Abstract
Background
Gastroesophageal reflux can cause high acidity in the esophagus and trigger heartburn and pain. However, because of the esophageal mucosal barrier, the acidity at the nerve terminals of pain-mediating C-fibers in esophageal mucosa is predicted to be substantially lower. We hypothesized that the esophageal DRG C-fibers are activated by mild acid (compared to acidic reflux), and express receptors and ion channels highly sensitive to acid.
Methods
Extracellular single unit recordings of activity originating in esophageal DRG C-fiber nerve terminals were performed in the innervated esophagus preparation ex vivo. Acid was delivered in a manner that bypassed the esophageal mucosal barrier. The expression of mRNA for selected receptors in esophagus-specific DRG neurons was evaluated using single cell RT-PCR.
Key Results
Mild acid (pH=6.5–5.5) activated esophageal DRG C-fibers in a pH-dependent manner. The response to mild acid at pH=6 was not affected by the TRPV1 selective antagonist iodo-resiniferatoxin. The majority (70–95%) of esophageal DRG C-fiber neurons (TRPV1-positive) expressed mRNA for acid sensing ion channels (ASIC1a, ASIC1b, ASIC2b and/or ASIC3), two-pore-domain (K2P) potassium channel TASK1, and the proton-sensing G-protein coupled receptor OGR1. Other evaluated targets (PKD2L1, TRPV4, TASK3, TALK1, G2A, GPR4 and TDAG8) were expressed rarely.
Conclusions & Inferences
Guinea pig esophageal DRG C-fibers are activated by mild acid via a TRPV1-independent mechanism, and express mRNA for several receptors and ion channels highly sensitive to acid. The high acid sensitivity of esophageal C-fibers may contribute to heartburn and pain in conditions of reduced mucosal barrier function.
Keywords: esophagus, acid sensing, nociceptor, pain
INTRODUCTION
Afferent nerves with their neurons in the spinal dorsal root ganglia (DRG) are considered to be the main pathway mediating pain, heartburn and other esophageal sensations(1). Acid refluxing from the stomach is arguably the most important noxious mediator initiating painful sensations from the esophagus (2). However, the mechanisms underlying acid sensing in the nerve terminals of esophageal spinal DRG C-fibers are incompletely understood.
The esophageal mucosa is highly resistant to acid. Experimental data show that strong acid (pH=1) in the esophageal lumen does not appreciably change the pH in the healthy mucosa as measured by interstitial pH in the esophageal basal epithelial layer(3). Indeed, in healthy subjects acid infusion is only weakly effective in inducing heartburn or pain (4–6). In patients with gastroesophageal reflux diseases (GERD) only a moderate reduction in esophageal mucosal resistance compared to healthy subjects has been reported (5–8). Yet in GERD patients acidic reflux or esophageal infusion of acid causes sensations of heartburn and pain. This raises the hypothesis that esophageal DRG C-fibers have mechanisms for activation by mild acid. At present the acid sensitivity of esophageal DRG C-fibers is unknown.
The capsaicin receptor TRPV1 is expressed by most C-fibers and can be activated by acid (9). In fact, TRPV1 is frequently considered to be the primary acid transducer in visceral nociceptors(10). Nonetheless, a relatively strong acidity (pH≤5.0, compared to pH>=7.3 in extracellular fluid) is required to fully activate TRPV1 (9, 10). On the other hand, a number of receptors and ion channels can be activated or modulated by relatively mild acid (pH=6–7, compared to pH>=7.3 in extracellular fluid) in a manner that leads to sensory activation. These receptors and ion channels include acid sensing ion channels (ASICs), certain two-pore-domain (K2P) potassium channels, certain TRP channels, and proton-sensing G-protein coupled receptors(10–12). At present it is not known whether esophageal DRG C-fibers express such receptors and ion channels. Here we addressed the hypothesis that esophageal DRG C-fibers express receptors and ion channels other than TRPV1 that are highly sensitive to acid. Moreover, we hypothesized that the non-TRPV1 mechanisms may be primarily responsible for activation of esophageal DRG C-fibers induced by mild acid (pH=6.0).
MATERIALS AND METHODS
The experiments described in this study were approved by the Johns Hopkins Animal Use and Care Committee.
Extracellular recordings from DRG neurons projecting into the esophagus were performed as described previously (13, 14). Single fiber recordings of nerve activity originating in esophageal DRG C-fiber terminals were performed in the isolated superfused ex vivo spinally-innervated guinea pig esophagus preparation. Esophagus with adjacent tissue (at the level of spinal ganglia approximately C8-Th5) that included a portion of left sympathetic trunk, and left spinal T1–T4 DRG ganglia were carefully dissected. Caution was made to preserve spinal afferent nerve pathways. The esophagus was secured dorsal side up in the tissue chamber. The DRG ganglia with short portions of their spinal nerves were pulled through a small hole into separately-perfused Sylgard-lined recording chamber and pinned. The hole was then sealed with vaseline. The tissue and recording chambers were separately superfused (4–6 ml/min) with Krebs solution (118mM NaCl, 5,4mM KCl, 1mM NaH2PO4, 1.2mM MgSO4, 1.9 mM CaCl2, 25 mM NaHCO3, 11mM dextrose, gassed with 95%O2/5%CO2, pH=7.4, 35°C) containing indomethacin (3 μM) and atropin (1μM). The silver/silver chloride return electrode and earth pellet were placed in the recording chamber. The aluminosilicate glass microelectrode (2 MΩ) filled with 3M sodium chloride was micromanipulated into the T2 or T3 DRGganglion. The recorded signal was amplified (Microelectrode AC amplifier 1800, A-M Systems) and filtered (low cut-off, 0.3 kHz; high cut-off, 1 kHz) and analyzed on Apple computer using the software TheNerveOfIt (sampling frequency 33 kHz; PHOCIS, Baltimore, MD, US). The dorsal surface of the entire esophagus was systematically searched with a concentric stimulation electrode delivering 90V pulses (duration 1ms, frequency 1–2Hz, stimulator model 215/I, Hugo Sachs Electronik, March-Hugstetten, Germany and stimulus isolation unit SIU5, Grass Instruments, West Warwick, RI). If the electrical pulse evoked an action potential the mechanosensitive receptive field was searched and identified by using focal mechanical compression and von Frey probes.
Acidic solutions were prepared by replacing bicarbonate in the Krebs solution by HEPES and adding sodium D-gluconate to maintain sodium concentration and osmolarity (in mM: 118 NaCl, 5.4 KCl, 1 NaH2PO4, 1.2 MgSO4, 1.9 CaCl2, 20 HEPES, and 11.1 dextrose, 18 Na D-gluconate). The pH was adjusted to desired value (acidic solutions 5.5, 6.0, 6.5 or control solution 7.4) by NaOH (1M) or HCl (1M). The tissue was continuously superfused with Krebs solution. In order to bypass the mucosal barrier the tissue was exposed to acid in superfusing fluid via serosal surface. This approach has been show to effectively alter the pH in the mucosa (3). The pH was measured in samples from superfusing fluid. Only one acidic solution per fiber was studied. In experiments with I-RTX, the tissue was superfused with the following solutions containing either I-RTX (1μM) or vehicle (DMSO 0.01%): Krebs solution (15 min), control solution (pH=7.4, 15 min), acidic solution (pH=6.0, 15min), Krebs solution (15 min) and Krebs solution containing capsaicin (0.1μM, 15 min). The change of solutions was rapid (completed within ≈10s).
Nerve activity (action potential discharge) was monitored continuously. The sustained aspect of activation evoked by acid and capsaicin was quantified as maximum 60s bin discharge defined as the maximal number of action potentials recorded in any 60s interval during the superfusion with the acid or capsaicin. The peak activation was quantified as peak frequency (Hz) defined as the maximal number of action potentials recorded in any 1s interval. Unpaired t-tests was used as appropriate, and the significance was defined as P< 0.05.
Retrograde labeling of the afferent neurons projecting into the guinea pig esophagus by DiI (0.1% in 10% DMSO in sterile saline) was performed as described previously (13, 15, 16). Spinal T2 and T3 DRG ganglia were harvested 10–15 days after the injections. Single cell RT-PCR was performed on individual neurons as described previously (17–19). The sensory ganglia were dissected, enzymatically dissociated, and the neurons were plated on poly-D-lysine/laminin-coated coverslips. Coverslips with dissociated neurons were perfused with cold PBS, and the DiI-labeled neurons identified under fluorescent microscope were individually harvested by glass-pipette into separate PCR tubes, immediately frozen and stored at −80°C. Only the neurons free of debris or attached cells were collected. Samples of bath solution were collected for no-template (bath) controls. RT-PCR: First strand cDNA was synthesized from single neurons by using the Super-Script(tm) III CellsDirect cDNA Synthesis System (Life Technologies) according to the manufacturer’s recommendations (combination of poly(dT) and random hexamer primers was used). 2μl of each sample were used for PCR amplification by the HotStar Taq Poymerase Kit (Qiagen) according to the manufacturer’s recommendations (final volume 20μl). PCR: activation 95°C/15min, 50 cycles of denaturation 94°C/ 30s, annealing 60°C/30s and extension 72°C /60s, final extension 72°C/10min. Custom-synthesized primers (Life Technologies) were used (Tab. 1). Products were visualized in ethidum-bromide stained 1.5% agarose gels. The figures were constructed by using Microsoft PowerPoint and Apple Preview. Controls. The primers were designed by using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (20) based guinea pig sequences and verified by using the guinea pig genome (http://genome.ucsc.edu/(21)). The ASCI primers have been evaluated previously (see Fig. 1 in (19)). Table 1 shows that most primers used in the study were intron-spanning. For most primers no genomic product can be amplified because its predicted size >1,000 bp is not achievable with the extension time of 30s used for PCR. For several targets (OGR1, G2A, GPR4, TDAG8 and TASK1) it was not possible to design intron-spanning primers because the available coding sequence was located on a single exon (Tab. 1). In these instances it is theoretically possible that the primers would amplify genomic DNA if the treatment with DNAse during the sample processing was insufficient. Nonetheless, collectively G2A, GPR4 and TDAG8 were detected only in 15% (9/64) of samples arguing against significant amplification of genomic DNA in our experiments. Furthermore, in 6 samples of DRG processed as RT- control (without adding reverse transcriptase) no OGR1 was detected. No product was amplified from negative (bath) controls (n=39) indicating negligible probability of false positive results. In our previous studies we have also reported negligible false positive rates (<1%, n=640 negative controls) (18, 19).
Table 1.
The primer sequences for single cell RT-PCR
| Target | Primer | Sequence (5′ to 3′) | Product size (bp) | Genomic size (bp) | Sequence |
|---|---|---|---|---|---|
| β-actin | Forward | TGGCTACAGTTTCACCACCA | 212 | 212 | NM_001172909.1 |
| Reverse | GGAAGGAGGGCTGGAAGA | ||||
| TRPV1 | Forward | CCAACAAGAAGGGGTTCACA | 168 | >1,000 | NM_001172652.1 |
| Reverse | ACAGGTCATAGAGCGAGGAG | ||||
| ASIC1 | Forward | CAGGAAGTGCCAGAAAGAGG | 151 | 286 | XM_00347578 |
| Reverse | CGGATGGTGAGGTAGGATGT | XM_00500647 | |||
| ASIC1a | Forward | GCAGGACACGACATACGAGA | 376 | >1,000 | XM_00347578 |
| Reverse | GCGGAGGCAGGTAGATGAG | ||||
| ASIC1b | Forward | GTGCCCCTTGCTCCTCCT | 240 | >1,000 | XM_00500647 |
| Reverse | ATTGCCTGTCCCACCCTTC | ||||
| ASIC2 | Forward | GGAGTTGGGCTTTGGGGTAG | 382 | >1,000 | XM_003469484 |
| Reverse | CTTGGCTGATGTTTTGCTTGG | ||||
| ASIC2a | Forward | GTTCAAGGGGCAGGAGTGT | 338 | >1,000 | XM_003469484* |
| Reverse | GTGGGGGCAGGTATGTGAG | ||||
| ASIC2b | Forward | GCTGCTCTCCTGCAAATACC | 293 | >1,000 | XM_003469484 |
| Reverse | CTACCCCAAAGCCCAACTC | ||||
| ASIC3 | Forward | CGGATGGGACAGTGCTACA | 260 | 485 | XM_003469663.2 |
| Reverse | TGGCAGGACACAAAAGTCTG | XM_005003488.1 | |||
| OGR1 (GPR68) | Forward | CTACATCAGCGTGGGCTTC | 200 | 200 | XM_005004868.1 |
| Reverse | AGACGAGGTGTTGGTCATTGT | ||||
| G2A (GPR132) | Forward | GTCGTTGTCATCTTCCTGGTC | 232 | 232 | scaffold_111* |
| Reverse | GGTGGAGTCTGGTCACTTCCT | ||||
| GPR4 | Forward | CCATCCTCTACTGCCTGGTC | 155 | 155 | scaffold_80* |
| Reverse | CACGCTGCTTCTCTTGGAA | ||||
| TDAG8 (GPR65) | Forward | GCAAGCAAAGAAGGAAAATGA | 139 | 139 | XM_003472463.1 |
| Reverse | GCACAAGGCAGCAGAGAAA | ||||
| TASK1 (KCNK3) | Forward | GGTACAAGAGCCGCGAGAA | 186 | 186 | XM_003472862.2 |
| Reverse | TGGACACGGAGCTGATGG | ||||
| TASK3 (KCNK9) | Forward | GAAACTCAAGGCGGAAGAGA | 391 | >1,000 | NM_001172977.1 |
| Reverse | GCCCATGCAGGAGAAGAAG | ||||
| TALK1 (KCNK16) | Forward | CCAAGGCAACTCCACCAA | 147 | 987 | scaffold_15* |
| Reverse | CCCACCAGGGCATAGAAG | ||||
| TRPV4 | Forward | ATCATCCTCACCTTCGTGCT | 191 | >1,000 | XM_003477915 |
| Reverse | TTGCCCACAGTCACCATCT | ||||
| PKD2L1 | Forward | GGCAAATCCGAACAGTGAAG | 294 | 986 | XM_003479572.2 |
| Reverse | CCAGGAACTCAAAGTCAGCA |
The sequences of guinea pig G2A, GPR4 and TALK1 were deduced from the CavPor3 assembly of the guinea pig genome (Feb. 2008 draft, UCSC Genome Bioinformatics Group) by using human, rat and/or mouse adenosine receptor sequences and compared with database sequences if available.
Mouse studies
The neurons retrogradely labeled from the mouse esophagus used for the analysis of ASIC3 expression were all obtained in our previous study as described therein (18) and the mouse β-actin, TRPV1 and ASIC3 primers used were evaluated before (Tab. 1 in (19)).
RESULTS
Electrophysiological Studies
We performed extracellular recordings of the nerve activity originating from the nerve terminals of spinal DRG C-fibers in the esophagus. Only one fiber was studied in each esophageal preparation. Among 43 nerve fiber studied we found 41 to be C-fibers based on conduction velocity <1 m/s. Seventeen out of 18 of esophageal C- fibers tested with capsaicin (0.1–1μM) responded with action potential discharge. In order to bypass the esophageal mucosal barrier the acid and drugs were delivered to the serosal surface of the esophagus as described in the Methods.
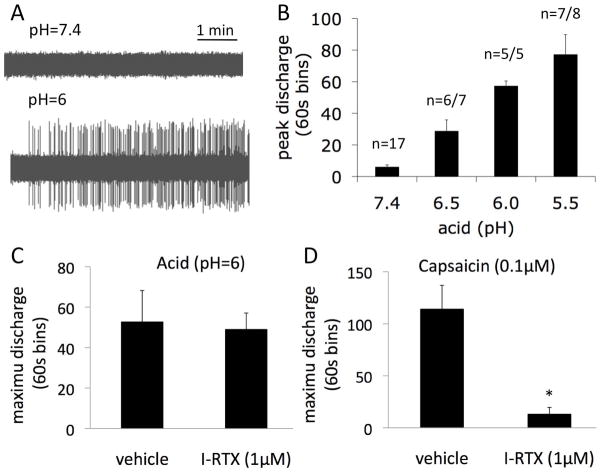
We investigated whether DRG C-fibers can be activated by mildly acidic stimuli, similar to acidic conditions that may be reached in the esophageal mucosa during an acidic reflux event. Based on direct pH measurement studies (3), a healthy epithelium will provide a sufficient barrier to prevent the diffusion of nearly all the protons if intraluminal esophageal pH is close to 1. We speculated that it is unlikely that pH <5.5 would be reached even when a pathology that leads to some barrier breakdown is present. We found that acid stimulated DRG C-fiber nerve terminals in the concentration (pH)-dependent manner (Fig. 1A–B). Acid activated C-fibers at concentrations as mild as pH=6.5. At pH=6.0 the peak frequency of the action potential discharge was 4±1Hz. The maximum 60s bin discharge that better describes more sustained activation was 57±3 action potentials/60s at pH=6.0. For comparison, a near maximally effective concentration (0.1μM) of capsaicin evoked activation that averaged 9±1Hz and 114±23 action potentials/60s, respectively (n=7, Fig. 1D). Thus, mild acid (pH=6.0) produces a response in esophageal DRG C-fibers that is approximately 40% that of the maximal noxious stimulus.
Figure 1. Mild acid stimulates esophageal DRG C-fibers by a mechanism independent of TRPV1.
(A) Examples of electrophysiological traces of baseline activity and the response to modest acid (pH=6.0). (B) pH-dependency of the activation of esophageal DRG C-fibers by acid. (C) The response to mild acid (pH=6) was not inhibited by pretreatment with the TRPV1 receptor selective antagonist I-RTX (1μM) although I-RTX nearly abolished the response to capsaicin in the same experiment (vehicle n=7, I-RTX n=5). *P<0.05.
The vast majority (95%) of esophageal DRG C-fibers were responsive to the TRPV1 agonist capsaicin. TRPV1 is a major candidate for sensory acid sensing. Nonetheless, human and guinea pig TRPV1 are not fully activated by acid until the pH≤5.0 (22). These observations raise the question whether there is a substantial TRPV1-independent mechanism that contributes to the response of esophageal C-fibers to mild acid (pH=6.0). In order to test the hypothesis we used the selective TRPV1 antagonist I-RTX as we have previously shown that I-RTX effectively inhibits the response to stronger acid in guinea pig tracheal C-fibers (23–25). Pretreatment with I-RTX (1μM) had no effect on the response to acid (pH=6.0, Fig. 1C). In contrast, as expected, I-RTX (1μM) virtually abolished (>90% inhibition) the response to capsaicin showing the effective block of TRPV1 in DRG C-fibers (Fig. 1D).
These studies indicate that non-TRPV1 acid sensors are largely responsible for the action potential discharge evoked by mild acid (pH=6.0) in DRG C-fibers innervating the esophagus. In order to obtain initial insight into what receptors and ion channels are likely to play a role in transduction of mild acid, we evaluated the gene expression of candidate receptors and ion channels in esophageal-specific DRG neurons.
Expression of mRNA for receptors and ion channels sensitive to acid
We investigated the expression of receptors and ion channels that are reported to be sensitive to acid near physiological pH (e.g. (10)). The expression of these targets was evaluated using single cell RT-PCR on spinal DRG neurons retrogradely labeled from the guinea pig esophagus.
TRPV1
In total 56 DRG T1–T2 labeled neurons were analyzed. The majority of the esophagus-labeled neurons (48/56) expressed the C-fiber marker TRPV1. This was consistent with our electrophysiological studies showing that the vast majority of spinal afferent neurons innervating the esophagus are C-fibers that are responsive to capsaicin (see above).
ASICs
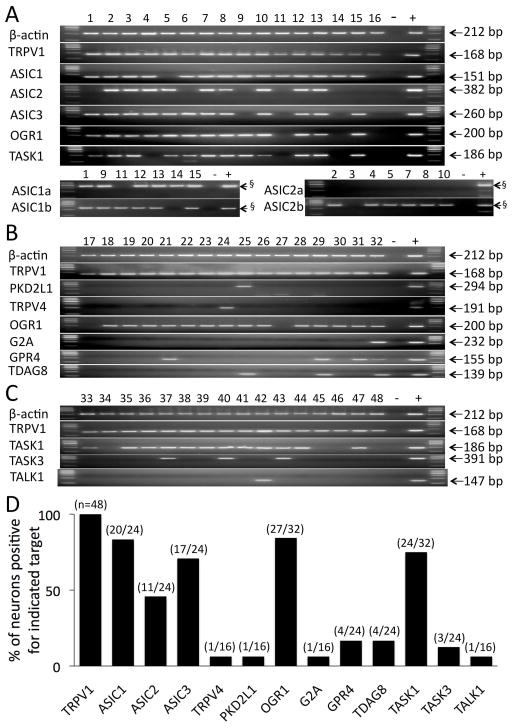
ASIC genes were frequently expressed in the spinal afferent neurons innervating the guinea pig esophagus. We evaluated 24 esophageal specific C-fiber (TRPV1 expressing) neurons for ASIC expression. ASIC1 and ASIC3 were found in 70–80% of neurons and ASIC2 was found in ~50% of the neurons (Fig. 2A). We noted that 23 out of 24 TRPV1-positive neurons expressed at least one ASIC gene and 15/24 expressed a combination of at least 2 ASIC genes. Both ASIC1 splice variants (ASIC1a and ASIC1b) were expressed, but only one ASIC2 splice variant (ASIC2b) was expressed in these DRG neurons (Fig. 2A).
Figure 2. Spinal DRG C-fiber (TRPV1-positive) neurons innervating the esophagus express multiple receptors and ion channels highly sensitive to acid.
Individual TRPV1-positive DRG neurons labeled from the esophagus are numbered. −, negative control, +, positive control. (A) Expression of ASICs. ASIC1a and ASIC1b were evaluated in ASIC1-positive neurons, and the ASIC2a and ASIC2b in ASIC2-positive neurons. OGR1 and TASK1 were also evaluated in the same neurons (see text). (B) Expression of selected TRP channels (PKD2L1 and TRPV4) and the proton-sensing GPCRs. (C) Expression of selected two pore domain (K2P) potassium channels. (D) Summary of expression results. The number of neurons positive for indicated target relative to the total number of neurons evaluated for that target is denoted as a fraction in parenthesis for each target. § The product sizes expected for ASIC1a, ASIC1b, ASIC2a and ASIC2b were 376, 240, 293 and 260 bp, respectively.
We have recently noted that although guinea pig vagal esophageal C-fibers extensively express ASIC3, ASIC3 is virtually absent in these nerves in the mouse (19). We therefore investigated ASIC3 expression in mouse DRG neurons retrogradely labeled from the esophagus. We found that the mouse DRG TRPV1-positive neurons innervating the esophagus did not express ASIC3 (0/10) (Fig. 3). ASIC3 was also rare in the TRPV1-negative mouse esophageal DRG neurons (3/38). In these experiments we used validated mouse ASIC3 primers that provided reproducible detection of ASIC3 in the mouse neurons in our previous study (19), and we used the whole DRG RNA as a positive control (3/3 positive).
Figure 3. Mouse spinal DRG C-fiber (TRPV1-positive) neurons innervating the esophagus do not express ASIC3.
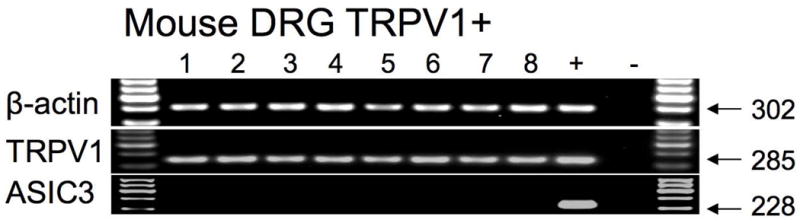
In control experiments in our previous study ASIC3 was reproducibly detected in a subset of mouse vagal TRPV1-negative neurons (Fig. 5 in (19)).
PKD2L1 and TRPV4
These TRP channels were rarely expressed in TRPV1-positive DRG neurons innervating the guinea pig esophagus (1/16 and 1/16, respectively, Fig. 2B).
Proton-sensing GPCRs
Among this family of potential acid sensors only OGR1 was extensively expressed (27/32), while other proton-sensing GPSRs G2A, GPR4 and TDAG8 were expressed relatively rarely in TRPV1-positive DRG neurons innervating the guinea pig esophagus (1/16, 4/24 and 4/24, respectively, Fig. 2B).
Two-pore-domain (K2P) potassium channels family members
Among this family of potential acid sensors, TASK1 was expressed in 75% of the neurons (24/32) while TASK3 and TALK1 were expressed rarely in TRPV1-positive DRG neurons innervating the guinea pig esophagus (3/24 and 1/16, respectively, Fig. 2C).
After initial experiments suggested that TASK1 and OGR1 are frequently expressed in esophageal DRG C-fibers, we evaluated their co-expression with ASICs in the same neurons (n=16) (Fig. 2A). Of these 16 neurons, 15 expressed ASICs, 12 expressed TASK1, and 13 expressed OGR1. Among these 16 neurons, 7 expressed all 5 targets, and additional 6 expressed at least 3 targets. Thus our expression data indicate that the majority of esophageal DRG C-fibers in the guinea pig express multiple receptors and/or channels that are potentially involved in sensing acid.
DISCUSSION
Our electrophysiological studies revealed that esophageal DRG C-fiber nerve terminals are highly sensitive to acid in the guinea pig. Acid at pH=6.5 was sufficient to induce a significant activation of esophageal DRG C-fibers (Fig. 1B). The magnitude of mild acid (pH=6.0)-induced action potential discharge was >40% of the activation induced by near maximally effective concentration of capsaicin (0.1μM). The response to acid (pH=6.0) was not affected by inhibiting TRPV1 with the selective antagonist I-RTX (which abolished the response to capsaicin) (Fig 1D). These data demonstrate the presence of a robust TRPV1-independent acid-sensing mechanism(s) in esophageal DRG C-fibers. Nonetheless, it is not possible to exclude the role for TRPV1 that may be redundant to TRPV1-independent mechanisms.
The pH in healthy esophageal mucosa is not significantly affected by a strong acid within the lumen (3). In the absence of direct measurement, the mucosal pH remains speculative in the conditions of reduced mucosal barrier function such as in patients with GERD. Nonetheless, if the concentration of acid in the mucosa was to change 10-fold from its normal values (i.e. from pH=7 to 6) a substantial increase in permeability to acid would be needed given the robust buffering capacity of extracellular fluid. Our electrophysiological data support the hypothesis that even mild acid such as this can lead to action potential discharge by a TRPV1-independent mechanism. It has been reported previously that a mild acid (around pH=6.0) is sufficient to evoke pain when infused into skin in humans (26, 27). Consistent with predictions from our experiments, the studies in humans also found only a minor role for TRPV1 in acid-induced pain in skin (28).
Characterizing precisely which of the large number of receptors and ion channels sensitive to acid are most important in acid-induced esophageal DRG C-fiber activation requires knowledge about what receptors and ion channels are expressed in these C-fibers. Obtaining such knowledge was the goal of the second part of our study. We found that esophageal DRG C-fibers express several receptors and ion channels highly sensitive to acid, namely ASICs, K2P potassium channel TASK1, and the proton-sensing GPCR OGR1. The majority (>75%) of DRG C-fibers expressed a combination of at least 3 of these receptors and ion channels.
ASIC channels were expressed in nearly all esophageal DRG C-fiber neurons (Fig. 2A). ASICs are excitatory cation channels formed by trimeric (homomeric or heteromeric) association of ASIC subunits (ASIC1a, ASIC1b, ASIC2a, ASIC2b, ASIC3 encoded by three ASIC genes)(29–32). The acid-sensing properties of ASICs such as the acid sensitivity (pH threshold) depend on the ASIC subunit composition (33). We found that esophageal DRG C-fiber neurons express ASIC1a, ASIC1b, ASIC2b, ASIC3 but not the ASIC2b subunit (see below). ASICs channels that can be formed by combinations of these 4 subunits are highly sensitive to acid (half-maximum pH activation pH50=6.5–6) (33). We noted that over 70% of esophageal DRG C-fiber neurons expressed ASIC3 (Fig. 1A, D). This is relevant because ASIC3 is responsible for the sustained component of acid-induced currents in homo/heteromultimeric ASIC channels (33). This sustained component is likely to contribute to sustained acid-induced action potential discharge such as that observed in C-fiber nerve terminals (Fig. 1A).
We have recently reported that in the vagal system the placodes-derived (vagal nodose) and the neural crest-derived (vagal jugular) esophageal C-fibers differ in the ASIC expression profile in that ASIC2a subunit is only expressed in the vagal placodes-derived nodose C-fibers (19). In the present study we found that similarly to vagal jugular C-fibers, the esophageal DRG C-fibers (neural crest-derived) also lacked ASIC2a. This is consistent with our previous findings that the spinal DRG C-fibers are similar to vagal jugular esophageal C-fibers, but different from the placodes-derived vagal nodose C-fibers (reviewed in (34)).
ASIC channels may play a less important role in sensing esophageal acid by C-fibers in the mouse than in the guinea pig. We found that unlike guinea pig esophageal DRG C-fiber neurons, mouse esophageal DRG neurons did not express ASIC3 (Fig. 2A and 3). We have previously found the same difference between the guinea pig and mouse in vagal esophageal C-fibers (19). Others reported that ASIC currents in mouse sensory neurons are less frequent, smaller and less sensitive to acid than those in rat (35–37). In this respect the situation in the guinea pig appears to be more similar to the rat (present data and (19)). In any case, these data indicate that there are substantial differences in certain aspects of ASIC expression between the species. It is noteworthy that Harrington et al (38) reported a TRPV1 component in the activation of spinal pathways (measured by pERK expression) induced by esophageal infusion of acid+pepsin. Whether the TRPV1 was activated by acid or indirectly by other mediators released due to the action of pepsin cannot be discerned from this study.
Two-pore-domain potassium (K2P) channels are constitutively open causing background potassium (leak) currents that stabilize resting membrane potential and counterbalance depolarization (39). Inhibition of K2P channels leads to membrane depolarization and initiation of action potentials. Of the K2P channels, TASK1, TASK3 and TALK1 can be inhibited by extracellular acid (12). We found that esophageal DRG C-fiber neurons expressed TASK1, but not TASK3 and TALK1. This is consistent with previous reports of TASK1 expression in DRG neurons and their preferential expression in the TRPV1-positive DRG neurons in the rat (40, 41). TASK1 is highly sensitive to acid and is nearly completely blocked at pH=6.5 (12, 42, 43). Our finding that TASK3 was rarely expressed in the C-fiber neurons is in agreement with the observation of the selective TASK3 expression in large diameter (putative A-fiber) TRPV1-negative DRG neurons (40, 41).
Proton-sensing GPCRs are highly sensitive to acid (activated at pH=6.4–6.8) and are expressed in DRG (44–47). We found that esophageal DRG C-fiber neurons expressed OGR1, but not other proton-sensing GPCRs. Our finding is in agreement with the report that OGR1 is preferentially expressed in the small- and medium-diameter neurons that were often TRPV1-positive and expressed C-fiber marker peripherin (45). OGR1 has been reported to couple to Gq/11 leading to activation of the phospholipase C (PLC)/Ca2+ signaling pathway(44). The activation of this pathway was reported to lead to nerve activation by multiple mechanisms including TRP channels(48).
The expression data were obtain by single cell RT-PCR. This technique provides a distinct advantage for addressing the hypotheses in our study. Firstly, it allows for detection of multiple targets (up to 20) in individual neurons (e.g. simultaneous detection of receptors and ion channels from different families in the same neuron, Fig. 2). Secondly, because of the nature of PCR technique and a careful design of the primers (Tab. 1) PCR has high selectivity for differentiation of the targets with relatively high sequence homology such as ASIC splice variants or closely related K2P channels. Nevertheless, it must be emphasized that RT-PCR detects only mRNA, which may not necessarily reflect functional protein expression. However, previous studies have shown that the single cell RT-PCR results correlated well with the functional response for a number of receptors and ion channels including MrgA3 (17), the adenosine A1 and A2A receptors (13, 49), PAR1 receptors (50) and ion channels TRPV1, TRPA1, P2X2 and P2X3 (14, 51, 52), or with immunohistochemical detection of protein GFRα3 (53). In our previous studies by using single cell RT-PCR we also observed segregation of markers between placodes-derived (P2X2 and TrkB) and neural crest-derived (TrkA, PPT-A and GFRα3) TRPV1-positive neurons innervating the mouse esophagus (18). Thus while caution is required, the single cell RT-PCR results correlated well with the protein expression or function in previous studies.
Considered together with our previous study (19), the present data suggest the conclusion that modest acidification of the esophageal mucosa can lead to activation of both spinal and vagal C-fibers, and that this likely involves complex TRPV1-independent mechanisms. It is not possible from current data to determine the contribution of specific channels to acid-induced action potential discharge, but the most likely candidates include ASIC3 and AISC1, TASK1, and ORG1. As the figure 2A illustrates the vast majority of esophageal C-fiber neurons expressed mRNA for a combination of these acid sensing channels and receptors.
Regrettably, potent and selective inhibitors of these acid sensing targets are presently unavailable. In preliminary studies we found the ASIC blocker diminazene (54) inhibited acid-induced action potential discharge in the C-fibers, but only at a concentration (100 μM) that also inhibited the capsaicin-induced responses and induced mechanical and electrical desensitization in some C-fibers indicating non-selective inhibitory effects (data not shown). Another ASIC blocker amiloride does not inhibit sustained currents mediated by ASIC3 (55), and amiloride analogues inhibit voltage-gated sodium channels responsible for action potential formation in guinea pig nerve fibers (56). Some ASIC subunits can be modulated by certain toxins from venoms (31), however, these tools were not available for our studies. Pharmacological agonist and antagonists that specifically target TASK1 and ORG1 are currently, to our best knowledge, unavailable. Inhibition of highly sensitive esophageal acid-sensing mechanisms may be an alternative or additive approach to acid suppression therapy in the treatment of GERD. Our results present the caveat however, that such a strategy may not yield to a single antagonist, but may require a combination approach.
KEY MESSAGE.
Because of the esophageal mucosal barrier, the acidity at the nerve terminals of esophageal pain-mediating C-fibers is predicted to be relatively mild compared to acidic gastroesophageal reflux.
We evaluated the response to mild acid by single fiber recordings and the expression of mRNA for acid-sensitive receptors and ion channels by single cell RT-PCR in the guinea pig esophageal DRG C-fibers.
Guinea pig esophageal DRG C-fibers are activated by mild acid via a TRPV1-independent mechanism, and express mRNA for several receptors and ion channels highly sensitive to acid (ASICs, TASK1 and OGR1).
Acknowledgments
FUNDING
This study was supported by Department of Education grant VEGA 1/0424/12 (Slovakia) and BioMed Martin (ITMS: 26220220187) co-funded by EU (Slovakia). M.K. received support from NIH DK074480 (US).
Footnotes
DISCLOSURES
No conflicts of interest declared.
FR performed retrograde tracing, cells picking, extracellular recording and data analysis, PB performed scRT-PCR, MK directed the study and wrote the manuscript.
A part of this study was presented at Joint International Neurogastroenterology and Motility Meeting NGM 2012 in Bologna (Neurogastroenterol Motil 2012; 24: suppl 2, p. 17).
References
- 1.Page AJ, Blackshaw LA. Roles of gastro-oesophageal afferents in the mechanisms and symptoms of reflux disease. Handb Exp Pharmacol. 2009;194:227–257. doi: 10.1007/978-3-540-79090-7_7. [DOI] [PubMed] [Google Scholar]
- 2.Orlando RC. Pathophysiology of gastroesophageal reflux disease. J Clin Gastroenterol. 2008;42:584–588. doi: 10.1097/MCG.0b013e31815d0628. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka S, Chu S, Hirokawa M, Montrose MH, Kaunitz JD. Direct measurement of acid permeation into rat oesophagus. Gut. 2003;52:775–783. doi: 10.1136/gut.52.6.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halicka J, Banovcin P, Jr, Halickova M, et al. Acid infusion into the esophagus increases the number of meal-induced transient lower esophageal sphincter relaxations (TLESRs) in healthy volunteers. Neurogastroenterol Motil. 2014;26:1469–1476. doi: 10.1111/nmo.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weijenborg PW, Smout AJ, Verseijden C, et al. Hypersensitivity to acid is associated with impaired esophageal mucosal integrity in patients with gastroesophageal reflux disease with and without esophagitis. Am J Physiol Gastrointest Liver Physiol. 2014;307:G323–329. doi: 10.1152/ajpgi.00345.2013. [DOI] [PubMed] [Google Scholar]
- 6.Farre R, Blondeau K, Clement D, et al. Evaluation of oesophageal mucosa integrity by the intraluminal impedance technique. Gut. 2011;60:885–892. doi: 10.1136/gut.2010.233049. [DOI] [PubMed] [Google Scholar]
- 7.Jovov B, Que J, Tobey NA, Djukic Z, Hogan BL, Orlando RC. Role of E-cadherin in the pathogenesis of gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:1039–1047. doi: 10.1038/ajg.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weijenborg PW, Rohof WO, Akkermans LM, Verheij J, Smout AJ, Bredenoord AJ. Electrical tissue impedance spectroscopy: a novel device to measure esophageal mucosal integrity changes during endoscopy. Neurogastroenterol Motil. 2013;25:574–e458. doi: 10.1111/nmo.12106. [DOI] [PubMed] [Google Scholar]
- 9.Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 10.Holzer P. Acid sensing by visceral afferent neurones. Acta Physiol (Oxf) 2011;201:63–75. doi: 10.1111/j.1748-1716.2010.02143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckler KJ. Two-pore domain k(+) channels and their role in chemoreception. Adv Exp Med Biol. 2010;661:15–30. doi: 10.1007/978-1-60761-500-2_2. [DOI] [PubMed] [Google Scholar]
- 12.Lesage F, Barhanin J. Molecular physiology of pH-sensitive background K(2P) channels. Physiology (Bethesda) 2012;26:424–437. doi: 10.1152/physiol.00029.2011. [DOI] [PubMed] [Google Scholar]
- 13.Ru F, Surdenikova L, Brozmanova M, Kollarik M. Adenosine-induced activation of esophageal nociceptors. Am J Physiol Gastrointest Liver Physiol. 2011;300:G485–493. doi: 10.1152/ajpgi.00361.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brozmanova M, Ru F, Surdenikova L, Mazurova L, Taylor-Clark T, Kollarik M. Preferential activation of the vagal nodose nociceptive subtype by TRPA1 agonists in the guinea pig esophagus. Neurogastroenterol Motil. 2011;23:e437–445. doi: 10.1111/j.1365-2982.2011.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu S, Ru F, Ouyang A, Kollarik M. 5-Hydroxytryptamine selectively activates the vagal nodose C-fibre subtype in the guinea-pig oesophagus. Neurogastroenterol Motil. 2008;20:1042–1050. doi: 10.1111/j.1365-2982.2008.01136.x. [DOI] [PubMed] [Google Scholar]
- 16.Yu S, Undem BJ, Kollarik M. Vagal afferent nerves with nociceptive properties in guinea-pig oesophagus. J Physiol. 2005;563:831–842. doi: 10.1113/jphysiol.2004.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Tang Z, Surdenikova L, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–1365. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surdenikova L, Ru F, Nassenstein C, Tatar M, Kollarik M. The neural crest- and placodes-derived afferent innervation of the mouse esophagus. Neurogastroenterol Motil. 2012;24:e517–525. doi: 10.1111/nmo.12002. [DOI] [PubMed] [Google Scholar]
- 19.Dusenkova S, Ru F, Surdenikova L, et al. The expression profile of Acid Sensing Ion Channel (ASIC) subunits ASIC1a, ASIC1b, ASIC2a, ASIC2b and ASIC3 in the esophageal vagal afferent nerve subtypes. Am J Physiol Gastrointest Liver Physiol. 2014 doi: 10.1152/ajpgi.00129.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozen S, Skaletsky J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 21.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savidge J, Davis C, Shah K, et al. Cloning and functional characterization of the guinea pig vanilloid receptor 1. Neuropharmacology. 2002;43:450–456. doi: 10.1016/s0028-3908(02)00122-3. [DOI] [PubMed] [Google Scholar]
- 23.Kollarik M, Undem BJ. Mechanisms of acid-induced activation of airway afferent nerve fibres in guinea-pig. J Physiol. 2002;543:591–600. doi: 10.1113/jphysiol.2002.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Undem BJ, Kollarik M. Characterization of the vanilloid receptor 1 antagonist iodo-resiniferatoxin on the afferent and efferent function of vagal sensory C-fibers. J Pharmacol Exp Ther. 2002;303:716–722. doi: 10.1124/jpet.102.039727. [DOI] [PubMed] [Google Scholar]
- 25.Wahl P, Foged C, Tullin S, Thomsen C. Iodo-resiniferatoxin, a new potent vanilloid receptor antagonist. Mol Pharmacol. 2001;59:9–15. doi: 10.1124/mol.59.1.9. [DOI] [PubMed] [Google Scholar]
- 26.Steen KH, Issberner U, Reeh PW. Pain due to experimental acidosis in human skin: evidence for non-adapting nociceptor excitation. Neurosci Lett. 1995;199:29–32. doi: 10.1016/0304-3940(95)12002-l. [DOI] [PubMed] [Google Scholar]
- 27.Birklein F, Weber M, Ernst M, Riedl B, Neundorfer B, Handwerker HO. Experimental tissue acidosis leads to increased pain in complex regional pain syndrome (CRPS) Pain. 2000;87:227–234. doi: 10.1016/S0304-3959(00)00286-4. [DOI] [PubMed] [Google Scholar]
- 28.Jones NG, Slater R, Cadiou H, McNaughton P, McMahon SB. Acid-induced pain and its modulation in humans. J Neurosci. 2004;24:10974–10979. doi: 10.1523/JNEUROSCI.2619-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deval E, Gasull X, Noel J, et al. Acid-sensing ion channels (ASICs): pharmacology and implication in pain. Pharmacol Ther. 2010;128:549–558. doi: 10.1016/j.pharmthera.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Sluka KA, Winter OC, Wemmie JA. Acid-sensing ion channels: A new target for pain and CNS diseases. Curr Opin Drug Discov Devel. 2009;12:693–704. [PMC free article] [PubMed] [Google Scholar]
- 31.Wemmie JA, Taugher RJ, Kreple CJ. Acid-sensing ion channels in pain and disease. Nat Rev Neurosci. 2013;14:461–471. doi: 10.1038/nrn3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherwood TW, Frey EN, Askwith CC. Structure and activity of the acid-sensing ion channels. Am J Physiol Cell Physiol. 2012;303:C699–710. doi: 10.1152/ajpcell.00188.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesselager M, Timmermann DB, Ahring PK. pH Dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem. 2004;279:11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- 34.Kollarik M, Ru F, Brozmanova M. Vagal afferent nerves with the properties of nociceptors. Auton Neurosci. 2010;153:12–20. doi: 10.1016/j.autneu.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattori T, Chen J, Harding AM, et al. ASIC2a and ASIC3 heteromultimerize to form pH-sensitive channels in mouse cardiac dorsal root ganglia neurons. Circ Res. 2009;105:279–286. doi: 10.1161/CIRCRESAHA.109.202036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland SP, Benson CJ, Adelman JP, McCleskey EW. Acid-sensing ion channel 3 matches the acid-gated current in cardiac ischemia-sensing neurons. Proc Natl Acad Sci U S A. 2001;98:711–716. doi: 10.1073/pnas.011404498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leffler A, Monter B, Koltzenburg M. The role of the capsaicin receptor TRPV1 and acid-sensing ion channels (ASICS) in proton sensitivity of subpopulations of primary nociceptive neurons in rats and mice. Neuroscience. 2006;139:699–709. doi: 10.1016/j.neuroscience.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 38.Harrington AM, Brierley SM, Isaacs NJ, Young RL, Ashley Blackshaw L. Identifying spinal sensory pathways activated by noxious esophageal acid. Neurogastroenterol Motil. 2013;25:e660–668. doi: 10.1111/nmo.12180. [DOI] [PubMed] [Google Scholar]
- 39.Enyedi P, Czirjak G. Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol Rev. 2010;90:559–605. doi: 10.1152/physrev.00029.2009. [DOI] [PubMed] [Google Scholar]
- 40.Talley EM, Solorzano G, Lei Q, Kim D, Bayliss DA. Cns distribution of members of the two-pore-domain (KCNK) potassium channel family. J Neurosci. 2001;21:7491–7505. doi: 10.1523/JNEUROSCI.21-19-07491.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollema-Mays SL, Centeno MV, Ashford CJ, Apkarian AV, Martina M. Expression of background potassium channels in rat DRG is cell-specific and down-regulated in a neuropathic pain model. Mol Cell Neurosci. 2013;57:1–9. doi: 10.1016/j.mcn.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duprat F, Lesage F, Fink M, Reyes R, Heurteaux C, Lazdunski M. TASK, a human background K+ channel to sense external pH variations near physiological pH. EMBO J. 1997;16:5464–5471. doi: 10.1093/emboj/16.17.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopes CM, Gallagher PG, Buck ME, Butler MH, Goldstein SA. Proton block and voltage gating are potassium-dependent in the cardiac leak channel Kcnk3. J Biol Chem. 2000;275:16969–16978. doi: 10.1074/jbc.M001948200. [DOI] [PubMed] [Google Scholar]
- 44.Ludwig MG, Vanek M, Guerini D, et al. Proton-sensing G-protein-coupled receptors. Nature. 2003;425:93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- 45.Huang CW, Tzeng JN, Chen YJ, Tsai WF, Chen CC, Sun WH. Nociceptors of dorsal root ganglion express proton-sensing G-protein-coupled receptors. Mol Cell Neurosci. 2007;36:195–210. doi: 10.1016/j.mcn.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Okajima F. Regulation of inflammation by extracellular acidification and proton-sensing GPCRs. Cell Signal. 2013;25:2263–2271. doi: 10.1016/j.cellsig.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 47.Tomura H, Mogi C, Sato K, Okajima F. Proton-sensing and lysolipid-sensitive G-protein-coupled receptors: a novel type of multi-functional receptors. Cell Signal. 2005;17:1466–1476. doi: 10.1016/j.cellsig.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbaum T, Simon SA. TRPV1 Receptors and Signal Transduction. In: Liedtke WB, SH, editors. TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades. Boca Raton: CRC Press; 2007. pp. 1–30. 2011/01/05 edn. [Google Scholar]
- 49.Chuaychoo B, Lee MG, Kollarik M, Pullmann R, Jr, Undem BJ. Evidence for both adenosine A1 and A2A receptors activating single vagal sensory C-fibres in guinea pig lungs. J Physiol. 2006;575:481–490. doi: 10.1113/jphysiol.2006.109371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwong K, Nassenstein C, de Garavilla L, Meeker S, Undem BJ. Thrombin and trypsin directly activate vagal C-fibres in mouse lung via protease-activated receptor-1. J Physiol. 2010;588:1171–1177. doi: 10.1113/jphysiol.2009.181669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nassenstein C, Kwong K, Taylor-Clark T, et al. Expression and function of the ion channel TRPA1 in vagal afferent nerves innervating mouse lungs. J Physiol. 2008;586:1595–1604. doi: 10.1113/jphysiol.2007.148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwong K, Kollarik M, Nassenstein C, Ru F, Undem BJ. P2X2 receptors differentiate placodal vs. neural crest C-fiber phenotypes innervating guinea pig lungs and esophagus. Am J Physiol Lung Cell Mol Physiol. 2008;295:L858–865. doi: 10.1152/ajplung.90360.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nassenstein C, Taylor-Clark TE, Myers AC, et al. Phenotypic distinctions between neural crest and placodal derived vagal C-fibres in mouse lungs. J Physiol. 2010;588:4769–4783. doi: 10.1113/jphysiol.2010.195339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Qiu L, Li M, et al. Diarylamidines: high potency inhibitors of acid-sensing ion channels. Neuropharmacology. 2010;58:1045–1053. doi: 10.1016/j.neuropharm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yagi J, Wenk HN, Naves LA, McCleskey EW. Sustained currents through ASIC3 ion channels at the modest pH changes that occur during myocardial ischemia. Circ Res. 2006;99:501–509. doi: 10.1161/01.RES.0000238388.79295.4c. [DOI] [PubMed] [Google Scholar]
- 56.Carr MJ, Gover TD, Weinreich D, Undem BJ. Inhibition of mechanical activation of guinea-pig airway afferent neurons by amiloride analogues. Br J Pharmacol. 2001;133:1255–1262. doi: 10.1038/sj.bjp.0704197. [DOI] [PMC free article] [PubMed] [Google Scholar]