and reproductive state (reproductive summer vs. non-reproductive winter) (Additional file 3), consistent with the above results (Figure 4D). Furthermore, the Spearman correlation matrix showed higher within-tissue differences in the VMN than in the H (Additional file 3). Significantly differentially expressed transcripts were separated into 109 K-means clusters, from which we identified clusters showing seasonal and daily expression patterns (Figure 8A) with upregulated expression in: all summer reproductive samples (“All-Summer”: SMV, SNV, SMH and SNH), summer VMN samples (“VMN-Summer”: SMV and SNV), or SNV (“Peak-SNV”).
Figure 8.

K-means clusters showing seasonal and daily patterns of gene expression. A) Representative K-means clusters showing upregulation in all summer sample groups, VMN summer groups, and peak expression in summer night VMN (SNV; see Figure 1C for other abbreviations). Total transcript numbers for each K-means cluster expression pattern are shown next to the cluster type names used in the text. B) FPKM expression levels are plotted for our candidate ion channels exhibiting peak expression in SNV. chrnb2: neuronal acetylcholine receptor subunit beta-2; gabrp: gamma-aminobutyric acid receptor subunit pi; cx30: connexin 30/gap junction beta-6; kcnma1 (or Kca1.1): calcium-activated potassium channel subunit alpha-1.
Candidate genes
We found evidence for daily and seasonal regulation of candidate genes for specific ion channel subtypes as well as steroidogenic enzymes and receptors belonging to major hormone signaling pathways previously implicated in regulating seasonal and daily cycles of vocal motor excitability and behavior in midshipman.
Seasonal comparisons
We first focused on finding candidate genes in All-Summer and VMN-Summer clusters (Figure 8A) that would contribute to known VMN neurophysiological properties (Figure 9). Neurotransmission related transcripts in All-Summer clusters included the gap junction alpha 1 (cx43) that was unique to All-Summer when compared to All-Winter clusters (Table 5). Among the ion channel subunit genes upregulated in VMN-Summer clusters were cx30, excitatory ionotropic glutamate receptors, inhibitory glycine receptors, voltage-gated potassium channel subunits, and a transient receptor potential cation channel subfamily M member 7 (trpm7) (Table 5). Interestingly, trpm7, which encodes a protein with both channel and kinase functions and is involved in regulating intracellular Ca2+ and Mg2+ levels, has been implicated in Guamanian ALS and Parkinson’s dementia [99].
Figure 9.

Summary of proposed candidate genes as molecular basis of known VMN properties. A) Known VMN neurophysiological properties. Extreme temporal precision of motoneuron firing is shown by corresponding traces from an intracellular recording (bottom, red) and VN recording (top, purple) (adapted from [8]). Numbers 1–5 correspond to intrinsic neuronal and network VMN properties listed in C. B) Schematic of known VMN neuroanatomical properties. Most of the motoneuron somata appear gray with subsets of red and black somata to highlight properties that are representative of the entire VMN. Glial expression of aromatase [88] and androgen receptor beta [120] is depicted in blue and orange. One of the black somata in the left VMN also depicts each motoneuron’s dendritic arbor that branches throughout each of the midline pair of motor nuclei and a single unbranched axon that exits via the ipsilateral vocal nerve (VN) (see [8]). The subsection of dense GABAergic innervation by cells lying outside of the VMN is also representative of the entire VMN. Abbreviations: AR: androgen receptor; ER: estrogen receptor; GABA: gamma-aminobutyric acid. C) For the suite of VMN properties, we identified corresponding transcripts that were significantly upregulated in the VMN compared to the surrounding hindbrain, and provide substantiating evidence from previous midshipman studies (see reference list for numbered citations). Abbreviations: ar-a: androgen receptor alpha; esr: estrogen receptor; cyp19a1a: aromatase; nrc3c1: glucocorticoid receptor; cx: connexin (gap junction); AP: action potential; kcn: voltage-gated potassium channels; AHP; afterhyperpolarization; gabra/GABAaR: GABAA receptor; ICC: immunocytochemistry; chrm2: muscarinic acetylcholine receptor m2. *indicates kcn subunits known to regulate the function of subunits listed in the row above.
Table 5.
Functionally important candidate genes showing daily and seasonal regulation
| Category | Cluster pattern | Blast2GO description |
|---|---|---|
| Neurotransmission | Peak-SNV | Anoctamin-10-like isoform x1 |
| Peak-SNV | Anoctamin-10-like isoform x4 | |
| Peak-SNV | ATP-sensitive inward rectifier potassium channel 8-like | |
| Peak-SNV | Calcium-activated potassium channel subunit alpha-1-like (kcnma1/Kca1.1) isoform x1 | |
| Peak-SNV | Calcium-activated potassium channel subunit alpha-1-like (kcnma1/Kca1.1) isoform x4 | |
| Peak-SNV | GABA receptor subunit pi-like (gabrp) | |
| All-Summer | Gap junction alpha 1 (cx43) | |
| VMN-Summer, Peak-SNV | Gap junction beta 6 (cx30) | |
| VMN-Summer | Glutamate ionotropic kainate 4 | |
| All-Summer | Glycine receptor subunit beta | |
| VMN-Summer | Glycine receptor subunit beta-like | |
| VMN-Summer, Peak-SNV | Neuronal acetylcholine receptor subunit alpha-3-like | |
| Peak-SNV | Neuronal acetylcholine receptor subunit beta-2-like (chrnb2) | |
| All-Summer | Potassium voltage-gated channel subfamily c member 4-like (kcnc4/Kv3.4) | |
| VMN-Summer | Potassium voltage-gated channel subfamily h member 5-like | |
| VMN-Summer | Potassium voltage-gated channel subfamily kqt member 2-like (kcnq2/Kv7.2) | |
| All-Summer | Sodium channel protein type 8 subunit alpha-like | |
| VMN-Summer | Sodium channel subunit beta-4-like | |
| VMN-Summer | Transient receptor potential cation channel subfamily m member 7-like (trpm7) | |
| VMN-Summer | Two pore calcium channel protein 1-like | |
| Steroid receptors and metabolic enzymes | VMN-Summer | 3-keto-steroid reductase-like |
| All-Summer | Androgen receptor alpha (ar-a) | |
| VMN-Summer | Androgen-induced gene 1 | |
| All-Summer | Cholesterol 25-hydroxylase-like protein member 1-like | |
| Peak-SNV | Estrogen receptor alpha (esr1) | |
| VMN-Summer | Glucocorticoid receptor (nr3c1) | |
| VMN-Summer | Hydroxysteroid dehydrogenase-like protein 2 | |
| VMN-Summer | Neutral cholesterol ester hydrolase 1-like | |
| VMN-Summer | Oxysterol-binding protein 10 isoform 1 | |
| VMN-Summer | Oxysterol-binding protein 8-like | |
| Peak-SNV | Oxysterol-binding protein 9-like isoform 3 | |
| Peak-SNV | Sterol 26- mitochondrial-like | |
| Peptide hormones and receptors | VMN-Summer, All-VMN | Calcitonin gene-related peptide precursor |
| Peak-SNV | Growth hormone receptor | |
| All-Summer | Growth hormone-regulated tbc protein 1-a-like | |
| VMN-Summer | Insulin gene enhancer protein isl-2a-like | |
| All-Summer | Insulin receptor-like | |
| VMN-Summer | Insulin-like growth factor-binding protein 1-like | |
| VMN-Summer | Opioid growth factor receptor-like | |
| VMN-Summer | Parathyroid hormone/parathyroid hormone-related peptide receptor-like | |
| Melatonin | All-Summer | Acetylserotonin o-methyltransferase-like |
Functionally important candidate genes found in All-Summer, VMN-summer, and Peak-SNV clusters. Different transcripts with the same sequence description found in both VMN and H clusters are not listed. Additionally, duplicate descriptions from multiple isoforms within each cluster type were reported only once. Genes mentioned in the text or shown in figures have gene symbols in parentheses.
All-Summer clusters also contained a potassium voltage-gated channel subfamily c member 4-like (kcnc4/Kv3.4) transcript (Table 5). Like the functional attributes of the kcnc3/Kv3.3 subunit discussed in the tissue comparisons section, potassium channels composed of KCNC4/Kv3.4 subunits could contribute to VMN’s ability to fire repetitively at high frequencies (Table 5, Figure 9). As also mentioned earlier, KCNC4/Kv3.4 currents are modulated by KCNS/Kv9 subunits, transcripts of which are highly expressed in VMN (Table 4), when coassembled in heteromeric channels [72,73]. Strikingly, in motoneurons of an ALS mouse model carrying the human sod1 mutation, kcnc4/Kv3.4 was significantly downregulated relative to expression in wild type mice [100], supporting its important role in motoneuronal function.
Steroid signaling transcripts, such as ar-a (a different isoform than the one significantly upregulated in VMN in tissue comparisons), were found in All-Summer clusters (Table 5), indicating a summer-dependent increase in androgen sensitivity within both VMN and surrounding H. VMN’s androgen sensitivity is a shared trait with other vocal vertebrates. It has been well documented that songbird syringeal motoneurons within the nXIIts concentrate high levels of androgens [101]. Both midshipman VMN and songbird nXIIts are sexually dimorphic in size, with males having larger motoneuronal somata and neuropil volume than females [68,102,103]. Furthermore, the songbird nXIIts responds to androgen treatment by increasing the size of motoneuronal somata, synaptic density, and the number of synaptic vesicles [103,104], which could increase their ability to drive vocalizations during the breeding season. Similarly, juvenile male midshipman treated with androgens exhibit increased size of VMN somata [105]. We also found a glucocorticoid receptor (nr3c1) in VMN-Summer clusters, indicating a summer-dependent increase in glucocorticoid sensitivity specifically in the VMN (Table 5). The presence of both androgen and glucocorticoid receptors is consistent with androgen- and cortisol-induced lengthening of hindbrain vocal CPG output [33,34].
The neurotransmission and neuroendocrine mechanisms highlighted by our candidate genes could influence each other to bring about seasonal changes in VMN function and midshipman vocal behavior. For example, ultrastructure studies in songbirds and rodents have shown that estrogen and androgens can increase gap junction expression (e.g. [106-108]). Testosterone treatment in female songbirds induced an increase in the number of soma-somatic gap junctions in HVC, a telencephalic song control nucleus, correlated with an increased repetition rate and decreased variability in frequency modulation of male-like song [106]. Steroid-dependent seasonal regulation of gap junction abundance in vocal control nuclei influencing vocal behavior could also apply to midshipman fish.
Daily comparisons
We found nine neurotransmission transcripts within Peak-SNV clusters (Table 5) that were all unique from those found in Peak-SNH clusters. These included an isoform of cx30, GABAAreceptor subunit pi-like (gabrp), and two calcium-activated potassium channel subunit alpha-1 (kcnma1/Kca1.1) isoforms (Table 5, Figure 8B). While cx30 and gabrp were both found in transcripts upregulated in the VMN compared to H (Table 4), they are different Trinity-predicted isoforms. Similar to between-tissue comparisons that highlighted the importance of cholinergic action in VMN, we found acetylcholine receptors, including neuronal acetylcholine receptor beta-2-like (chrnb2) in Peak-SNV clusters (Table 5, Figure 8B). Steroid signaling transcripts in Peak-SNV clusters included an estrogen receptor alpha (esr1) isoform (Table 5), shown by in-situ to be expressed in vocal motoneurons [94]. We propose that seasonal and/or daily variation in the expression levels of neurotransmission and steroid hormone related transcripts support modulation of known VMN firing properties that, in turn, translates directly into changes in vocal behavior at times of high vocal activity.
Conclusions
The vocal motor system of midshipman fish exemplifies a simple vertebrate model with which we can identify genetic components supporting neural network function leading to a single behavior, in this case vocalization that serves a social communication function in multiple vertebrate lineages. This VMN transcriptome project provides a global view of molecular pathways responsible for neuronal function and hormone modulation in the vocal motoneurons of midshipman and vertebrate in general. We identified a suite of candidate genes whose functions underlie previously identified VMN firing properties, as well as novel candidates whose functions regulate aspects of neuronal excitability that remain to be studied in VMN (summarized in Figure 9).
The interpretive power of our results benefits directly from a large body of corroborating evidence in the midshipman model system, from synaptic ultrastructure to intracellular neurophysiology, anatomical localization of steroid signaling pathways, and hormonal modulation of VMN output and vocal behavior (Figure 9). The new results presented here guide future molecular, anatomical, and pharmacological investigations for determining how specific metabolic, hormonal, and neurotransmission pathways sculpt neurophysiological events that pattern vocal behavior.
Our findings have broad relevance to other vertebrate taxa given the highly conserved nature of vocal mechanisms, shared demands for temporal precision in sound production, and the equally widespread occurrence of seasonal and daily variation in vocal behaviors (see discussions in [3,39]). We propose that candidate genes and pathways identified here shape neurophysiological characters of motoneuronal populations driving the sound producing superfast muscles in toadfish [65], birds [4,5], bats [6], and rattlesnakes [109]. The hypothesis that fish use the same fundamental molecular machinery as other vertebrates for controlling acoustic communication is challenging but testable. Furthermore, contrasting our results to other motor systems will yield insight into the molecular machinery utilized by neuromuscular systems varying in speed, synchrony, and precision. Finally, molecular insights into motoneuronal function will also be relevant for motoneuron dysfunction, such as diseases characterized by the loss of resilience to oxidative stress.
Methods
Tissue collection
Midshipman fish have two male reproductive morphs: type I males that build and guard nests, acoustically court females and are the focus of this study, and type II males that sneak spawn [22,26]. We collected tissue samples from type I males at different time points in the summer reproductive and winter non-reproductive seasons (see Figure 1D). None of the fish used were actively vocalizing when sacrificed, although we cannot completely discount recently produced isolated grunting or growling behavior (see below). All procedures were approved by the Institutional Animal Care and Use Committee at Cornell University.
Reproductive fish: In May 2011, type I males were hand collected from rocky intertidal zones in northern California. Fish were then held for 1–2 days at the University of California Bodega Marine Laboratory in large outdoor tanks with artificial shelters and flow-through seawater. We are confident that the fish used in this study were not engaged in extended vocal behaviors such as humming for several reasons. First, although the fish could have been producing isolated agonistic grunts or growls in this setting, we did not observe the inflated swim bladders associated with humming (AHB unpub obs). Second, males from California do not take up residence and engage in any defense or courtship vocal behaviors until three weeks in captivity (NYF and AHB upub obs); fish used in this study were only held in large communal tanks for 1–2 days. Future transcriptome experiments using tissues from fish that were recorded and confirmed to be actively humming will provide insight into genes upregulated during vocal behavior.
The ambient light:dark cycle was approximately 14.25 h light: 9.75 h dark, with sunrise at approximately 6:00 am and sunset at 8:15 pm. The “summer night (SN)” males (n = 6) were sacrificed at the middle of the dark phase (middark), between 12:00 am and 2:00 am; and the “summer morning (SM)” males (n = 6) were sacrificed after sunrise, between 7:00 am and 9:40 am. Fish were deeply anesthetized under 0.025% benzocaine (Sigma-Aldrich, St. Louis, MO) and exsanguinated from the heart. The brain was removed after craniotomy, transected at the midbrain-hindbrain boundary, and stored in tubes containing RNAlater solution (Life Technologies, Carlsbad, CA) overnight in 4°C followed by −20°C. The samples were shipped overnight to Cornell University (Ithaca, NY) on dry ice and stored in −20°C until use.
Non-reproductive fish: Midshipman migrate offshore to deeper waters in the winter non-reproductive season [110]. In January 2011, type I males were obtained by otter trawl (R/V John H. Marin, Moss Landing Marine Laboratories) in Monterey Bay, offshore from Moss Landing, CA at depths of 115–150 meters. These fish, referred to as “winter night (WN)”, were kept under running seawater in large indoor tanks with ambient lighting through skylights, and sacrificed within 24 h of collection at middark (n = 5; 11:30 pm to 1:00 am) following the same tissue collection procedures as used for reproductive fish.
cDNA library preparation and sequencing
The paired VMN located at the midline contain approximately 4000 homogeneous motoneurons, presynaptic input, and surrounding glial cells that can be isolated in toto (Figures 1C and 9) [8,45,46]). Immediately before RNA extraction, we used fine forceps and minutien pins to isolate the paired VMN from the surrounding hindbrain (H) as previously described [46]. Separation of VMN from the rest of H, which included premotor populations and other motor populations, was verified in sectioned material. Total RNA was extracted from VMN and H tissues using the Trizol reagent (Life Technologies) following the manufacturer’s protocol. RNA concentrations were quantified using a Qubit fluorometer (Life Technologies), and equal masses of RNA were pooled by tissue and collection time. Reproductive summer night (SN) and summer morning (SM) VMN and H groups (SNV, SNH, SMV, SMH) were each pooled from 6 individuals, non-reproductive winter night (WN) VMN (WNV) was pooled from 4 individuals, and winter night H (WNH) was pooled from 5 individuals (Figure 1D). Each pool contained 2 μg of total RNA. We treated the pooled RNA samples with DNase I (Ambion) to eliminate potential genomic DNA contamination.
Figure 2 provides an overview of this study’s key procedures and analyses with reference to relevant figures and tables. We closely followed the protocol of Zhong et al. [111] for strand-specific cDNA library construction, which included poly(A) RNA isolation and chemical fragmentation, first-strand cDNA synthesis, second-strand cDNA synthesis with dUTP, end-repair, dA-tailing, Y-shape barcoded adapter ligation, DNA purification and size selection. The second-strand DNA was digested with uracil DNA glycosylase to leave first-strand cDNA with differential adapters on the 5′ and 3′ ends. We then enriched our cDNA library using 14 cycles of PCR, and verified the size by agarose gel. Examining 40 ng of each cDNA library with an Agilent Bioanalyzer at Cornell’s Biotechnology Resource Center showed size peaks between 251 bp and 257 bp. Finally, 20 ng of cDNA from each sample group were combined with 20 ng from each of 4 other groups of cDNA from saccular epithelia of the inner ear into a single pool of 2000 ng pooled cDNA library; the auditory tissues contributed to a companion study [49]. The completed cDNA library was submitted for paired-end sequencing by Illumina HiSeq2000 at the Biotechnology Resource Center’s Genomics Facility.
Transcriptome assembly and annotation
Read pairs were discarded if either read did not pass Illumina’s quality control. Adaptor sequences and low quality nucleotides were removed from the ends using Trimmomatic tools and reads less than 10 bases after trimming were dropped [112]. Over 90% of raw reads passed quality filtering and trimming, leaving 20.2 ± 2.4 million (mean ± S.D.) reads per group. Forward and reverse reads from all sample groups, including those from the saccular epithelia, were concatenated and transferred to the Blacklight system at the Pittsburgh Supercomputing Center where the full transcriptome was assembled with the Trinity software package (version r2013-02-15) [113], one of the most robust de novo assembly methods [114]. In order to limit occurrences of fusion transcripts we used the “jaccard_clip” option and set the “min_kmer_cov” to 2.
We used Trinity’s TransDecoder utility to discard assembled transcripts with open reading frames (ORF) of less than 50 amino acids [113]. To assess the completeness of our transcriptome, we submitted our filtered transcriptome to the CEGMA software [52]. CEGMA looks for the presence of 248 highly-conserved core eukaryotic genes (CEGs) and a larger set of 458 CEGs [52]. We used Blast2GO to annotate our assembled transcriptome based on the NCBI non-redundant protein database [53]. All transcripts were submitted to blastx using an E-value cutoff of 1e-10 via Blas2GO; those without blastx hits were then subjected to blastn a 1e-10 E-value cutoff. Mapping and annotation steps were performed with default settings (i.e., E-value cutoff = 1e-6). InterProScan and GO-Enzyme Codes were run on all transcripts.
With Trinity-supported companion programs (i.e. bowtie and RSEM) we mapped the individual reads back onto the assembled transcriptome and estimated the abundance of transcripts from each tissue group [113]. Lowly supported transcripts, accounting for less than 1% of total reads of a gene (IsoPct), were discarded. The length distribution statistics of surviving transcripts were obtained through PRINSEQ [115]. As an additional assessment of assembly quality, we submitted the final set of surviving transcripts to TransDecoder and counted the number of transcripts with complete ORFs (containing start and stop codons). The assembled transcriptome and sample group reads were submitted to the NCBI Transcriptome Shotgun Assembly and Sequence Read Archive databases under BioProject accession PRJNA269550.
Because four ear tissue groups were included in the transcriptome assembly, our analyses of the vocal system excluded transcripts with read count sums of zero across all six brain sample groups (see Figure 1D). We focused on transcripts (isoforms) rather than predicted genes (also called “gene components” by Trinity, [113]) because the single sequence available for each transcript allows straight-forward annotations and the potential to identify differential regulation of alternate isoforms of a single gene.
VMN vs. H tissue comparisons
Tissue-based normalization
TMM normalization (Figure 4), used by default Trinity analysis pipeline [113], as well as several methods evaluated in [116], was insufficient in correcting the nonlinear skew observed in VMN vs H comparisons, likely due to systematic sequencing biases caused by differential tissue RNA population composition (see [56,116] for discussion). However, a cyclic loess method called fastlo [55], originally designed for microarray data, worked well to correct our nonlinear skew (Figure 4). To allow proper fastlo normalization, we added 1 to all raw counts prior to normalizing.
Differential expression using edgeR
For the comparison between tissues, we treated the three time points sampled as biological replicates, estimated transcript-wise dispersion values, followed by differential expression analysis with edgeR’s exact test [117]. Differentially expressed transcripts (FDR < 0.05, largely corresponding to fold change > 4; see blue lines in Figure 7A) showing upregulation in VMN or H were then subjected to downstream, GO term enrichment and KEGG pathway mapping analyses. Furthermore, we selected nine candidate genes of functional interest upregulated in VMN samples for validation by sequencing and qPCR (see below).
Gene category-based analyses
To gain a sense of the relative contribution of gene functional categories to VMN function (Figure 2), we extracted fastlo-normalized expression levels for transcripts (regardless of whether they were differentially expressed) that were classified under: 1) “neurotransmission”: ion channels and neurotransmitter receptors; 2) “neuroendocrine”: steroid hormone signaling genes such as receptors and biosynthetic enzymes, and thyroid hormone related genes; 3) “neuropeptide”: neuropeptides and their receptors, as well as peptide hormones and their receptors; 4) “neuromodulator”: neurochemicals other than steroids and neuropeptides that are known within this system, namely genes related to catecholamine, serotonin, and melatonin biosynthesis and signaling; 5) “cellular respiration”: cellular respiration-related genes; and 6) “antioxidant”: antioxidant enzymes and proteins. Transcript IDs and descriptions in each gene category are listed in Additional file 1: Table S4.
We compared the tissue expression levels of gene categories to assess the relative importance of each gene category for VMN vs. H (Figure 5). We first examined whether H expressed more genes by dichotomizing the data into 1 if the total tissue expression was > 0 and 0 if the total was 0 (not expressed), followed by a chi square test (Figure 5A). To compare total expression levels for each gene category, we summed expression levels across the three VMN or H sample groups for expressed genes (tissue sum > 0) (Figure 5B). We then Box-Cox transformed this dataset in order to statistically compare H vs. VMN tissue expression by performing one-way ANOVA for each gene category and setting the Bonferroni corrected alpha level at 0.008 using JMP9.0 (SAS Institute Inc., Cary, NC, USA) (Figure 5B). The non-parametric Wilcoxon rank sum test for tissue comparisons followed by Bonferroni correction produced the same results. Finally, to evaluate correlation patterns across sample groups for each gene category, we performed Spearman’s correlation for pair-wise comparisons between all six-tissue groups in R [118] (Figure 6).
Daily and seasonal variation in gene expression
For daily and seasonal gene expression comparisons, we used Trinity’s downstream analysis pipeline with default parameters [113]. Transcripts that were differentially expressed in at least one pairwise sample comparison were identified using the Trinity-supported edgeR Bioconductor package [57], We ran the TMM normalized FPKM matrix through Trinity-supported differential expression analyses to generate hierarchically clustered heatmaps of transcripts with at least four-fold differential expression and false discovery rate (FDR) corrected P values of <0.001 (23,855 out of 77,285 transcripts) (Additional file 3). Spearman correlation matrices for pairwise sample group comparisons were also generated (Additional file 3). We generated 109 K-means clusters based on the rule of thumb for determining cluster number [119] with 23,855 differentially expressed genes from which we could extract sequence ID, annotation, and expression levels. We combined clusters exhibiting similar expression patterns of day-night and seasonal variation across sample groups. Candidate genes with functions directly related to neural transmission (e.g. ion channels) and neuromodulation (e.g. steroid hormones) were identified and considered for potential actions in sculpting VMN firing properties.
Quantitative PCR (qPCR) validation of gene expression
We used qPCR on a subset of transcripts to confirm the accuracy of our assembled transcriptome and RSEM predicted differential transcript abundance among our sample groups. First, to validate within-tissue gene expression ratios we selected 19 transcripts chosen based on the following criteria: 1) each transcript was the only transcript in its gene component; 2) the SNH/SMH FPKM ratios of all the transcripts fell within a broad range (0.05-100); 3) the primers we chose based on the assembled transcript sequences produced only the predicted PCR product (Additional file 1: Table S6 contains gene descriptions and primer sequences). These criteria allowed for straightforward qPCR analyses from H samples, which provided more cDNA for analysis than VMN samples. Specificity of the qPCR primers was verified by sequencing at the Cornell Genomics Core facility. We selected the reference gene based on its very low coefficient of variance (0.10) in FPKM values across all sample groups and being the only transcript for its gene component. The reference gene (Blast2Go annotation “btb poz domain-containing protein kctd6”) was confirmed by qPCR to show no apparent or significant expression differences between SNH and SMN (P = 0.26). Each qPCR was performed in triplicate on the same, pooled cDNA sample produced for RNA-seq as well as no template controls. Each reaction contained 5 μl of 2x Power SYBR Green PCR Master Mix (Life Technologies, Carlsbad, CA), 1 μl each of forward and reverse primers (50 nM final concentration for the reference gene and 100 nM for target genes), 1 μl dH2O, and 2 μl of the appropriate pooled cDNA. The qPCR reaction was run on an ABI Viia7 system at the Cornell Genomics Core facility. We used the standard curve method to extrapolate copy numbers, which were normalized against our reference gene. Pearson’s correlations were calculated for qPCR and RNA-seq predicted SNH/SMH expression ratios (Figure 3D).
Next, to validate VMN vs. H tissue comparisons with qPCR, we chose nine functionally important candidate genes that were upregulated in the VMN (Additional file 1: Table S6), regardless of how many isoforms were predicted by Trinity. We used the same, pooled SMV and SMH samples that were submitted for RNAseq. SMV and SMH samples were chosen due to cDNA availability. The qPCR procedures and reference gene were the same as above for SNH/SMH comparisons. Pearson’s correlations were calculated for qPCR vs. fastlo-normalized SMV/SMH ratios, which were averaged across all isoforms for each Trinity predicted gene component (Figure 7C), as well as for qPCR vs fastlo-normalized values separately analysed for SMV and SMH (Additional file 2). Student’s paired t test was performed in R to compare qPCR measured SMV and SMH expression levels using log-transformed data (Figure 7D).
Availability of supporting data
The assembled transcriptome and reads from each sample supporting the results of this article are available in the NCBI Transcriptome Shotgun Assembly and Sequence Read Archive databases under BioProject accession number [PRJNA269550].
Acknowledgements
We sincerely thank Silin Zhong and the Jim Giovannoni lab (USDA-ARS, Cornell University) for help with cDNA library construction. Cornell University BioHPC Lab resources and the Bioinformatics Facility, especially Dr. Qi Sun, provided us with the expertise, guidance, and computing resources to complete our project. We are grateful to Lynn M. Johnson at Cornell Statistical Consulting Unit, James Booth and Martin Wells at Cornell Department of Statistics for their generous help with data analysis and tissue normalization. We would also like to thank Brian Haas from the Broad Institute for his help with using Trinity, Cameron Finucane for programming support, as well as Richard Harrison, Alon Keinan and especially Ben Matthews for comments on early drafts of the manuscript. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number OCI-1053575. Research support, in part, came from NSF IOS 1120925 (AHB) and Cornell University’s Center for Vertebrate Genomics (NYF).
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- ANOVA
Analysis of variance
- ATP
Adenosine triphosphate
- BLAST
Basic local alignment search tool
- CEGMA
Core eukaryotic genes mapping approach
- CEGs
Core eukaryotic genes
- CPG
Central pattern generator
- FDR
False discovery rate
- FPKM
Fragments per kilobase of transcript per million reads mapped
- GABA
Gamma-aminobutyric acid
- GO
Gene ontology
- H
Surrounding hindbrain without VMN
- HVC
Letter based name for a telencephalic song control nucleus in songbirds
- KEGG
Kyoto encyclopedia of genes and genomes
- Kv
Voltage-gated potassium channel
- nXIIts
Tracheosyringeal division of the hypoglossal motor nucleus
- ORFs
Open reading frames
- qPCR
Quantitative PCR
- RNA-seq
RNA sequencing
- SMH
Reproductive summer morning hindbrain
- SMV
Reproductive summer morning VMN
- SNH
Reproductive summer night hindbrain
- SNV
Reproductive summer night VMN
- TCA
Tricarboxylic acid
- TMM
Trimmed mean of M values
- VMN
Vocal motor nucleus
- WNH
Non-reproductive winter night hindbrain
- WNV
Non-reproductive winter night VMN
Additional files
GO terms enriched in transcripts upregulated in VMN. GO category notations: C = cellular component; F = molecular function; P = biological process. FDR = false discovery rate adjusted P values. GO terms related to cellular respiration are counted in column D. Table S2. GO terms enriched in transcripts upregulated in surrounding hindbrain (H). GO category notations: C = cellular component; F = molecular function; P = biological process. FDR = false discovery rate adjusted P values. GO terms related to cellular respiration are counted in column D. Table S3. KEGG pathways mapped to upregulated transcripts in VMN or H. Names and the number of sequences are listed for each KEGG pathway. Table S4. Functional gene category transcripts. List of sequence IDs and Blast2GO sequence descriptions for transcripts under each functional gene category. Expression levels for these transcripts are plotted in Figure 5. Table S5. Functionally important candidate genes upregulated in H. Table S6. Sequences and primers used for qPCR validation. Forward (F) and reverse (R) qPCR primer sequences are listed for each Trinity predicted isoform or gene component.
QPCR validation of RNAseq candidate gene expression. A) SMV qPCR values (copy numbers normalized by a reference gene) for nine candidate genes are significantly correlated with RNA-seq predicted values (FPKM). B) SMH qPCR values (copy numbers normalized by a reference gene) for nine candidate genes are significantly correlated with RNA-seq predicted values (FPKM).
A) Heatmap of hierarchically clustered expression levels (median centered FPKM +1) of significantly differentially expressed transcripts based on the TMM-normalized dataset. Each line is a transcript, and each column is a sample group. Sample groups are hierarchically clustered based on their spearman correlation coefficients. B) Heatmap of pairwise Spearman correlation coefficients. Hindbrain sample groups show higher correlation of transcript expression patterns than VMN sample groups. See Figure 1C for explanation of sample group abbreviations.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NYF, DJF and AHB designed experiments; NYF and DJF conducted experiments; NYF, DJF and AHB analyzed data and wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Ni Y Feng, Email: nf82@cornell.edu.
Daniel J Fergus, Email: danieljfergus@gmail.com.
Andrew H Bass, Email: ahb3@cornell.edu.
References
- 1.Bass AH, Gilland EH, Baker R. Evolutionary origins for social vocalization in a vertebrate hindbrain-spinal compartment. Science. 2008;321:417–21. doi: 10.1126/science.1157632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kingsbury MA, Kelly AM, Schrock SE, Goodson JL. Mammal-like organization of the avian midbrain central gray and a reappraisal of the intercollicular nucleus. PLoS ONE. 2011;6:e20720. doi: 10.1371/journal.pone.0020720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass AH, Chagnaud BP. Shared developmental and evolutionary origins for neural basis of vocal-acoustic and pectoral-gestural signaling. Proc Natl Acad Sci USA. 2012;109(Suppl 1):10677–84. doi: 10.1073/pnas.1201886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elemans C, Spierts I, Müller UK, van Leeuwen JL. Bird song: superfast muscles control dove’s trill. Nature. 2004;431:146. doi: 10.1038/431146a. [DOI] [PubMed] [Google Scholar]
- 5.Elemans CPH, Mead AF, Rome LC, Goller F. Superfast Vocal Muscles Control Song Production in Songbirds. PLoS ONE. 2008, 3:e2581. [DOI] [PMC free article] [PubMed]
- 6.Elemans CPH, Mead AF, Jakobsen L, Ratcliffe JM. Superfast Muscles Set Maximum Call Rate in Echolocating Bats. Science. 2011;333:1885–8. doi: 10.1126/science.1207309. [DOI] [PubMed] [Google Scholar]
- 7.Amador A, Perl YS, Mindlin GB, Margoliash D. Elemental gesture dynamics are encoded by song premotor cortical neurons. Nature. 2013;495:59–64. doi: 10.1038/nature11967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chagnaud BP, Zee MC, Baker R, Bass AH. Innovations in motoneuron synchrony drive rapid temporal modulations in vertebrate acoustic signaling. J Neurophysiol. 2012;107:3528–42. doi: 10.1152/jn.00030.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg JH, Farries MA, Fee MS. Basal ganglia output to the thalamus: still a paradox. Trends Neurosci. 2013;36:695–705. doi: 10.1016/j.tins.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi A, Kaczmarek LK, Kelley DB. Functional specialization of male and female vocal motoneurons. J Neurosci. 2003;23:11568–76. doi: 10.1523/JNEUROSCI.23-37-11568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: neurochemical specializations of the oscine song system. PLoS ONE. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovell PV, Carleton JB, Mello CV. Genomics analysis of potassium channel genes in songbirds reveals molecular specializations of brain circuits for the maintenance and production of learned vocalizations. BMC Genomics. 2013;14:470. doi: 10.1186/1471-2164-14-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitney O, Pfenning AR, Howard JT, Blatti CA, Liu F, Ward JM, et al. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346:1256780. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron. 2012;73:537–52. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, et al. The genome of a songbird. Nature. 2010;464:757–62. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser BA, Janowitz I, Thairu M, Travis J, Hughes KA. Phenotypic and genomic plasticity of alternative male reproductive tactics in sailfin mollies. Proc R Soc B Biol Sci. 2014;281:20132310. doi: 10.1098/rspb.2013.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rittschof CC, Bukhari SA, Sloofman LG, Troy JM, Caetano-Anollés D, Cash-Ahmed A, et al. Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. Proc Natl Acad Sci USA. 2014;111:17929–34. [DOI] [PMC free article] [PubMed]
- 18.Schunter C, Vollmer SV, Macpherson E, Pascual M. Transcriptome analyses and differential gene expression in a non-model fish species with alternative mating tactics. BMC Genomics. 2014;15:1–13. doi: 10.1186/1471-2164-15-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanogo YO, Band M, Blatti C, Sinha S, Bell AM. Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proc R Soc B Biol Sci. 2012;279:4929–38. [DOI] [PMC free article] [PubMed]
- 20.Greenfield DW, Winterbottom R, Collette BB. Review of the Toadfish genera (Teleostei: Batrachoididae) Proc Calif Acad Sci. 2008;59:665–710. [Google Scholar]
- 21.Rice AN, Bass AH. Novel vocal repertoire and paired swimbladders of the three-spined toadfish, Batrachomoeus trispinosus: insights into the diversity of the Batrachoididae. J Exp Biol. 2009;212:1377–91. doi: 10.1242/jeb.028506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brantley RK, Bass AH. Alternative male spawning tactics and acoustic signals in the plainfin midshipman fish porichthys notatus Girard (Teleostei, Batrachoididae) Ethology. 1994;96:213–32. doi: 10.1111/j.1439-0310.1994.tb01011.x. [DOI] [Google Scholar]
- 23.McKibben JR, Bass AH. Behavioral assessment of acoustic parameters relevant to signal recognition and preference in a vocal fish. J Acoust Soc Am. 1998;104:3520–33. doi: 10.1121/1.423938. [DOI] [PubMed] [Google Scholar]
- 24.Ibara RM, Penny LT, Ebeling AW, van Dykhuizen G, Cailliet G. Predators and prey in fishes. Dordrecht: Springer Netherlands; 1983. The mating call of the plainfin midshipman fish, Porichthys notatus; pp. 205–12. [Google Scholar]
- 25.Rubow TK, Bass AH. Reproductive and diurnal rhythms regulate vocal motor plasticity in a teleost fish. J Exp Biol. 2009;212:3252–62. doi: 10.1242/jeb.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McIver EL, Marchaterre MA, Rice AN, Bass AH. Novel underwater soundscape: acoustic repertoire of plainfin midshipman fish. J Exp Biol. 2014;217:2377–89. doi: 10.1242/jeb.102772. [DOI] [PubMed] [Google Scholar]
- 27.Bass AH, Marchaterre MA, Baker R. Vocal-acoustic pathways in a teleost fish. J Neurosci. 1994;14:4025–39. doi: 10.1523/JNEUROSCI.14-07-04025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodson JL, Bass AH. Vocal–acoustic circuitry and descending vocal pathways in teleost fish: Convergence with terrestrial vertebrates reveals conserved traits. J Comp Neurol. 2002;448:298–322. doi: 10.1002/cne.10258. [DOI] [PubMed] [Google Scholar]
- 29.Kittelberger JM, Bass AH. Vocal motor and auditory connectivity of the midbrain periaqueductal gray in a teleost fish. J Comp Neurol. 2013;521:791–812. doi: 10.1002/cne.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chagnaud BP, Baker R, Bass AH. Vocalization frequency and duration are coded in separate hindbrain nuclei. Nat Commun. 2011;2:346–11. doi: 10.1038/ncomms1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bass AH. Central pattern generator for vocalization: is there a vertebrate morphotype? Curr Opin Neurobiol. 2014;28:94–100. doi: 10.1016/j.conb.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass AH, Baker R. Sexual dimorphisms in the vocal control system of a teleost fish: Morphology of physiologically identified neurons. J Neurobiol. 1990;21:1155–68. doi: 10.1002/neu.480210802. [DOI] [PubMed] [Google Scholar]
- 33.Remage-Healey L, Bass AH. A rapid neuromodulatory role for steroid hormones in the control of reproductive behavior. Brain Res. 2006;1126:27–35. doi: 10.1016/j.brainres.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 34.Remage-Healey L, Bass AH. Rapid, hierarchical modulation of vocal patterning by steroid hormones. J Neurosci. 2004;24:5892–900. doi: 10.1523/JNEUROSCI.1220-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remage-Healey L, Bass AH. Estradiol interacts with an opioidergic network to achieve rapid modulation of a vocal pattern generator. J Comp Physiol A. 2009;196:137–46. doi: 10.1007/s00359-009-0500-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remage-Healey L, Bass AH. Plasticity in brain sexuality is revealed by the rapid actions of steroid hormones. J Neurosci. 2007;27:1114–22. doi: 10.1523/JNEUROSCI.4282-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodson JL, Bass AH. Rhythmic midbrain-evoked vocalization is inhibited by vasoactive intestinal polypeptide in the teleost Porichthys notatus. Brain Res. 2000;865:107–11. doi: 10.1016/S0006-8993(00)02232-0. [DOI] [PubMed] [Google Scholar]
- 38.Goodson JL, Bass AH. Forebrain peptides modulate sexually polymorphic vocal circuitry. Nature. 2000;403:769–72. doi: 10.1038/35001581. [DOI] [PubMed] [Google Scholar]
- 39.Feng NY, Bass AH. Melatonin action in a midbrain vocal-acoustic network. J Exp Biol. 2014;217:1046–57. doi: 10.1242/jeb.096669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturdy CB, Wild JM, Mooney R. Respiratory and telencephalic modulation of vocal motor neurons in the zebra finch. J Neurosci. 2003;23:1072–86. doi: 10.1523/JNEUROSCI.23-03-01072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rübsamen R, Betz M. Control of echolocation pulses by neurons of the nucleus ambiguus in the rufous horseshoe bat, Rhinolophus rouxi. I. Single unit recordings in the ventral motor nucleus of the laryngeal nerves in spontaneously vocalizing bats. J Comp Physiol A. 1986;159:675–87. doi: 10.1007/BF00612040. [DOI] [PubMed] [Google Scholar]
- 42.Riede T, Goller F. Peripheral mechanisms for vocal production in birds - differences and similarities to human speech and singing. Brain Lang. 2010;115:69–80. doi: 10.1016/j.bandl.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hage SR, Jürgens U. On the role of the pontine brainstem in vocal pattern generation: a telemetric single-unit recording study in the squirrel monkey. J Neurosci. 2006;26:7105–15. doi: 10.1523/JNEUROSCI.1024-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hage SR, Gavrilov N, Salomon F, Stein AM. Temporal vocal features suggest different call-pattern generating mechanisms in mice and bats. BMC Neurosci. 2013;14:99. doi: 10.1186/1471-2202-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bass A, Andersen K. Inter- and intrasexual dimorphisms in the vocal control system of a teleost fish: motor axon number and size. Brain Behav Evol. 1991;37:204–14. doi: 10.1159/000114359. [DOI] [PubMed] [Google Scholar]
- 46.Fergus DJ, Bass AH. Localization and divergent profiles of estrogen receptors and aromatase in the vocal and auditory networks of a fish with alternative mating tactics. J Comp Neurol. 2013;521:2850–69. doi: 10.1002/cne.23320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bass AH, Remage-Healey L. Central pattern generators for social vocalization: Androgen-dependent neurophysiological mechanisms. Horm Behav. 2008;53:659–72. doi: 10.1016/j.yhbeh.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forlano PM, Sisneros JA, Rohmann KN, Bass AH. Neuroendocrine control of seasonal plasticity in the auditory and vocal systems of fish. Front Neuroendocrinol. 2015;37:129–45. [DOI] [PMC free article] [PubMed]
- 49.Fergus DJ, Feng NY, Bass AH. Gene expression patterns in auditory hair cell epithelium: insights into the molecular basis of seasonal plasticity in frequency sensitivity [abstract]. Washington DC: Society for Neuroscience, 183.02/SS28;2014.
- 50.Balakrishnan CN, Mukai M, Gonser RA, Wingfield JC, London SE, Tuttle EM, et al. Brain transcriptome sequencing and assembly of three songbird model systems for the study of social behavior. PeerJ. 2014;2:e396. doi: 10.7717/peerj.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–7. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 52.Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37:289–97. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huth TJ, Place SP. De novo assembly and characterization of tissue specific transcriptomes in the emerald notothen, Trematomus bernacchii. BMC Genomics. 2013;14:805. doi: 10.1186/1471-2164-14-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20:2778–86. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- 56.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brantley RK, Bass AH. Cholinergic neurons in the brain of a teleost fish (Porichthys notatus) located with a monoclonal antibody to choline acetyltransferase. J Comp Neurol. 1988;275:87–105. doi: 10.1002/cne.902750108. [DOI] [PubMed] [Google Scholar]
- 59.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu G, Fang Y-Z, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–92. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 61.Delage E, Puyaubert J, Zachowski A, Ruelland E. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: Convergences and divergences among eukaryotic kingdoms. Prog Lipid Res. 2013;52:1–14. doi: 10.1016/j.plipres.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Ophir AG, SCHRADER SB, Gillooly JF. Energetic cost of calling: general constraints and species-specific differences. J Evol Biol. 2010;23:1564–9. doi: 10.1111/j.1420-9101.2010.02005.x. [DOI] [PubMed] [Google Scholar]
- 63.Gillooly JF, Ophir AG. The energetic basis of acoustic communication. Proc R Soc B Biol Sci. 2010;277:1325–31. doi: 10.1098/rspb.2009.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walsh PJ, Mommsen TP, Bass AH. Biochemical and molecular aspects of singing in batrachoidid fishes. In Biochemistry and Molecular Biology of Fishes. Volume 4. Metabolic Biochemistry. Edited by Hochachka PW, Mommsen TP; Amsterdam: Elsevier; 1995:279–289.
- 65.Rome LC. Design and function of superfast muscles: new insights into the physiology of skeletal muscle. Annu Rev Physiol. 2006;68:193–221. doi: 10.1146/annurev.physiol.68.040104.105418. [DOI] [PubMed] [Google Scholar]
- 66.Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 2007;8:45–56. doi: 10.1038/nrn2044. [DOI] [PubMed] [Google Scholar]
- 67.Pereda AE. Electrical synapses and their functional interactions with chemical synapses. Nat Rev Neurosci. 2014;15:250–63. doi: 10.1038/nrn3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bass AH, Marchaterre MA. Sound-generating (sonic) motor system in a teleost fish (Porichthys notatus): Sexual polymorphisms and general synaptology of sonic motor nucleus. J Comp Neurol. 1989;286:154–69. doi: 10.1002/cne.902860203. [DOI] [PubMed] [Google Scholar]
- 69.Judge SIV, Bever CT. Potassium channel blockers in multiple sclerosis: neuronal Kv channels and effects of symptomatic treatment. Pharmacol Ther. 2006;111:224–59. doi: 10.1016/j.pharmthera.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 70.Gutman GA, Chandy KG, Adelman JP, Aiyar J, Bayliss DA, Clapham DE, et al. International Union of Pharmacology. XLI. Compendium of voltage-gated ion channels: potassium channels. Pharmacol Rev. 2003;55:583–6. doi: 10.1124/pr.55.4.9. [DOI] [PubMed] [Google Scholar]
- 71.Ding S, Matta SG, Zhou F-M. Kv3-like potassium channels are required for sustained high-frequency firing in basal ganglia output neurons. J Neurophysiol. 2011;105:554–70. doi: 10.1152/jn.00707.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richardson FC, Kaczmarek LK. Modification of delayed rectifier potassium currents by the Kv9.1 potassium channel subunit. Hear Res. 2000;147:21–30. doi: 10.1016/S0378-5955(00)00117-9. [DOI] [PubMed] [Google Scholar]
- 73.Stocker M, Hellwig M, Kerschensteiner D. Subunit assembly and domain analysis of electrically silent K+ channel alpha-subunits of the rat Kv9 subfamily. J Neurochem. 1999;72:1725–34. doi: 10.1046/j.1471-4159.1999.721725.x. [DOI] [PubMed] [Google Scholar]
- 74.Brown D. M-currents: an update. Trends Neurosci. 1988;11:294–9. doi: 10.1016/0166-2236(88)90089-6. [DOI] [PubMed] [Google Scholar]
- 75.Grove CL, Szabo TM, McIntosh JM, Do SC, Waldeck RF, Faber DS. Fast synaptic transmission in the goldfish CNS mediated by multiple nicotinic receptors. J Physiol. 2011;589:575–95. doi: 10.1113/jphysiol.2010.197608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMahon LL, Yoon KW, Chiappinelli VA. Electrophysiological evidence for presynaptic nicotinic receptors in the avian ventral lateral geniculate nucleus. J Neurophysiol. 1994;71:826–9. doi: 10.1152/jn.1994.71.2.826. [DOI] [PubMed] [Google Scholar]
- 77.Bellingham MC, Berger AJ. Presynaptic depression of excitatory synaptic inputs to rat hypoglossal motoneurons by muscarinic M2 receptors. J Neurophysiol. 1996;76:3758-70. [DOI] [PubMed]
- 78.Miles GB, Hartley R, Todd AJ, Brownstone RM. Spinal cholinergic interneurons regulate the excitability of motoneurons during locomotion. Proc Natl Acad Sci USA. 2007;104:2448–53. doi: 10.1073/pnas.0611134104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zagoraiou L, Akay T, Martin JF, Brownstone RM, Jessell TM, Miles GB. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64:645–62. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hellstr MJ, Oliveira ALR, Meister BR, Cullheim S. Large cholinergic nerve terminals on subsets of motoneurons and their relation to muscarinic receptor type 2. J Comp Neurol. 2003;460:476–86. doi: 10.1002/cne.10648. [DOI] [PubMed] [Google Scholar]
- 81.Muennich EAL, Fyffe REW. Focal aggregation of voltage-gated, Kv2.1 subunit-containing, potassium channels at synaptic sites in rat spinal motoneurones. J Physiol. 2004;554:673–85. doi: 10.1113/jphysiol.2003.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown DA, Passmore GM. Neural KCNQ (Kv7) channels. Br J Pharmacol. 2009;156:1185–95. doi: 10.1111/j.1476-5381.2009.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caulfield MP, Birdsall NJ. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol Rev. 1998;50:279–90. [PubMed] [Google Scholar]
- 84.Luetje CW, Piattoni M, Patrick J. Mapping of ligand-binding sites of neuronal nicotinic acetylcholine-receptors using chimeric alpha-subunits. Mol Pharmacol. 1993;44:657–66. [PubMed] [Google Scholar]
- 85.McIntosh JM, Dowell C, Watkins M, Garrett JE, Yoshikami D, Olivera BM. Alpha-conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J Biol Chem. 2002;277:33610–5. doi: 10.1074/jbc.M205102200. [DOI] [PubMed] [Google Scholar]
- 86.Chambers MS, Atack JR, Broughton HB, Collinson N, Cook S, Dawson GR, et al. Identification of a novel, selective gaba aα5 receptor inverse agonist which enhances cognition. J Med Chem. 2003;46:2227–40. doi: 10.1021/jm020582q. [DOI] [PubMed] [Google Scholar]
- 87.Remage-Healey L, Bass AH. From social behavior to neural circuitry: Steroid hormones rapidly modulate advertisement calling via a vocal pattern generator. Horm Behav. 2006;50:432–41. doi: 10.1016/j.yhbeh.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 88.Forlano PM, Deitcher DL, Myers DA, Bass AH. Anatomical distribution and cellular basis for high levels of aromatase activity in the brain of teleost fish: aromatase enzyme and mRNA expression identify glia as source. J Neurosci. 2001;21:8943–55. doi: 10.1523/JNEUROSCI.21-22-08943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Labrie F, Luu-The V, Lin SX, Simard J, Labrie C, El-Alfy M, et al. Intracrinology: role of the family of 17 beta-hydroxysteroid dehydrogenases in human physiology and disease. J Mol Endocrinol. 2000;25:1–16. doi: 10.1677/jme.0.0250001. [DOI] [PubMed] [Google Scholar]
- 90.Kusakabe M, Nakamura I, Young G. 11beta-hydroxysteroid dehydrogenase complementary deoxyribonucleic acid in rainbow trout: cloning, sites of expression, and seasonal changes in gonads. Endocrinology. 2003;144:2534–45. doi: 10.1210/en.2002-220446. [DOI] [PubMed] [Google Scholar]
- 91.Genova RM, Marchaterre MA, Knapp R, Fergus D, Bass AH. Glucocorticoid and androgen signaling pathways diverge between advertisement calling and non-calling fish. Horm Behav. 2012;62:426–32. doi: 10.1016/j.yhbeh.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 92.Forlano PM, Kim SD, Krzyminska ZM, Sisneros JA. Catecholaminergic connectivity to the inner ear, central auditory, and vocal motor circuitry in the plainfin midshipman fish Porichthys Notatus. J Comp Neurol. 2014;522:2887–927. doi: 10.1002/cne.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stacey N. Hormones, pheromones and reproductive behavior. Fish Physiol Biochem. 2003;28:229–35. doi: 10.1023/B:FISH.0000030540.99732.2c. [DOI] [Google Scholar]
- 94.Forlano PM, Deitcher DL, Bass AH. Distribution of estrogen receptor alpha mRNA in the brain and inner ear of a vocal fish with comparisons to sites of aromatase expression. J Comp Neurol. 2005;483:91–113. doi: 10.1002/cne.20397. [DOI] [PubMed] [Google Scholar]
- 95.Davies KJ. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000;50:279–89. doi: 10.1080/15216540051081010. [DOI] [PubMed] [Google Scholar]
- 96.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 97.Cozzolino M, Pesaresi MG, Gerbino V, Grosskreutz J, Carrì MT. Amyotrophic Lateral Sclerosis: New insights into underlying molecular mechanisms and opportunities for therapeutic intervention. Antioxid Redox Signal. 2012;17:1277–330. doi: 10.1089/ars.2011.4328. [DOI] [PubMed] [Google Scholar]
- 98.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance part I. Crit Rev Biochem Mol. 1995;30:445–520. [DOI] [PubMed]
- 99.Hermosura MC, Nayakanti H, Dorovkov MV, Calderon FR, Ryazanov AG, Haymer DS, et al. A TRPM7 variant shows altered sensitivity to magnesium that may contribute to the pathogenesis of two Guamanian neurodegenerative disorders. Proc Natl Acad Sci USA. 2005;102:11510–5. doi: 10.1073/pnas.0505149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bandyopadhyay U, Cotney J, Nagy M, Oh S, Leng J, Mahajan M, et al. RNA-seq profiling of spinal cord motor neurons from a presymptomatic sod1 als mouse. PLoS ONE. 2013;8:e53575. doi: 10.1371/journal.pone.0053575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arnold AP, Nottebohm F, Pfaff DW. Hormone concentrating cells in vocal control and other areas of the brain of the zebra finch (Poephila guttata) J Comp Neurol. 1976;165:487–511. doi: 10.1002/cne.901650406. [DOI] [PubMed] [Google Scholar]
- 102.Bass AH, Horvath BJ, Brothers EB. Nonsequential developmental trajectories lead to dimorphic vocal circuitry for males with alternative reproductive tactics. J Neurobiol. 1996;30:493–504. doi: 10.1002/(SICI)1097-4695(199608)30:4<493::AID-NEU5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 103.DeVoogd TJ, Pyskaty DJ, Nottebohm F. Lateral asymmetries and testosterone-induced changes in the gross morphology of the hypoglossal nucleus in adult canaries. J Comp Neurol. 1991;307:65–76. doi: 10.1002/cne.903070107. [DOI] [PubMed] [Google Scholar]
- 104.Clower RP, Nixdorf BE, DeVoogd TJ. Synaptic plasticity in the hypoglossal nucleus of female canaries: structural correlates of season, hemisphere, and testosterone treatment. Behav Neural Biol. 1989;52:63–77. doi: 10.1016/S0163-1047(89)90170-2. [DOI] [PubMed] [Google Scholar]
- 105.Bass AH, Forlano PM. Neuroendocrine mechanisms of alternative reproductive tactics: the chemical language of reproductive and social plasticity. Cambridge: Cambridge University Press; 2008. pp. 109–31. [Google Scholar]
- 106.Gahr M, Garcia-Segura LM. Testosterone-dependent increase of gap-junctions in HVC neurons of adult female canaries. Brain Res. 1996;712:69–73. doi: 10.1016/0006-8993(95)01448-9. [DOI] [PubMed] [Google Scholar]
- 107.Matsumoto A, Arnold AP, Zampighi GA, Micevych PE. Androgenic regulation of gap junctions between motoneurons in the rat spinal cord. J Neurosci. 1988;8:4177–83. doi: 10.1523/JNEUROSCI.08-11-04177.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perez J, Tranque PA, Naftolin F, Garcia-Segura LM. Gap junctions in the hypothalamic arcuate neurons of ovariectomized and estradiol-treated rats. Neurosci Lett. 1990;108:17–21. doi: 10.1016/0304-3940(90)90699-A. [DOI] [PubMed] [Google Scholar]
- 109.Rome LC, Syme DA, Hollingworth S, Lindstedt SL, Baylor SM. The whistle and the rattle: the design of sound producing muscles. Proc Natl Acad Sci USA. 1996;93:8095–100. doi: 10.1073/pnas.93.15.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sisneros JA, Forlano PM, Knapp R, Bass AH. Seasonal variation of steroid hormone levels in an intertidal-nesting fish, the vocal plainfin midshipman. Gen Comp Endocrinol. 2004;136:101–16. doi: 10.1016/j.ygcen.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 111.Zhong S, Joung J-G, Zheng Y, Chen Y-R, Liu B, Shao Y, et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc. 2011;2011:940–9. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 112.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:btu170–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li B, Fillmore N, Bai Y, Collins M, Thomson JA, Stewart R, et al. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014;15:553. [DOI] [PMC free article] [PubMed]
- 115.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glusman G, Caballero J, Robinson M, Kutlu B, Hood L. Optimal scaling of digital transcriptomes. PLoS ONE. 2013;8:e77885. doi: 10.1371/journal.pone.0077885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–7. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 118.Team RC . R: a Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 119.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. New York: Academic Press; 1979.
- 120.Forlano PM, Marchaterre M, Deitcher DL, Bass AH. Distribution of androgen receptor mRNA expression in vocal, auditory, and neuroendocrine circuits in a teleost fish. J Comp Neurol. 2010;518:493–512. doi: 10.1002/cne.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]


