Abstract
The circadian (~24 h) clock is continuously entrained (reset) by ambient light so that endogenous rhythms are synchronized with daily changes in the environment. Light-induced gene expression is thought to be the molecular mechanism underlying clock entrainment. mRNA translation is a key step of gene expression, but how clock entrainment is controlled at the mRNA translation level is not understood. Here we report that a light- and circadian clock-regulated MAPK/MNK pathway leads to phosphorylation of the cap-binding protein eIF4E in the mouse suprachiasmatic nucleus (SCN) of the hypothalamus, the locus of the master circadian clock in mammals. Phosphorylation of eIF4E specifically promotes translation of Period (Per) 1 and 2 mRNAs and increases the abundance of basal and inducible PER proteins, which facilitates circadian clock resetting and precise timekeeping. Together, these results highlight a critical role for light-regulated translational control in the physiology of the circadian clock.
Keywords: Circadian clock, translational control, light, entrainment, Period, eIF4E, phosphorylation
INTRODUCTION
Circadian (~24 h) rhythmicity is a fundamental property of nearly all living organisms1. In mammals, the master circadian clock is located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus2. The SCN generates ~24-h rhythms that orchestrate a variety of physiological and behavioral processes3. The circadian rhythms allow animals to predict and prepare for upcoming environmental changes, which is critical for survival. Rhythmic gene expression constitutes the molecular basis of circadian oscillation. The molecular clockwork is composed of transcriptional-translational feedback loops4. The primary negative feedback loop is driven by rhythmic activation of Period (Per) and Cryptochrome (Cry) transcription by the CLOCK (or NPAS2) and BMAL1 heterodimers. Per and Cry mRNAs encoded proteins form multimer complexes. As they accumulate in the cytoplasm, the PER/CRY complexes translocate into the nucleus and interact with the CLOCK/BMAL1 heterodimers to repress their own gene transcription.
The circadian clock is entrained (reset) by external cues so that the endogenous rhythms are continuously synchronized with the daily changes in the environment5. Light is the dominant signal for clock entrainment. The phasing of the SCN clock is tightly regulated by the ambient light/dark (LD) cycles. At night, transient light exposure causes a rapid resetting of the clock6. Although the precise molecular mechanisms whereby photic input drives clock entrainment have not been resolved, it is thought that rapid induction of gene expression drives the resetting process7. Previous studies have revealed multiple signaling mechanisms by which Per transcription is induced by light8–10. For example, light stimulates the mitogen- and stress-activated protein kinase (MSK) 111, which in turn leads to activation of transcription factors such as the cAMP response element binding protein (CREB). CREB activates the expression of early response genes that harbor cAMP response elements (CRE) in their promoters, including c-Fos and Per112. Little is known, however, about how mRNA translation, a key step in gene expression, is coordinated in the clock entrainment process.
Control of mRNA translation occurs predominantly at the initiation step, which under most circumstances is rate-limiting in protein synthesis13. Translation initiation begins with the recognition of the m7GpppN (where N is any nucleotide) structure (mRNA 5’ cap), which is present at the 5’-end of all nuclear transcribed mRNAs, by the eukaryotic translation initiation factor 4E (eIF4E)11. eIF4E binds to the 5’ cap as a subunit of the eIF4F complex together with eIF4A and eIF4G13,14. As a rate-limiting translation factor, eIF4E is a major target for translational control. Its abundance and activities are intricately controlled by several mechanisms. For example, the mTOR-regulated eIF4E binding proteins (4E-BPs) prevent the eIF4E binding to eIF4G, and thus the formation of the eIF4F complex and its binding to the 5’ cap13,14. We have recently reported a key role for 4E-BP1-mediated translational control in the circadian clock15. In addition, the activities of eIF4E can be regulated by phosphorylation. In response to extracellular signals, the p38 and p42/44 mitogen-activated protein kinases (MAPKs) activate the MAPK-interacting serine/threonine-protein kinases (MKNKs or MNKs) including MNK1 and MNK216, which in turn phosphorylate eIF4E at a single amino acid, Ser20917.
To study the role of eIF4E phosphorylation in the circadian clock, we have used a unique mouse model, the EIF4ES209A/S209A mouse (referred to hereafter as the KI mouse), in which Serine 209 of eIF4E is mutated to alanine and therefore cannot be phosphorylated18. Using a combination of biochemical, molecular and behavioral approaches, we found that phosphorylation of eIF4E is regulated by light and the circadian clock, and that it plays an important role in the SCN clock physiology by specifically promoting translation of Per1 and Per2 mRNAs.
RESULTS
A light and clock-regulated MAPK/MNK/p-eIF4E pathway
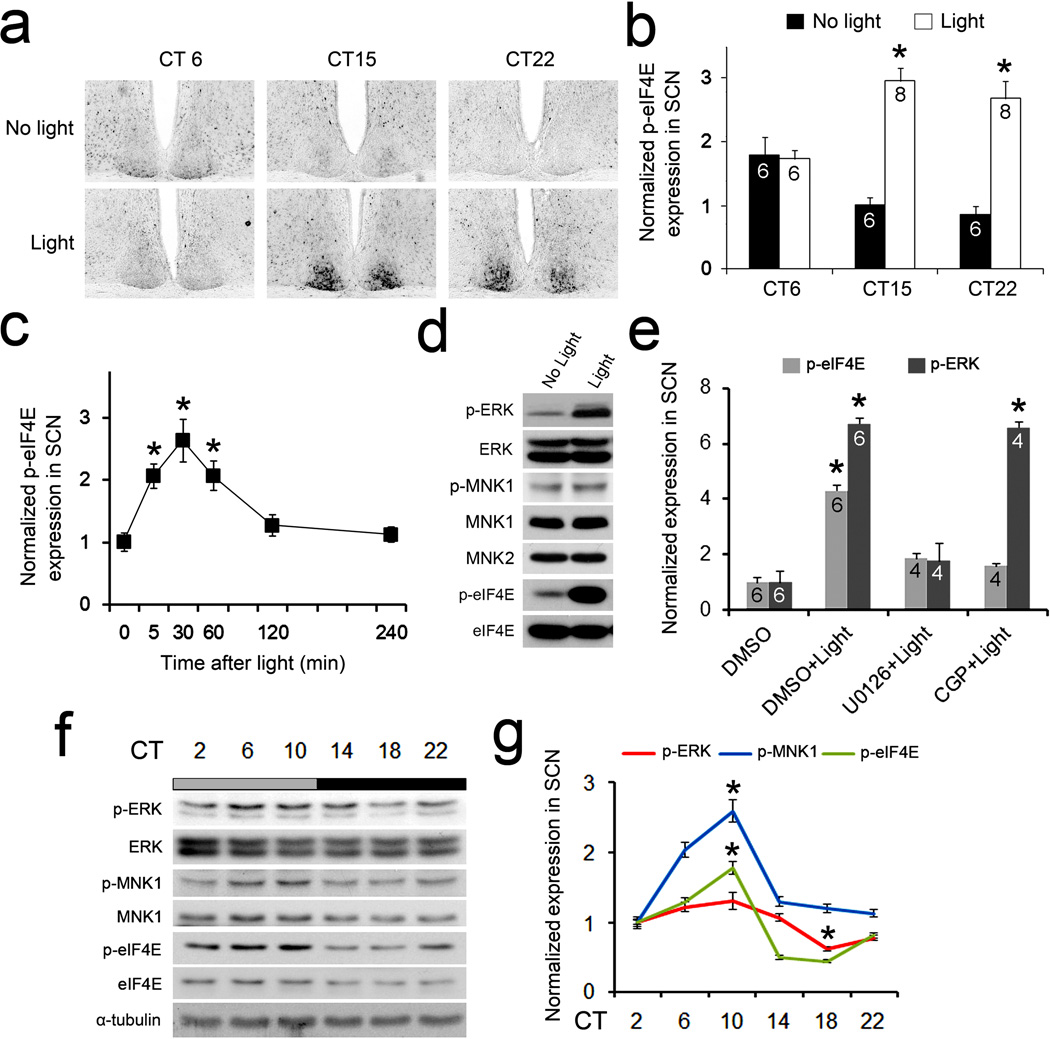
We first characterized the expression of phosphorylated eIF4E (at Ser209, p-eIF4E) by immunostaining. In mice kept under constant dark (DD) conditions, eIF4E was modestly phosphorylated in the SCN at circadian time (CT) 6, 15 and 22 (Fig.1a,b, No light). Strikingly, a light pulse (55 lx, 15 min) induced a ~3-fold increase of eIF4E phosphorylation at subjective night (CT 15 and CT 22), but not during the subjective day (CT 6) (Fig.1a,b, Light). The photic induction of eIF4E phosphorylation was rapid (within 30 min) and lasted for about 2 h (Fig.1c). These results demonstrate that light stimulates phosphorylation of eIF4E in the SCN and that the effect is phase-dependent, suggesting that eIF4E phosphorylation may be involved in photic entrainment of the circadian clock.
Figure 1. Light and circadian clock-regulated phosphorylation of eIF4E in the SCN.
(a) Representative microscopic images of coronal SCN sections show light-induced phosphorylation of eIF4E (at Ser209) in the SCN. Mice were exposed to light (55 lx, 15 min) during the day (CT6) or at night (CT15 and CT22) and sacrificed 30 min after light. Quantitation of intensities of p-eIF4E labeling is shown in (b). See “Methods” for the quantitation method.*P<0.05 vs. “No Light” (one-way ANOVA, F=2.97, P=0.044). (c) Time course of light-induced p-eIF4E expression in the SCN. A light pulse (55 lx, 15 min) was applied at CT15 and mice were sacrificed at indicated time points. *P<0.05 vs.0 min (one-way ANOVA, F=10.141, P=0.000). (d) Light-induced phosphorylation of ERK, MNK1 and eIF4E in the SCN. Western blots on SCN lysates before and 30 min after light exposure (55 lx, 15 min) at CT15 are shown. The blots of ERK, MNK1 and eIF4E were also shown as control. Full-length blots/gels are presented in Supplementary Figure 6. (e) Light-induced p-eIF4E and p-ERK expression in the SCN.MEK inhibitor U0126 (10 mM), MNK1 inhibitor CGP57380 (10 mM) or vehicle DMSO was infused to the lateral ventricle 30 min before light exposure. *P<0.05 vs. “DMSO” (one-way ANOVA, F=103.220, P=0.000). (f) Phosphorylation of ERK, MNK1 and eIF4E in the SCN over a 24-h period. Full-length blots/gels are presented in Supplementary Figure 6. (g) Phosphorylated proteins are quantified according to the levels of total proteins. *P<0.05 vs. CT2 (one-way ANOVA, F=18.964, P=0.000). The values are presented as the mean ± standard error of the mean (SEM).
eIF4E is phosphorylated by MNKs through the p38 and p42/44(ERK) MAPK signaling cascades16. Interestingly, previous studies have revealed a prominent role for the ERK MAPK pathway in synaptic plasticity and photic entrainment of the circadian clock19,20. We detected activities of the MAPK/MNK/eIF4E pathway by Western blotting for phosphorylated ERK (p-ERK, at Thr202/Tyr204), MNK1 (at Thr 250) and eIF4E (at Ser209) in the SCN. ERK, MNK1/2 and eIF4E were all abundantly expressed in the SCN (Fig.1d). Consistent with previous results19, a light pulse at night led to a marked increase of ERK phosphorylation in the SCN (Fig.1d). Light also induced a modest increase of MNK1 phosphorylation and a robust increase in the phosphorylation of eIF4E (Fig.1d and Supplementary Fig.1a,b). These results indicate that the MAPK/MNK pathway is activated by light in the SCN.
To investigate whether photic induction of eIF4E phosphorylation is dependent on activation of the ERK/MNK pathway, we infused the MEK inhibitor U012621 or the MNK inhibitor CGP5738022 into the SCN before light exposure. Light-induced p-ERK expression was blocked by U0126 but not by CGP57380 (Fig.1e). In contrast, photic induction of p-eIF4E was abolished by both U0126 and CGP57380, indicating that phosphorylation of eIF4E is downstream of sequential activation of ERK and MNKs. These results suggest that the ERK/MNK pathway couples light to the eIF4E phosphorylation.
We next studied whether activities of the MAPK/MNK/eIF4E pathway is under circadian control. Consistent with previous results19, we detected rhythmic ERK phosphorylation in the SCN (P<0.0001, bonferroni corrected RAIN, Fig.1f,g). Moreover, the levels of p-MNK1 and p-eIF4E also exhibited significant rhythmicity (P<0.0001, bonferroni corrected RAIN) with peaks at CT10 during a 24-h period (Fig.1f,g). The expression of p-MNK1 and p-eIF4E followed a temporal pattern similar to that of p-ERK. These data indicate that the circadian clock controls the activity of the MAPK/MNK1/eIF4E pathway. We also observed significant circadian day/night variations in the levels of MNK1 (P<0.01) and eIF4E (P<0.005) but not ERK (P=0.355, bonferroni corrected RAIN, Supplementary Fig.1c). The p-ERK and p-eIF4E exhibited similar temporal patterns when animals were placed in 12h/12h light/dark (LD) as compared to DD (Supplementary Fig.1d). Together, these results indicate that a light- and circadian clock-regulated MAPK/MNK pathway leads to eIF4E phosphorylation in the SCN.
eIF4E phosphorylation facilitates clock phase resetting
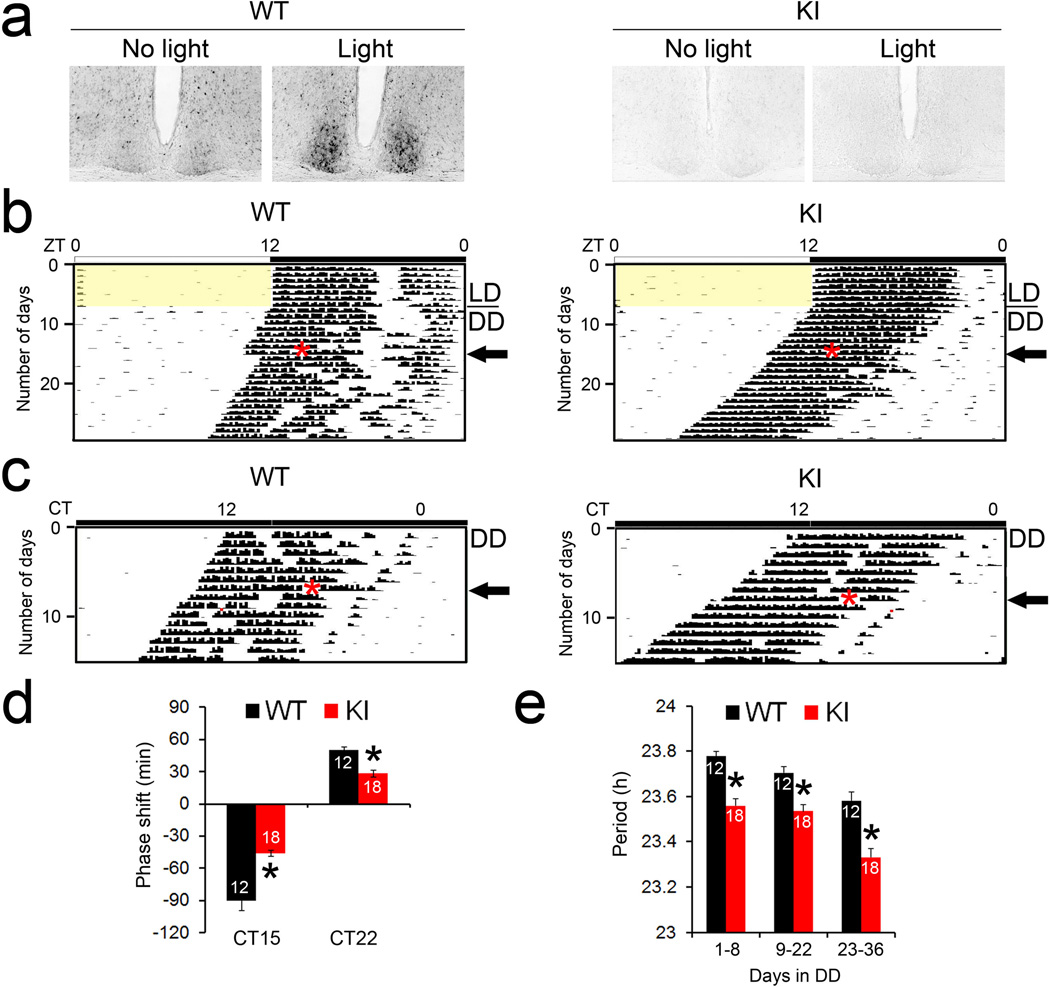
To further corroborate the function of eIF4E phosphorylation in the circadian clock, we utilized the non-phosphorylatable eIF4E KI mice. Importantly, the level of total eIF4E is unchanged in the mice and they do not exhibit any gross defects18. As expected, light-induced and basal phosphorylation of eIF4E was eliminated in the SCN of the KI mice (Fig. 2a). Next, we studied circadian behavior of the mice by monitoring their wheel-running activities. Mice were entrained to a 12h/12h LD cycle for 7 d and then transferred to DD. Both the KI mice and their WT littermates were entrained to the LD cycle and exhibited free-running rhythms in DD (Fig. 2b,c). On the eighth day in DD, mice were exposed to a light pulse (55 lx, 15 min) in the early night (CT15, 3 h after the onset of wheel-running behavior). As expected, the light pulse induced a phase delay in the onset of wheel-running in the WT mice. Significantly, however, light-induced phase delay was decreased by ~50% in the KI mice (P<0.05 vs. WT, Student’s t-test, Fig.2b,d). Fourteen days after the light pulse at CT15, a second light pulse (55 lx, 15 min) was applied to the same mice in the late night (CT22, 10 h after the onset of wheel running). At this time, light induced a modest phase advance in the WT mice. As was the case with the phase delay, the phase advance was attenuated by ~50% in the KI mice (P<0.05 vs. WT, Student’s t-test, Fig.2c,d).
Figure 2. Light-induced phase shift of circadian behavior is decreased in the eIF4E KI mice.
(a) Light-induced phosphorylation of eIF4E in the SCN. Entrained and dark-adapted mice were exposed to light (55 lx, 15 min) at CT15. Mice were sacrificed 30 min after light and SCN sections were immunolabeled for p-eIF4E. (b and c) Representative actograms of wheel-running behavior demonstrating light-induced circadian phase delay (b) and phase advance (c). For these experiments, animals were entrained to a 12 h/12 h light/dark (LD) cycle for 7 d and released into constant dark (DD) for 8 d before a light pulse (55 lx, 15 min) was applied at CT15 (top), as indicated by the red asterisk (b). 14 d after the first light pulse, a second light pulse (55 lx, 15 min) was applied at CT22 (bottom), as indicated by the red asterisk (c). The black arrows indicate the day when a light pulse was applied. Note that light at CT15 induced a significant phase delay (b) and light at CT22 induced a modest phase advance (c) in the WT mice (left). Compared to the WT mice, both the phase delay and the phase advance were decreased in the KI mice (right). (d) Quantitation of light-induced-phase shifts are shown. *P<0.05 vs WT (Student’s t-test, CT15: F=13.100, P=0.002; CT22: F=21.702, P=0.000). (e) Quantitation of circadian period in DD. The numbers of animals in each group are indicated on the histograms. *P<0.05 vs WT (Student’s t-test, D1-8: F=33.165, P=0.000; D9-22: F=17.843, P=0.000; D23–26: F=21.726, P=0.000). The values are presented as the mean ± standard error of the mean (SEM).
To determine whether the entrainment pathway upstream of eIF4E phosphorylation was compromised in the KI mice, we examined light-induced ERK phosphorylation and c-Fos expression, both of which are sensitive markers of photic input to the SCN. Light-induced ERK phosphorylation and c-Fos expression in the SCN were not different in the KI mice compared to the WT (Supplementary Fig. 2, P>0.05, ANOVA), indicating that the entraining pathway upstream of eIF4E is intact in these mice. Lastly, the free-running period of the KI mice was ~0.2 h shorter than the WT mice (P<0.05, ANOVA, Fig.2e). Together, these results demonstrate that phosphorylation of eIF4E facilitates photic phase resetting and modulates the period of circadian behavior.
eIF4E phosphorylation is critical for T-cycle entrainment
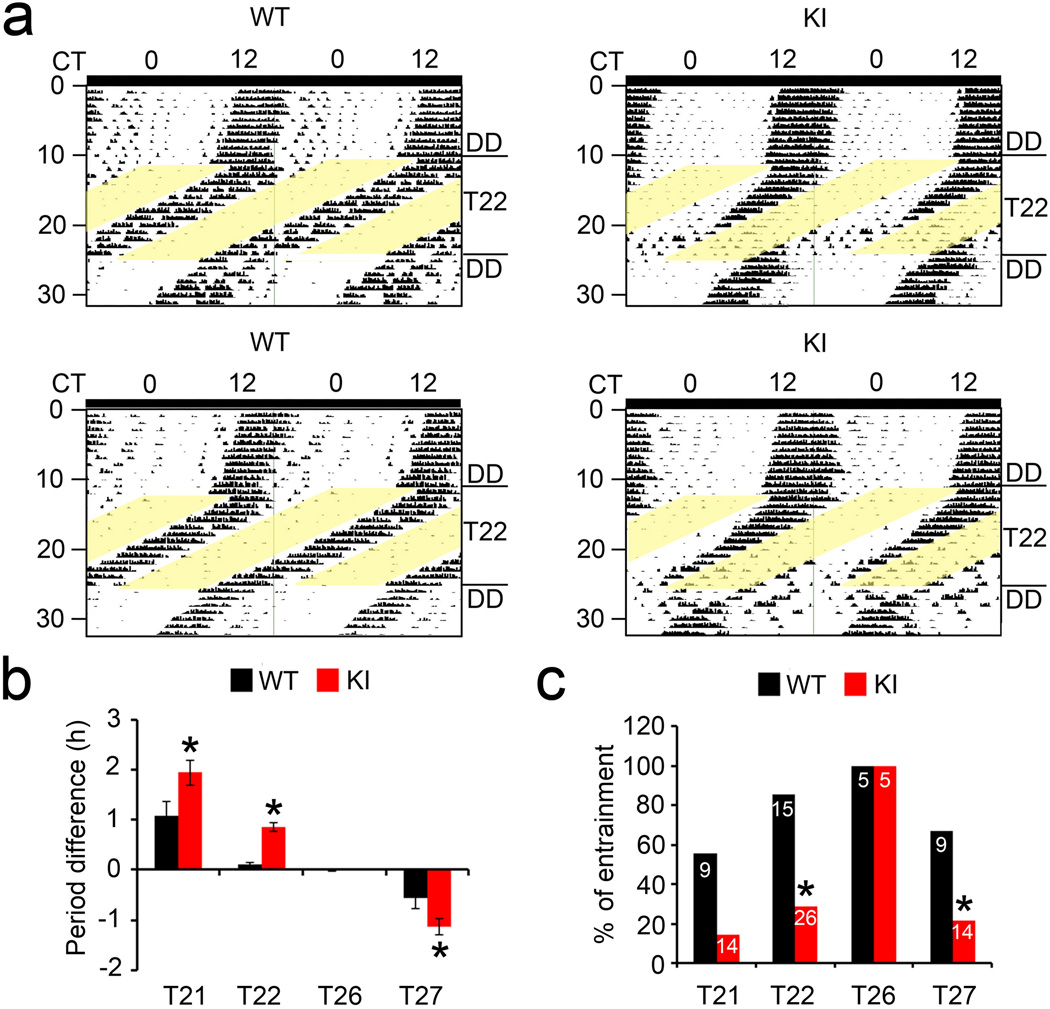
To further assess the role of eIF4E phosphorylation in photic phase resetting of the clock, we exposed the mice to various non-24 h LD cycles (T-cycles). As the endogenous period of mice is ~24 h, entrainment to a non-24-h T-cycle requires large daily phase shift of the circadian clock, and thus can be used as a rigorous test of the entraining capacity and range of a circadian clock23. Mice were first placed in LD or DD for 10 d and then transferred to a T21 (10.5 h/10.5 h LD), T22 (11 h/11 h LD), T26 (13 h/13 h LD) or T27 (13.5 h/13.5 h LD) cycle for at least 14 d. Whereas all mice retained circadian rhythmicity as assessed by X2 analysis in all lighting conditions, light entrainment was distinctly impaired. To assess this, the circadian period of the wheel-running rhythm was calculated in each T-cycle using activity onset in the first 5 d, and the difference between the period of the activity rhythm and the period of the T-cycle was determined. Representative actograms demonstrate that the WT mice were stably entrained to the T22 cycle whereas the KI mice failed to stably entrain. In the KI mice, the circadian rhythms of activity immediately detached from the imposed T22 cycle (Fig.3a).
Figure 3. Photic entrainment to non-24-h light/dark cycles (T-cycles) is impaired in the eIF4E KI mice.
(a) Representative actograms of wheel-running behavior demonstrating differential entrainment to a T22 cycle in two WT and two KI mice. Initially, animals were in DD for 11 d. A T22 (11h/11h LD) cycle started on the 12th d and lasted for 14 d. Animas were transferred to DD after the T-cycle for 7 d. (b) Quantitation of circadian periods of mice in various T-cycles. For these experiments, mice were first entrained in a 12h/12h LD for 9 d and put in DD for 10~14 d. Next, mice were transferred to various T-cycles (T21, T22, T26 and T27) for 14 d followed by 10 d in DD. To assess the efficacy of entrainment, the circadian period of the wheel-running rhythm was measured using the first 5d in each T-cycle (based on activity onset), and the difference between the period of the activity rhythm and the period of the T-cycle was determined. Positive values indicate that the animal activity period is longer than the T-cycle period and negative values indicate that the activity period is shorter than the T-cycle period. *P<0.05 vs WT (Student’s t-test, T21: F=5.448, P=0.03; T22: F=39.493, P=0.000; T26: F=1.389, P=0.272; T27: F=4.690, P=0.042). (c) The ratio of animals (%) entrained to various T-cycles. The numbers of animals used for each experiment are indicated on the histograms. *P<0.05 vs WT (X2 test, T21: X2=4.407, P=0.052; T22: X2=17.564, P=0.000; T26: X2=0, P=1.000; T27: X2=4.707, P=0.042). The values are presented as the mean ± standard error of the mean (SEM).
For T21, T26 and T27, representative actograms (Supplementary Fig.3) also show more stable entrainment in the WT mice as compared to the KI mice. On average, the difference in the period between the activity rhythm and the T-cycle was larger in the KI mice than in the WT mice (P<0.05, ANOVA, Fig.3b), indicating compromised entrainment in the KI animals. Thirteen out of fifteen WT mice were entrained to T22 by Day 14. In stark contrast, only 7 out of 26 KI mice were entrained to T22 (P<0.05 vs. WT, X2 test, Fig.3c). Five out of 9 WT mice were entrained to a T21 cycle whereas only 2 out of 14 KI mice were entrained (P=0.052, KI vs. WT, X2 test, Fig.3c, also Supplementary Fig.3). Similarly, 6 out of 9 WT mice were entrained to a T27 cycle, whereas only 3 out of 14 KI mice were entrained (P<0.05, KI vs. WT, X2 test, Fig.3c, also Supplementary Fig.3). However, all the WT and KI mice were entrained to T26 (P>0.05, KI vs. WT, X2 test, Fig.3c, also Supplementary Fig.3). As the range of T-cycles to which the KI mice can be entrained was narrowed, the limit of clock entrainment is decreased in these mice. These results further demonstrate that eIF4E phosphorylation is important for photic entrainment of the circadian clock.
eIF4E phosphorylation increases abundance of PER proteins
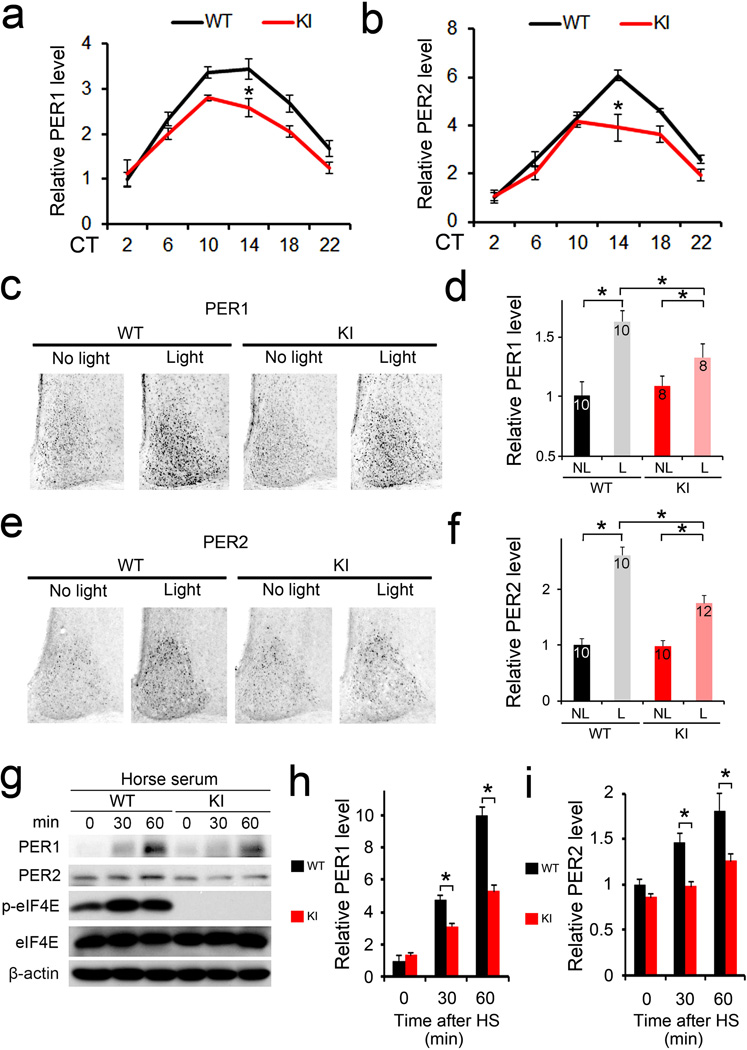
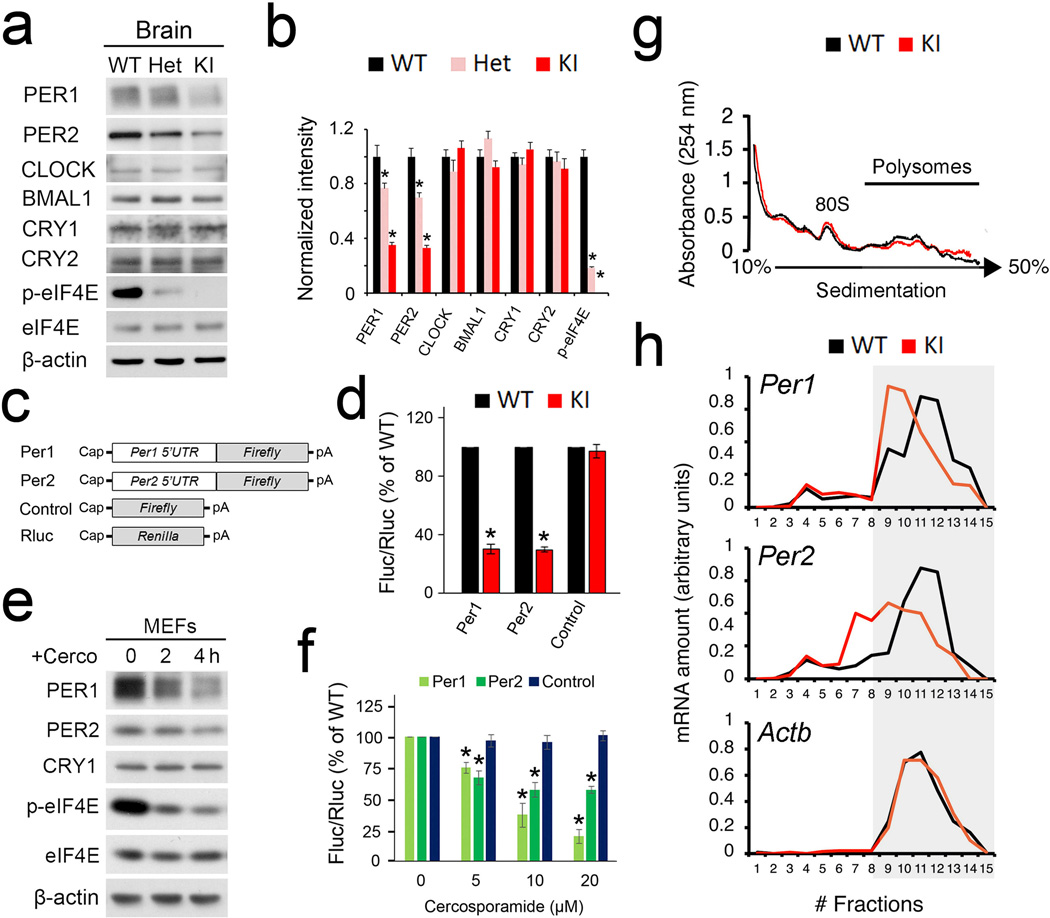
Per1 and Per2 are essential components of the autoregulatory negative feedback loop within the core clock machinery4. Furthermore, they are the only core clock genes that can be induced by light in the SCN8–10. It is thought that induction of Per gene expression underlies the clock resetting process10,24,25. Given the marked role of eIF4E phosphorylation in clock entrainment and a pivotal role of eIF4E in translation initiation, we reasoned that phosphorylation of eIF4E might regulate circadian entrainment through its control of PER protein synthesis. To address this question, we first determined the levels of PER1 and PER2 in the SCN by immunostaining using validated antibodies15,23,26. Consistent with previous reports26, the levels of PER1/2 proteins in the SCN exhibited circadian oscillations and reached a peak at around CT14 in both the WT and KI mice. Importantly, however, PER1/2 levels were significantly decreased in the SCN of KI mice compared to the levels in the WT mice (P<0.05 at CT14 vs. WT, ANOVA, Fig. 4a,b, also Supplementary Fig.4a,b). In a second experiment, a light pulse (15 min, 55 lux) given at CT15 induced a significant increase in PER1/2 at CT19 (4 h after the light pulse) in the WT mice (Fig.4c–f, Light vs. No light, P>0.05, ANOVA), consistent with published results27,28. Notably, this effect was reduced by ~50% in the KI mice (P<0.05 vs. WT, ANOVA).
Figure 4. Basal and inducible PER levels are decreased in the eIF4E KI tissue.
(a) and (b) Temporal profiles of PER1(a) and PER2(b) in the SCN over 24 h. Brains were harvested every 4 h throughout 24 h when mice were in DD and SCN sections were immunostained for PER1 or PER2. *P<0.05 vs WT (a: one-way ANOVA, F=22.698, n=6, P=0.000; b: one-way ANOVA, F=28.840, n=6, P=0.000) (c-f) Light-induced PER1 (c and d) and PER2 (e and f) expression in the SCN. Representative microscopic images of SCN immunostained for PER1 (c) or PER2 (e) are on the left. Quantitation of the staining for PER1 is shown in (d) and for PER2 is in (f). For these experiments, animals were entrained for 14 d and dark-adapted for 2 d. On the 3rd day in DD, mice were exposed to light (55 lx, 15 min) at CT15 and sacrificed at CT19. See “Methods” for the quantitation method. *P<0.05 vs WT (d: one-way ANOVA, F=8.049, P=0.000; f: one-way ANOVA, F=67.135, P=0.000). (g) Serum-induced PER expression in the mouse embryonic fibroblasts. 10% horse serum was added to the culture medium when the cells reached confluence. Representative Western blots are shown in (g). Full-length blots/gels are presented in Supplementary Figure 6. Quantitation of PER1 blots is in (h) and PER2 quantitation is in (i). Note that the induction of PER1 and PER2 were attenuated in the KI cells. *P<0.05 vs WT (h: one-way ANOVA, F=394.178, n=3, P=0.000; i: one-way ANOVA, F=36.576, n=3, P=0.000). The values are presented as the mean ± standard error of the mean (SEM).
Serum shock induces Per gene expression and is widely used to synchronize cellular clocks in vitro29. To test whether phosphorylation of eIF4E regulates PER induction in peripheral tissue, we used horse serum to induce PER proteins in mouse embryonic fibroblasts (MEFs). For Western blotting, we used validated anti-PER1/2 antibodies reported by Lee et al.30. We found that serum-stimulated expression of PER1 and PER2 proteins was attenuated by ~50% in the KI cells (P<0.05 vs. WT, ANOVA, Fig.4g–i), consistent with the decreased photic induction of PER in the SCN of KI mice (Fig.4c–f).
To determine the period of molecular PER oscillations, we crossed the KI mice with the PER2::Luciferase (PER2::LUC) reporter mice and monitored the PER2 bioluminescence rhythms of SCN explants as reported15. Consistent with the behavioral data, KI explants exhibited a shorter period (25.8±0.30 vs. 24.7±0.09, WT vs. KI, P<0.05, Student’s t-test, Supplementary Fig.4c, d, e). To complement the genetic approach, we applied the MNK inhibitor CGP57380 to the SCN explants. CGP57380 markedly decreased the amplitude of PER2::LUC rhythms by ~50% in WT explants but not in KI explants (P<0.05 vs. WT, Student’s t-test, Supplementary Fig. 4d, f), consistent with decreased PER levels in the SCN of KI mice (Fig. 4a,b). On the other hand, CGP57380 increased period length in both WT and KI explants (Supplementary Fig.4d,e), possibly due to its effects on other MNK targets independent of eIF4E phosphorylation16. Together, these results demonstrate that phosphorylation of eIF4E increases the abundance of basal and induced PER proteins, consistent with its role in facilitating circadian clock entrainment.
eIF4E phosphorylation promotes Period mRNA translation
To pursue the biochemical mechanisms by which eIF4E phosphorylation regulates PER abundance, we utilized forebrain lysates (because of the limited amount of material from the SCN). We first examined the levels of canonical clock proteins including PER1/2, CLOCK, BMAL1 and CRY1/2 in the brain by Western blotting. PER1/2 levels were decreased in the KI brain in a gene dosage-dependent manner, with the heterozygotes (Het) showing intermediate levels between WT and KI mice (PER1: F=130.360, P=0.000; PER2: F=135.16, P=0.000 vs. WT, n=3 for each group, one-way ANOVA, Fig. 5a,b). The levels of other clock proteins were not changed (CLOCK: F=4.154, P=0.074; BMAL1: F=4.160, P=0.074; CRY1: F=2.395, P=0.172; CRY2: F=0.979, P=0.429; p-eIF4E: F=511.440, P=0.000 vs. WT, n=3 for each group, one-way ANOVA, Fig. 5a,b), indicating specific regulation of PER levels by eIF4E phosphorylation. Notably, total eIF4E level was not changed in the KI brains (Fig.5a), which is consistent with unaltered global protein synthesis in the KI mice (Supplementary Fig.5a,b).
Figure 5. Phosphorylation of eIF4E promotes Period 1 and Period 2 mRNA translation.
(a) Representative Western blots of canonical clock proteins in the forebrain. Quantitation is shown in (b). *P<0.05 vs WT. (c) Schematic diagrams of the Firefly (Fluc) and Renilla (Rluc) luciferase reporter mRNAs. (d) Luciferase assay indicating translation levels of the reporter mRNAs in MEFs. Rluc mRNA was co-transfected with Fluc mRNAs as a transfection control. Error bars represent the standard error of mean of three independent experiments. *P<0.05 vs WT. (e) Western blots indicating that the MNK1 inhibitor cercosporamide decreased eIF4E phosphorylation and inhibited PER expression in MEFs. For (a) and (e), full-length blots/gels are presented in Supplementary Figure 6. (f) Cercosporamide inhibits translation of Per1 and Per2 5’UTR mRNA reporters in a dose dependent manner. *P<0.05 vs 0 µM. (g and h) Brain polysome profiling assay. (g) Polysome profiles from forebrain lysates. The positions of the 80S ribosome peaks and polysomes are indicated. Polysome to monosome ratios are shown to the right. (h) From top to bottom: on the left are qRT-PCR results of levels of Per1, Per2 and Actb mRNA extracted from the polysome fractions. Levels of total mRNAs from the brain are shown to the right. Note that for Per1 and Per2, a shift of mRNA distribution to the left (lighter gradient fractions) indicates decreased mRNA translation initiation in the KI brain. No differences were found in Per1, Per2 and Actb transcription between WT and KI mice. The values are presented as the mean ± standard error of the mean (SEM).
To investigate whether phosphorylation of eIF4E controls translation of Per1 and Per2 mRNAs through an mRNA 5’UTR-dependent mechanism, we created Per1 and Per2 mRNA 5’UTR fused to a luciferase reporter and transfected them into MEFs (Fig.5c). The translation activities of the Per1 and Per2 reporters were reduced by ~70% in the KI MEFs (Per1: F=294.000, P=0.000; Per2: F=291.84, P=0.000 vs. WT, n=3 for each group, Student’s t-test, Fig. 5d), which indicates that phosphorylation of eIF4E facilitate the translation of Per1 and Per2 5’UTR reporters. As control, expression of a capped mRNA luciferase reporter without a 5’UTR was not changed (F=0.521, P=0.510 vs.WT, n=3 for each group, Student’s t-test, Fig. 5d). These results demonstrate that the 5’UTRs are essential for control of Per mRNA translation by eIF4E phosphorylation. To complement the genetic approach, we used the MNK inhibitor cercosporamide31 to decrease eIF4E phosphorylation in WT MEFs. Cercosporamide inhibited phosphorylation of eIF4E and decreased the levels of PER1 and PER2 proteins in WT but not in the KI cells (Fig. 5e and Supplementary Fig.5c). Consistent with the Western data, translation of Per1 and Per2 mRNA 5’UTR reporters was suppressed by cercosporamide in a dose-dependent manner in WT but not in the KI cells (Per1: F=126.016, P=0.000; Per2: F=42.225, P=0.000; Control: F=1.091, P=0.407 vs. 0 µM, n=3 for each group, one-way ANOVA, Fig. 5f, also Supplementary Fig.5d).
Lastly, we performed polysome profiling to analyze mRNA translation of Per1 and Per2 in the brain. Forebrain lysates were fractionated by sucrose density gradient centrifugation. Actively translated mRNAs are associated with polysomes and sediment at the heavy-density fractions (Fig.5g). The polysome/monosome ratio was not different in the KI brains compared to that in the WT brains (KI vs. WT: 2.78±0.24 vs. 2.92±0.11, n=3, F=0.231, P=0.656, Student’s t-test), indicating that translation of most mRNAs is not affected. The amount of Per1, Per2 and Actb mRNAs in each gradient fraction was quantified by qRT-PCR, and the distribution of the mRNAs was compared between the WT and KI brains. In the KI brains, Per1 and Per2 mRNA distribution shifted towards the lighter-density fractions, as compared to WT brains (Fig.5h). Notably, the levels of total Per1 and Per2 mRNAs were similar (KI vs. WT: Per1: 0.32±0.06 vs.0.3±0.08, F=0.211, P=0.67; Per2: 0.21±0.11 vs.0.22±0.1, F=0.015, P=0.908; Actb: 0.65±0.1 vs.0.7±0.09, F=0.383, P=0.960, n=3 for each group, Student’s t-test), demonstrating repressed Per1 and Per2 mRNA translation in the KI brain and no detectable changes in transcription. The effect on Per1 and Per2 mRNA translation was specific, inasmuch as the distribution was not changed for mRNAs of Actb and other clock genes including Clock, Bmal1, Cry1 and Cry2 (Fig.5h and Supplementary Fig.5e). These data are consistent with decreased protein levels of PER1 and PER2, but not β-actin and other clock proteins in the brains of KI mice (Fig.5a,b). Together, these results demonstrate that phosphorylation of eIF4E specifically facilitates Per1 and Per2 gene expression by promoting their mRNA translation.
DISCUSSION
In the present study, we show that the abundance of the clock protein PER1 and PER2 is controlled at the mRNA translation initiation step through MAPK/MNK-dependent phosphorylation of the cap-binding protein, eIF4E. In the SCN, eIF4E phosphorylation is regulated by light and exhibits circadian rhythmicity under constant conditions. Thus, during the clock resetting process, when upregulation of PER proteins is required, eIF4E phosphorylation may function as a trigger to boost Per mRNA translation and increase PER protein levels. Under constant conditions, rhythmic phosphorylation of eIF4E may contribute to the precision of the circadian clock through timely modulation of PER protein synthesis.
The SCN clock is continuously entrained by the external light/dark cycle and light-induced gene expression is thought to be the molecular basis of resetting circadian behavior5. Mechanistically, light stimulates melanopsin-expressing retinal ganglion cells and evokes neurotransmitter release at the terminals of retinohypothlamic tract6,7. The neurotransmitters bind to the postsynaptic receptors and activate second messenger-mediated signal transduction cascades in the SCN neurons. These pathways ultimately impinge on the core clock machinery and regulate clock gene expression7. The level of PER proteins is a critical state variable of the primary autoregulatory feedback loop that generates circadian oscillations. An increase in the PER level disrupts the dynamic balance of the feedback loops and leads to phase resetting of the SCN clock7.
Light regulates Per gene expression in the SCN through multiple mechanisms. Previous studies have focused mostly on inducible gene transcription. For example, the transcription factor cAMP-response element-binding protein (CREB) is a key nexus whereby multiple intracellular signal transduction pathways converge11,32,33. CREB is activated by phosphorylation34 and in turn binds to cAMP-responsive elements (CREs) to promote CRE-mediated gene expression. Importantly, CREs have been identified in the promoter regions of Per genes12. CREB-dependent transcription is also regulated by the CREB coactivator CRTC (CREB-regulated transcription coactivator), also known as TORC (transducer of regulated CREB)35,36.
Much less is known on the mechanisms of post-transcriptional regulation in the clock resetting process. Recent work has revealed a role for MAPK signaling mediated microRNA expression, which modulates Per1 gene transcription and Per2 protein stability37,38. Also, light activates the mTOR pathway28, which in turn activates S6Ks and inhibits 4E-BPs. We have recently reported that 4E-BP1 regulates SCN network synchrony by repressing Vip mRNA translation in the SCN15. Here we show that as a downstream kinase of the ERK MAPK pathway, light-activated MNK phosphorylates eIF4E, promotes Per1 and Per2 mRNA translation and thereby facilitates photic entrainment of the circadian clock. Phosphorylation of eIF4E is an integral part of the molecular entrainment pathway that couples light to mRNA translation in the core clock machinery in the SCN. The mechanism enables the SCN to respond sufficiently to a phase-resetting light pulse at night and to entrain to a wide range of non-24-h light cycles, which might represent an evolutionary advantage.
Under clock free-running conditions, Per gene expression is also controlled at multiple levels. The production and degradation rate of PER proteins determine the length of a circadian cycle4. At the transcriptional level, rhythmic Per transcription is driven by the CLOCK: BMAL1 complexes through the E-box enhancers4. At the posttranscriptional level, Per mRNA processing is regulated by methylation39. PER cycling is also governed by post-translational modifications such as phosphorylation30,40. Recent work has highlighted the important role of translational control for clock gene expression41–45. For example, the RNA binding proteins Ataxin-2 (Atx2) and Twenty-four (Tyf) interact to activate Per translation in pacemaker neurons to sustain robust behavioral rhythms in Drosophila44–46. Here we show that Per1 and Per2 mRNA translation is controlled by rhythmic phosphorylation of the cap-binding protein eIF4E in mammals. All these mechanisms contribute to dynamic regulation of the PER level and precise timekeeping of the circadian clock.
The mRNA cap-binding protein eIF4E serves as a pivotal node for regulation of protein synthesis in the mammalian circadian clock by several signaling pathways. The MAPK and mTORC1 pathways converge on eIF4E to regulate circadian clock function by controlling protein synthesis. In our previous work15, we show that rhythmic mTORC1 regulated eIF4E activity by phosphorylating and inhibiting the eIF4E repressor protein 4E-BP1. 4E-BP1 represses the translation of Vip mRNA and decreases the abundance of Vip. Thus, mTORC1 promotes Vip translation and facilitates entrainment and synchrony of SCN cell network. In the current study, we show that MAPK pathway leads to eIF4E phosphorylation through MNKs in the photo-recipient SCN cells and facilitates light-induced PER protein synthesis. Also circadian clock-regulated eIF4E phosphorylation promotes basal PER protein synthesis. Thus, the highly regulated eIF4E activity is important for timely protein synthesis and proper function of the circadian clock.
METHODS
Animals
The eIF4ES209A/S209A (KI) mice and the littermates on a C57BL/6 background15 were maintained in the animal facility at McGill University in accordance with institutional guidelines. All procedures were approved by the Institutional Animal Care and Use Committee at McGill University.
Photic stimulation
Adult (8~10-week-old) C57BL/6 mice were entrained to a 12 h LD cycle for at least 2 weeks and then transferred to constant darkness for two consecutive 24 h cycles. After dark adaptation, animals received a light pulse (55 lux, 15 min) at one of three time points: the middle of the subjective day (CT 6), early subjective night (CT 15), or late subjective night (CT 22). CTs were calculated based on Zeitgeber time (ZT) and the tau of the mice (approximately 23 h 45 min) under free running conditions, with ZT 0 denoting light on and ZT 12 denoting light off. The “no light” control animal groups underwent the same handling conditions at the same time points.
Cannulation and infusion
Mice were cannulated in the lateral ventricles using the techniques as described28. The following coordinates were used to place the tip of a 24-gauge guide cannula into the lateral ventricle: posterior, 0.34 mm from bregma; lateral, 0.90 mm from the midline; dorsoventral, −2.15 mm from bregma. To disrupt MEK activity, U0126 (10 mM, 2 μl; Cell Signaling Tech., MA) was infused through the cannula. To disrupt MNK1 activity, CGP57380 (30 mM, 2 μl; Cell Signaling Tech., MA) was infused. Control animals were infused with the same volume of DMSO.
Brain tissue processing, immunostaining, and microscopic imaging analysis
Under indicated conditions, mice were sacrificed and brain tissue was harvested and processed. SCN coronal sections were immunostained for p-eIF4E, p-ERK, PER1 or PER2 as described28.The antibody information is shown in Supplementary Table 1. Bright-field microscopy images were captured using a digital camera mounted on an inverted Zeiss microscope (Oberkochen, Germany). The staining intensity in micrographs was quantified using Adobe Photoshop software (Adobe Systems Incorporated, San Jose, CA) as described12. Briefly, images of the SCN were acquired (10 ×), digitally outlined, and the mean pixel values were determined. A digital oval (150×200 pixels) was placed on the adjacent lateral hypothalamus, and the mean value was subtracted from the value of the SCN signal to generate a normalized SCN intensity value. Notably, the lateral hypothalamic immunolabeling values were not altered with treatment conditions. For all experiments, the data were averaged from three central SCN sections per animal, and these values were pooled to generate a mean value for each group. The lowest value in an experiment is normalized to be “1”.
Circadian behavioral assay
Eight- to 10-week-old male KI mice or WT littermates were individually housed in cages equipped with running wheels. Wheel rotation was recorded using the VitalView program (Mini Mitter, Bend, USA)20. For the light pulse experiments, mice were entrained to a 12 h/12 h LD cycle (55 lx) for 7 d and released into constant dark (DD) for 8 d. On the 9th day in DD a light pulse (15 min, 55 lx) was applied at CT15. 14 d after the first light pulse, a second light pulse (15 min, 55 lx) was applied at CT22. Animals were kept in DD for another 14 d after the second light pulse. The CTs were calculated for each mouse based on their individual actograms prior to the light pulse.
For the T-cycle experiments, mice were first entrained in a 12h/12h LD for 9 d and put in DD for 14 d. Next, mice were transferred to T21 (10.5h/10.5h LD), T22 (11h/11h LD), T26 (13h/13h LD) or T27 (13.5 h/13.5 h LD) for 14 d followed by 10 d in DD. The actograms of wheel-running activities were analyzed using ActiView (Mini Mitter, Bend, USA) and ClockLab software (Actimetrics, Wilmette, USA). Circadian period is determined by the chi-square periodogram using the ClockLab software. For unstably entrained animals, period is determined from the actograms of the first 5 d in a T cycle. Stable entrainment is defined if animals show a consistent phase angle of entrainment (the time gap between light offset and activity onset) for at least five consecutive days.
Explant culture, kinetic bioluminescence recording and data analysis
The eIF4E KI mice were crossed with mPER2::LUC transgenic reporter mice to obtain the eIF4E KI: mPER2::LUC mice. Explants of WT and KI SCN tissue were dissected and cultured as reported12. CGP57380 (10µM) was applied to the explants without changing the medium. Real-time circadian reporter assays were performed using a Lumicycle luminometer (Actimetrics, Inc.) as described12. Baseline-subtracted data (counts/sec) were plotted against time (days) in culture. For comparison, the first peak was aligned in the plotted data. The LumiCycle Analysis program (version2.31, Actimetrics, Inc.) was used to analyze rhythm parameters. For period length analysis, raw data were baseline fitted, and the baseline-subtracted data were fitted to a sine wave (damped), from which the period was determined. All samples showed persistent rhythms and goodness-of-fit of >90% was achieved. For amplitude analysis, baseline-subtracted data (polynomial order = 1; 3~6 days of recording data) were fitted to a sine wave, from which the amplitude was determined using Sin Fit.
Protein extraction and Western blotting analysis
The SCN tissue was excised under a dissection microscope and frozen on dry ice. SCN tissue was pooled from 10 mice per condition. Brain tissue was homogenized with a pestle grinder (Fisher Scientific Limited, Nepean, Canada) and lysed. Western blotting analysis was performed as described12. The antibody information is shown in Table 1.
Preparation of the 5’UTR reporter mRNAs and luciferase assay
PCR products encoding a T7 promoter followed by the 5’UTR of mouse Per1 or Per2 were generated using a cDNA library derived from mouse embryonic fibroblasts (MEFs), and inserted into pGAL3-Basic vector (Promega) between HindIII and NcoI sites. Resultant plasmids (pGAL3-Per1-5’UTR and pGAL3-Per2-5’UTR) were used as templates for in vitro transcription. Luciferase reporter mRNAs were generated using MAXIscript T7 in vitro Transcription kit (Ambion) according to the manufacturer’s protocol in the presence of the cap analog, and Poly (A) tail was added using the Poly (A) Tailing Kit (Ambion). WT and KI MEFs were seeded in a 24-well-plate and maintained at 37°C in 5% of CO2 atmosphere in Dulbecco' modified Eagle medium (DMEM), supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were transfected with firefly luciferase reporter mRNAs together with renilla luciferase mRNA as transfection control using Lipofectamine (Invitrogen). Different doses of cercosporamide (Sigma-Aldrich) were added to the culture medium after transfection. Cells were incubated and lysed for luciferase activity assay using Dual-Luciferase Reporter Assay System (Promega) according to the manufacture’s instruction.
Brain polysome profiling and real-time quantitative reverse transcription-PCR (qRT-PCR)
Brain polysome profiling was performed as described12. The polysome to monosome ratio was calculated as the area under the A254 absorbance curve, using the absorbance values processed with the definite integral command in MATLAB. RNA extraction and qRT-PCR were performed as reported12. The primer information is shown in Supplementary Table 2.
Statistical analysis
The data were collected and processed randomly. Data collection and analysis were not performed blind to the conditions of the experiments. The values are presented as the mean ± standard error of the mean (SEM) or percentage (%). Data distribution was assumed to be normal but this was not formally tested. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications11,37,39,40. Statistical analysis was performed using SPSS software (SPSS Inc, Chicago, IL). Mean values from multiple groups were compared via one-way ANOVA, followed by the Student-Newman-Keuls test for individual comparisons. Mean values from two groups were compared via Student’s t-test. Ratios of entrainment (%) of WT and KI mice were compared via the non-parametric X2 test. Circadian rhythmicity of protein abundance was determined using Rhythmicity Analysis Incorporating Non-parametric methods47(RAIN) performed in R (v3.1.1, R Foundation for Statistical Computing, Vienna, Austria) and bonferroni correction was applied. P < 0.05 was considered as statistically significant. A supplementary methods checklist is available.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Weaver, Isaac Edery, and Jane Stewart for advice and critical reading of the manuscript and Chadi Zakaria, Nathaniel Robichaud, Annie Sylvestre, Barry Robinson and Isabelle Harvey for excellent technical assistance. We are indebted to David Weaver for his gift of the anti-PER1 serum and Joseph Takahashi for his gift of the mPER2::LUC transgenic mice. This work was supported by Canadian Institute of Health Research (CIHR) Grants (MOP 114994 to N.S. and MOP 13625 to S.A.) and a National Institutes of Health Grant (NINDS R01NS054794 to A.L.). N.S. is a senior international research scholar of the Howard Hughes Medical Institute (HHMI). R.C. and N.Z. are recipients of the Fonds de recherche du Québec-Santé (FRQS) Postdoctoral Award and R.C. is a recipient of the Banting Postdoctoral Fellowship.
Footnotes
AUTHOR CONTRIBUTIONS: R.C.,S.A. and N.S. designed research; R.C.,N.Z.,C.G.G.,I.D.B., Y.T.,A.Y., and H.X. performed research; C.L. and K.S. contributed reagents/analytic tools; R.C.,I.D.B.,C.G.G.,A.Y. and H.X. analyzed data; R.C.,A.L.,S.A. and N.S. wrote the manuscript.
References
- 1.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert S, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- 6.Peirson S, Foster RG. Melanopsin: another way of signaling light. Neuron. 2006;49:331–339. doi: 10.1016/j.neuron.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Golombek D, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 8.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 9.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 10.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 11.Cao R, Butcher GQ, Karelina K, Arthur JS, Obrietan K. Mitogen- and stress-activated protein kinase 1 modulates photic entrainment of the suprachiasmatic circadian clock. Eur J Neurosci. 2013;37:130–140. doi: 10.1111/ejn.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 15.Cao R, et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79:712–724. doi: 10.1016/j.neuron.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases) Front Biosci. 2008;13:5359–5373. doi: 10.2741/3086. [DOI] [PubMed] [Google Scholar]
- 17.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furic L, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 20.Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 21.Favata MF, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 22.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21:5500–5511. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbour VL, Robinson B, Amir S. Variations in daily expression of the circadian clock protein, PER2, in the rat limbic forebrain during stable entrainment to a long light cycle. J Mol Neurosci. 2011;45:154–161. doi: 10.1007/s12031-010-9469-z. [DOI] [PubMed] [Google Scholar]
- 24.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 26.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, Silver R. Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur J Neurosci. 2004;19:1105–1109. doi: 10.1111/j.1460-9568.2004.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J Neurosci. 2010;30:6302–6314. doi: 10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 31.Konicek BW, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 2011;71:1849–1857. doi: 10.1158/0008-5472.CAN-10-3298. [DOI] [PubMed] [Google Scholar]
- 32.Tischkau SA MJ, Tyan SH, Buchanan GF, Gillette MU. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem. 2003;278:718–723. doi: 10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- 33.Gamble KL, Allen GC, Zhou T, McMahon DG. Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J Neurosci. 2007;27:12078–12087. doi: 10.1523/JNEUROSCI.1109-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 35.Jagannath A, et al. The CRTC1-SIK1 pathway regulates entrainment of the circadian clock. Cell. 2013;154:1100–1111. doi: 10.1016/j.cell.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakamoto K, et al. Clock and light regulation of the CREB coactivator CRTC1 in the suprachiasmatic circadian clock. J Neurosci. 2013;33:9021–9027. doi: 10.1523/JNEUROSCI.4202-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng H, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Saavedra M, et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–51. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Meng QJ, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima S, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morf J, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 43.Bradley S, Narayanan S, Rosbash M. NAT1/DAP5/p97 and Atypical Translational Control in the Drosophila Circadian Oscillator. Genetics. 2012 doi: 10.1534/genetics.112.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim C, et al. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Ling J, Yuan C, Dubruille R, Emery P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science. 2013;340:879–882. doi: 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.