Abstract
Pro-opiomelanocortin (POMC)- and agouti-related peptide (AgRP)-expressing neurons are oppositely regulated by caloric depletion and co-ordinately stimulate and inhibit homeostatic satiety, respectively. This bimodality is principally underscored by the antagonistic actions of these ligands at downstream melanocortin-4 receptors (MC4R) within the paraventricular nucleus of the hypothalamus. Although this population is critical to energy balance the underlying neural circuitry remains unknown. Enabled by mice expressing Cre-recombinase in MC4R neurons, we demonstrate bidirectional control of feeding following real-time activation and inhibition of PVHMC4R neurons and further identify these cells as a functional exponent of ARCAgRP neuron-driven hunger. Moreover, we reveal this function to be mediated by a PVHMC4R→lateral parabrachial nucleus (LPBN) pathway. Activation of this circuit encodes positive valence, but only in calorically depleted mice. Thus, the satiating and appetitive nature of PVHMC4R→LPBN neurons supports the principles of drive reduction and highlights this circuit as a promising target for anti-obesity drug development.
Introduction
The appropriate maintenance of energetic state is contingent upon the sensing of, and reaction to, homeostatic perturbation1,2. To this end, the interoceptive awareness of caloric sufficiency and the initiation of appropriate feeding-related behaviours is in part dependent upon the central melanocortin network3,4. This bimodal system is defined by the physiologically antagonistic actions of two non-overlapping population of neurons in the arcuate nucleus of the hypothalamus (ARC) and their opposing effects on the activity of second-order satiety-promoting neurons, presumably expressing cognate receptor isoforms5. Indeed, real-time chemo- or optogenetic activation of ARC agouti-related peptide (ARCAgRP) and pro-opiomelanocortin (ARCPOMC) neurons guides an increase and decrease in food intake, respectively,6–10 in a manner predicted to involve their reciprocal regulation of downstream melanocortin-4 receptor (MC4R) expressing neurons
The importance of the MC4R to energy balance regulation is supported by a wealth of pharmacological and genetic data that has clearly established its satiety-promoting and weight-loss inducing function11–17. Perhaps most cogently, inactivating germline Mc4r mutations in both mice and humans18,19 engender an extreme state of obesity underscored principally by increased food consumption12,13. Furthermore, despite broad expression across the mammalian neuraxis, conditional manipulations of Mc4r expression within genetically defined neuronal populations has identified the paraventricular nucleus of the hypothalamus (PVH) as the principle site of MC4R-reguated appetite, while more caudal populations, within the brainstem and spinal cord, underlie MC4R-regulated energy expenditure and glucose homeostasis11,15,20.
As a structure the PVH is critical to the maintenance of energy homeostasis21,22 and has recently been identified as a principle site of functional outflow for ARCAgRP neurons, which induce hunger via inhibition of satiety-promoting post-synaptic neurons, the identity of which has not been fully clarified7,23,24. While it may be predicted that these post-synaptic neurons express MC4R, the explicit contribution of PVHMC4R neurons to the real-time regulation of feeding behaviour and their place within the broader melanocortinergic network remains to be elucidated.
Technological advances in the field of neuroscience now permit real-time circuit-level interrogation of genetically-defined populations of neurons within the context of freely enacted behaviour. Indeed, such approaches have been successful in establishing the importance of ARCAgRP neurons to feeding behavior6,8. Despite a widely ramifying efferent profile, ARCAgRP neurons promote feeding via their projections to the PVH, anterior bed nucleus of the stria terminalis (aBNST), lateral hypothalamus (LH) and paraventricular thalamus (PVT)25. While the post-synaptic targets for orexigenic ARCAgRP efferents within these sites remain to be identified, MC4R-expressing neurons (as found in the PVH, LH and BNST) would seem a logical second-order population. Here, using ARCAgRP neuron driven hunger as a physiological framework in which to interrogate MC4R-regulated appetite, we now provide circuit-level analysis of this system and its salience to the real-time control of feeding behaviour.
Results
PVHMC4R, but not aBNSTMC4R or LHMC4R, neurons are a downstream target for ARCAgRP-driven hunger
To facilitate cell-specific manipulation of MC4R circuitry, we generated Mc4r-t2a-Cre knock-in mice expressing Cre-recombinase under control of endogenous Mc4r regulatory elements (Fig 1a). Cre-mediated expression of a germline tdTomato-reporter allele indicated the faithful recapitulation of central MC4R expression across the rostral-caudal extent of the neuraxis, including robust labelling in the PVH, LH and aBNST. (Fig 1b, Fig S1–S2).
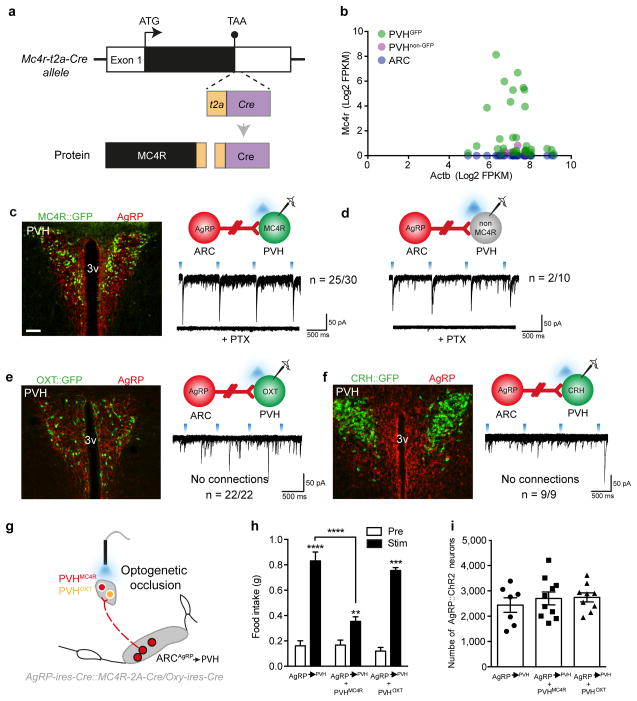
Figure 1. PVHMC4R neurons are a downstream target for ARCAgRP-driven hunger.
a, Schematic of Mc4r-t2a-Cre locus. b, Single cell RNA sequencing validation of individual PVHGFP (n=31), PVHnon-GFP (n=5) and ARC (n=30) neurons demonstrating endogenous Mc4r mRNA only in PVHGFP cells. c–f, CRACM from ARCAgRP neurons (red) onto putative post-synaptic PVH neurons demonstrating monosynaptic inhibitory input onto 83% of PVHMC4R (c), 20% of non-MC4R PVH (d), but not onto PVHOXT (e) or PVHCRH (f) neurons. g, In vivo optogenetic occlusion schematic involving concomitant stimulation of MC4R/OXT soma and ARCAgRP terminals in the PVH. h, ARCAgRP→PVH (n=7) ChR2-driven light-cycle food intake was significantly attenuated by simultaneous activation of PVHMC4R (n=10), but not PVHOXT (n=8), soma (Repeated measures ANOVA, main effect of treatment (F(1,22)=240.99, p<0.0001), main effect of genotype (F(2,22)=16.88, p<0.0001) and interaction (F(2,22)=25.95, p<0.0001); post-hoc: ARCAgRP→PVH (pre vs stim), ****p<0.0001; ARCAgRP→PVHMC4R (pre vs stim), **p=0.0036; ARCAgRP→PVHOXT (pre vs stim), ****p<0.0001; ARCAgRP→PVH vs ARCAgRP→PVHMC4R, ****p<0.0001). i, All mice in (h) exhibited comparable levels of ARCAgRP ChR2-mCherry transduction (One-way ANOVA, F(3,23)=0.41, p=0.66). Abbreviations, 3v, third ventricle; PTX, picrotoxin. Scale bar in panel c, 100 μm and relates to all images. All values presented as mean±SEM.
To determine whether PVHMC4R neurons lie downstream of ARCAgRP neurons, we mapped monosynaptic connectivity from pre-synaptic ARCAgRP neurons to putative post-synaptic MC4RPVH neurons using channelrhodopsin (ChR2)-assisted circuit mapping (CRACM) in Agrp-ires-Cre::Mc4r-t2a-Cre mice23,26. MC4R-expressing neurons were visualised via viral transduction with a Cre-dependent GFP reporter. We detected time-locked light-evoked picrotoxin-sensitive inhibitory post-synaptic currents (IPSCs) in approximately 83% of post-synaptic PVHMC4R neurons (Fig 1c), a far higher connection rate than observed when recording from unlabelled or single minded 1 (Sim1)-expressing PVH neurons7,24. Moreover, ARCAgRP input to non-MC4R expressing PVH neurons revealed only a 20% connection rate (Fig 1d) highlighting PVHMC4R neurons as an enriched population of ARCAgRP inhibited neurons. Interestingly, while a previous report found that oxytocin-expressing PVH (PVHOXT) neurons, demarked by a transgenic-reporter line, received monosynaptic input from ARCAgRP neurons,7 our CRACM analysis using a validated Oxt-ires-Cre knock-in mouse27 failed to detect light-evoked IPSCs in any recorded PVHOXT neurons (Fig 1e). Similarly, despite evidence of MC4R-expression on corticotropin releasing-hormone (CRH)-expressing PVH neurons28 we failed to detect light-evoked IPSCs on any recorded PVHCRH neuron (Fig 1f). Thus, MC4R-expressing neurons represent an identified subpopulation of the PVH to be monosynaptically inhibited by ARCAgRP neurons. While identifying monosynaptic connectivity between endogenous agonist producing ARCPOMC and PVHMC4R neurons would be desirable, the absence of classical fast neurotransmitter identity in the pre-synpatic cell precluded this analysis.
If ARCAgRP neurons promote food intake by inhibiting PVHMC4R neurons, then simultaneous stimulation of ARCAgRP→PVH terminals and PVHMC4R neurons should occlude the ARCAgRP→PVH driven feeding. Indeed, concomitant in vivo optogenetic activation of ChR2-expressing PVHMC4R soma and inhibitory ARCAgRP→PVH terminals in doubly-transduced Agrp-ires-Cre::Mc4r-t2a-Cre mice significantly attenuated light-evoked food consumption, compared to stimulation of ARCAgRP→PVH terminals alone (Fig 1g-h; Fig S3a-b). Importantly this effect was not a function of ARCAgRP::ChR2-mCherry transduction efficiency (Fig 1i). Contrasting the functional requirement for PVHMC4R neurons, repetition of this study in Agrp-ires-Cre::Oxt-ires-Cre mice demonstrated that co-activation of ChR2-expressing PVHOXT neurons failed to occlude ARCAgRP→PVH induced feeding (Fig 1g-i; Fig S3c), consistent with the absence of any direct synaptic connection between these two populations. Immunohistochemical analysis of endogenous PVHOXT expression in an Mc4r-t2a-Cre::tdTomato reporter line confirmed these cell types as spatially distinct populations (Fig S4).
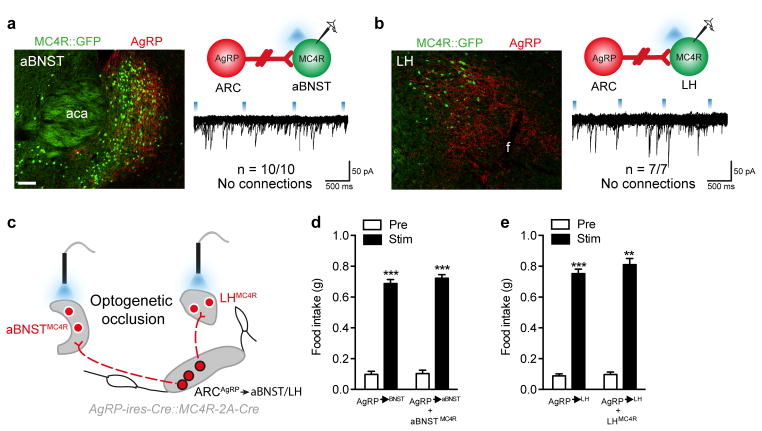
Further distinguishing the selective role of PVHMC4R neurons in ARCAgRP-driven feeding we found that neither aBNSTMC4R nor LHMC4R neurons were synaptically connected to ARCAgRP neurons, as evidenced by the absence of light-evoked IPSCs (Fig 2a-b). Concordantly, simultaneous optogenetic activation of ChR2-expressing ARCAgRP→aBNST terminals + aBNSTMC4R neurons or ARCAgRP→LH terminals + LHMC4R neurons failed to occlude ARCAgRP-driven food consumption at these sites (Fig 2c, d-e; Fig S3d-e). Thus despite ARCAgRP→LH and ARCAgRP→BNST efferents being sufficient to drive feeding behaviour25 these projections appear to promote food intake through an MC4R neuron-independent pathway.
Figure 2. aBNST and LHMC4R neurons are not a downstream target for ARCAgRP-driven hunger.
a–b, CRACM from ARCAgRP neurons (red) onto putative post-synaptic aBNSTMC4R (a) and LHMC4R (b) neurons (green) demonstrating the absence of monosynaptic inhibitory connections. c, In vivo optogenetic occlusion schematic involving concomitant stimulation of MC4R soma and ARCAgRP terminals in the aBNST and LH. d–e, ARCAgRP→aBNST (d; n=5) and →LH (e; n=6) ChR2-driven light-cycle food intake was unaffected by simultaneous activation of aBNSTMC4R (Repeated measures ANOVA, main effect of treatment (F(1,4)=1645, p<0.0001), no main effect of genotype (F(1,4)=2.132, p=0.22) and interaction (F(1,4)=0.38, p=0.58; post-hoc: ARCAgRP→aBNST (pre vs stim), ***p=0.0001; ARCAgRP→aBNSTMC4R (pre vs stim), ***p=0.0001) or LHMC4R (Repeated measures ANOVA, main effect of treatment (F(1,5)=359.9, p<0.0001), no main effect of genotype (F(1,5)=0.43, p=0.58) and interaction (F(1,5)=0.16, p=0.70); post-hoc: ARCAgRP→LH (pre vs stim), ***p=0.0009; ARCAgRP→LHMC4R (pre vs stim), **p=0.0013) soma Abbreviations, f, fornix; aca, anterior commissure anterior part. Scale bar in panel c, 100 μm and relates to all images. All values presented as mean±SEM.
PVHMC4R neurons regulate energy balance in real-time
Since hunger-promoting ARCAgRP neurons are inhibitory, synaptically connected PVHMC4R neurons would be predicted to signal satiety. Consistent with this notion, analysis of PVHMC4R neurons demonstrated a significant increase in FOS-immunoreactivity (a molecular correlate of neuronal activation) in sated (fed) versus hungry (fasted) mice (Fig 3a-b). To directly probe the sufficiency of these cells to induce satiety we virally-transduced PVHMC4R neurons with Cre-dependent stimulatory hM3Dq-DREADD receptors (Fig 3c; Fig S5a)8,29, facilitating their activation upon administration of the otherwise inert ligand clozapine-N-oxide (CNO; Fig S5b). Consistent with our hypothesis, acute chemogenetic activation of PVHMC4R neurons significantly reduced food intake in two paradigms of physiological hunger; that occurring naturally at the onset of the animals’ active period (dark-cycle feeding; Fig 3d) and that following an overnight fast (post-fast re-feeding; Fig 3e). Substantiating the neuroanatomical divergence in energy intake versus energy expenditure regulating MC4R-populations,11 direct activation of PVHMC4R neurons affected neither energy expenditure (Fig 3f) nor respiratory exchange ratio (Fig S6). Thus, artificial activation of PVHMC4R neurons promotes satiety (but not energy expenditure) and attenuates the homeostatic drive to consume food, even in the face of marked caloric insufficiency.
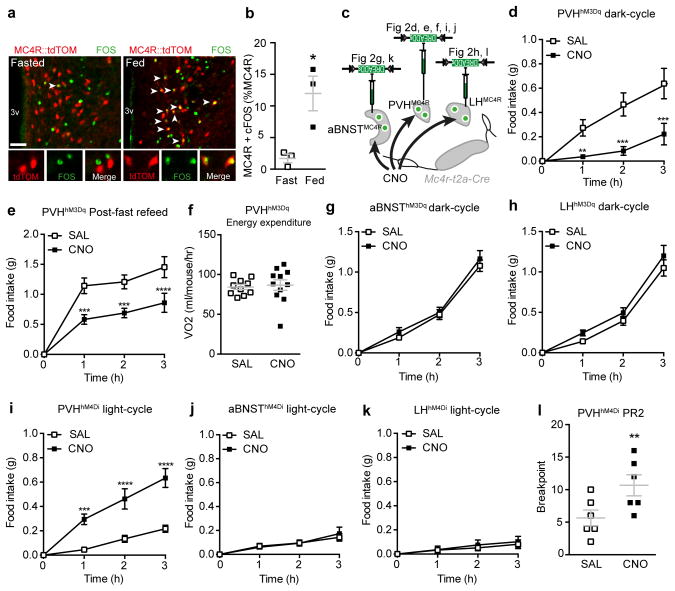
Figure 3. PVHMC4R neurons regulate homeostatic satiety in real-time.
a–b, PVHMC4R neurons (red) exhibit significantly increased FOS-immunoreactivity (green) in the fed versus fasted state (n=3 per group, Unpaired two-tailed t-test, t(4)=3.64, *p=0.02.). c, Chemogenetic manipulation of PVHMC4R, BNSTMC4R and LHMC4R via viral DREADD constructs, studies were within subject, all animals received both SAL and CNO. d–f, Chemogenetic activation of PVHMC4R neurons reduced ad libitum food intake during the dark-cycle (d; n=5, Repeated measures ANOVA, main effect of treatment (F(1,16)=79.30, p<0.0001), main effect of time (F(3,16)=8.43, p=0.001) and interaction (F(3,16)=10.57, p=0.0004): post-hoc, 1h, **p=0.0034; 2h, ****p<0.0001; 3h, ****p<0.0001) and re-feeding following an overnight fast (e; n=5, Repeated measures ANOVA, main effect of treatment (F(1,16)=71.90, p<0.0001), main effect of time (F(3,16)=26.13, p<0.0001) and interaction (F(3,16)=8.04, p=0.002): post-hoc, 1h, ***p=0.0001; 2h, ***p=0.0003; 3h, ****p<0.0001) but did not influence oxygen consumption (f; n=11, Paired two-tailed t-test, t(10)=0.08, p=0.94.). g–h, Chemogenetic activation of BNSTMC4R (g; n=6, Repeated measures ANOVA, main effect of time (F(3,20)=92.26, p<0.0001), no main effect of treatment (F(1, 20)=2.550, p=0.13), and interaction F(3, 20)=0.4909, p=0.69) or LHMC4R (h; n=8. Repeated measures ANOVA, main effect of time (F(3,28)=64.09 p<0.0001), no main effect of treatment (F(1, 28)=5.106, p=0.03), and interaction F(3, 28)=0.68, p=0.57) neurons did not influence dark-cycle food intake. i–k, Chemogenetic inhibition of PVHMC4R (i; n=9, Repeated measures ANOVA, main effect of treatment (F(1,32)=77.49, p<0.0001), main effect of time (F(3,32)=25.70, p<0.0001) and interaction (F(3,32)=10.06, p<0.0001); post-hoc: 1h, ***p=0.0004; 2h, ****p<0.0001; 3h, ****p<0.0001) but not BNSTMC4R (j; n=5. Repeated measures ANOVA, main effect of time (F(3,16)=7.19, p=0.003), no main effect of treatment (F(1, 16)=0.38, p=0.55), and interaction F(3, 16)=1.0, p=0.42) or LHMC4R (k; n=6, Repeated measures ANOVA, no main effect of treatment (F(1, 20)=2.57, p=0.12), time (F(3,20)=1.9, p=0.15) and interaction F(3, 20)=0.63, p=0.61) neurons increased light-cycle food intake. l, Chemogenetic inhibition of PVHMC4R neurons increases the motivation to work for a food reward in a progressive ration 2 (PR2) nosepoke assay (n=6, Paired two-tailed t-test, t(5)=4.04, p=0.01). Abbreviations, 3v, third ventricle; f, fornix; aca, anterior commissure anterior part. Scale bar in panel a, 100 μm. All values presented as mean±SEM.
Given recent evidence that BNST and LH neurons guide robust alterations in food consumption,30,31 we assessed the functional contribution of aBNSTMC4R and LHMC4R neurons to feeding behaviour. Chemogenetic stimulation of either population failed to influence dark-cycle food intake (Fig 3g-h; S5c-d), suggesting a dispensability of these cells to appetite control and substantiating their lack of involvement in ARCAgRP-driven feeding.
The necessity of PVHMC4R neurons for satiety was addressed by chemogenetic inhibition using hM4Di-DREADD receptors (Fig S5e-f)8,32. Feeding behaviour was assessed at a time of physiological satiety, the start of the light-cycle, when consumption is low following dark-cycle ad libitum access to food. CNO-mediated silencing of PVHMC4R, but not aBNSTMC4R or LHMC4R, neurons significantly increased food consumption (Fig 3i-k; Fig S5g-h). Furthermore, in an instrumental nosepoke assay chemogenetic silencing of PVHMC4R neurons in sated mice significantly increased the motivation to attain a food reward, as measured by the highest number of consecutive nosepokes an animal performed to procure a single food pellet (breakpoint; Fig 3l). Additionally, and congruent with the above results and previous findings,27,33 chemogenetic silencing of PVHOXT or PVHCRH neurons did not affect feeding behaviour (Fig S6d-e). Thus, selective silencing of PVHMC4R neurons re-established the motivation to feed, even within the context of caloric repletion, and demonstrates these cells to be necessary for the maintenance of satiety.
PVHMC4R→LPBN neurocircuit is responsive to nutritional state
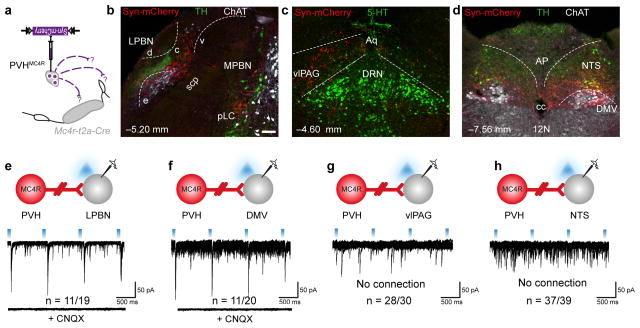
To deconvolute the neurocircuitry underlying satiety-promoting PVHMC4R neurons, a Cre-dependent synaptically-targeted fluorophore was used to map their neuroanatomical targets (Fig 4a).34 This revealed dense efferent fields within the median eminence, central LPBN (Fig 4b), nucleus of the solitary tract (NTS) and dorsal motor nucleus of the vagus (DMV; Fig 4c), with sparser projections to the ventrolateral periaqueductal grey (vlPAG; Fig 4d), pre-locus coeruleus/locus coeruleus (pLC/LC), rostral ventrolateral medulla (RVLM) and spinal cord (SC) (Fig S7). To assess the functional significance of these projections we conducted a comprehensive CRACM analysis of regional connectivity at feeding-related sites. We detected CNQX-sensitive light-evoked EPSCs in the majority of blindly recorded LPBN (11/19; Fig 4e) and DMV (11/20; Fig 4f) neurons, but few to no light-evoked EPSCs in the vlPAG (Fig 4g), pLC/LC (Fig S7) and NTS (Fig 4h).
Figure 4. Glutamatergic PVHMC4R neurons monosynaptically engage the LPBN.
a–d, Genetically-encoded efferent mapping from PVHMC4R neurons using a synaptically-targeted fluorophore revealed axonal terminals within the central LPBN (b), vlPAG (c), NTS and DMV (d). e–h, Investigation of monosynaptic connectivity using CRACM from PVHMC4R neurons (red) to unlabelled putative post-synaptic neurons (grey). 55% of cLPBN (e), 55% of DMV neurons (f), 7% of vlPAG (g) and 5% of NTS (h) receive CNQX-sensitive glutamatergic inputs from PVHMC4R neurons. Abbreviations, 4v, fourth ventricle; 5-HT, serotonin; 12N, hypoglossal nucleus; AP, area postrema; c, central LPBN; cc, central canal; ChAT, choline acetyltransferase d, dorsal LPBN; e, external LPBN; MPBN, medial parabrachial nucleus; pLC, pre-locus coeruleus; TH, tyrosine hydroxylase; v, ventral LPBN; scp, superior cerebellar peduncle. Co-ordinates in b-d represent distance from Bregma. Scale bar in panel b, 200 μm and relates to all images.
As a viscerosensory relay the LPBN has been implicated with appetite-suppression within a number of contexts35–38, suggesting that it is neuroanatomically positioned to signal satiety. To determine the physiological relevance of LPBN-projecting PVHMC4R neurons we visually isolated this subpopulation, by way of retrograde labelling from the LPBN (Fig 5a), and compared their relative ex vivo electrophysiological activity in opposing nutritional states. All PVHMC4R→LPBN neurons exhibited increased action potential firing in the fed versus fasted state, implicating this circuit in the promotion of satiety (Fig 5b-d). This responsiveness to caloric sufficiency would be expected to be contingent, in part, upon actions of the anorectic POMC-derived agonists. Indeed, we found that all PVHMC4R→LPBN neurons recorded were activated by a selective MC4R agonist (Fig 5e-g), suggesting that within a physiological context these neurons are likely to be activated by anorectic POMC neurons. Consistent with this idea we find that PVHMC4R neuron lie within the POMC→PVH efferent fibre field (Fig S8).
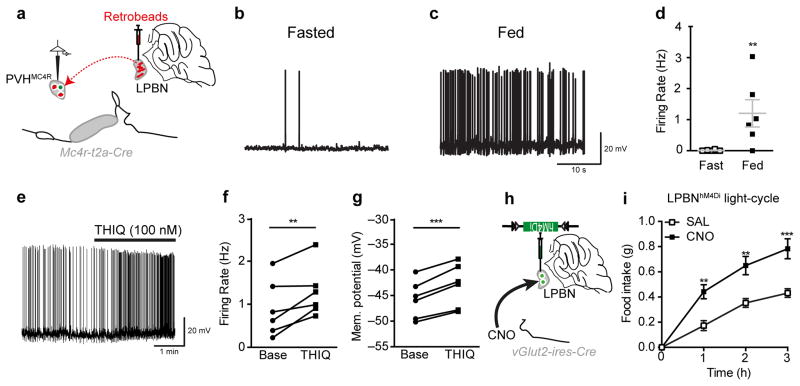
Figure 5. PVHMC4R→LPBN neurocircuit is responsive to nutritional state.
a, PVHMC4R→LPBN neuron isolation by retrograde labelling from the LPBN for electrophysiological current clamp recordings. b–d, PVHMC4R→LPBN neurons demonstrated a significantly increased action potential firing rate in the fed (n=8) versus fasted (n=6) state (Unpaired two-tailed t-test, t(12)=3.19, p=0.008). e–g, PVHMC4R→LPBN neurons (n=6) exhibited increased action potential firing rate (f; Paired two-tailed t-test, t(5)=3.50, **p=0.01) and membrane depolarization (g; Paired two-tailed t-test, t(5)=8.40, ***p=0.0004) in response to selective MC4R agonist THIQ. h–i, Chemogenetic silencing of LPBNvGLUT2 neurons resulted in a significant increase in light-cycle food consumption (n=4, Repeated measures ANOVA, main effect of treatment (F(1,12)=58.89, p<0.0001), main effect of time (F(3,12)=49.95, p<0.0001) and interaction (F(3,12)=3.12, p<0.0068; post-hoc: 1h, **p=0.0032; 2h, **p=0.015; 3h, ***p=0.0004). All values presented as mean±SEM.
Next, to confirm the real-time involvement of the PBN in promoting satiety we broadly silenced the structure through chemogenetic inhibition of Slc17a6 (vGLUT2)-expressing neurons,39 which are known to receive glutamatergic input from PVH neurons.15 We observed a significant increase in light-cycle food consumption upon CNO treatment (Fig 5h-i). While the LPBN gates multiple aversive inputs, many of which produce anorexia,35,36,38 the observed elevation in food intake upon PBNvGLUT2 neuron silencing in sated mice strongly suggests that a subset PBN neurons relay satiety.
PVHMC4R→LPBN neurocircuit is satiating and appetitive
To investigate the physiological relevance of this explicit PVHMC4R→LPBN neurocircuit we employed an in vivo optogenetic approach. Firstly, to ensure that any observed behavioural response could be attributed specifically to the PVHMC4R→LPBN circuit, the anatomical organization of these projections was evaluated using a rabies terminal-mapping strategy (Fig 6a).25 This approach revealed that only a subset of PVHMC4R neurons project to the LPBN (assessed via co-labelling of fluorescent signals within the PVH) and that these PVHMC4R→LPBN neurons do not collateralize to other PVHMC4R efferent sites, as EGFP signal was detected only in the LPBN terminal field.
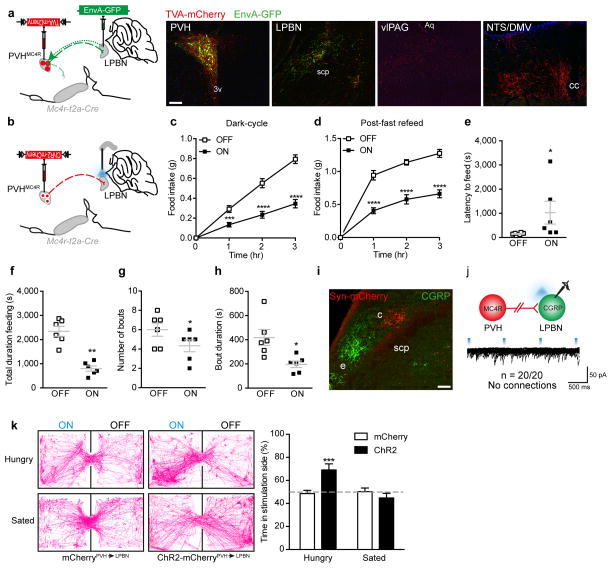
Figure 6. PVHMC4R→LPBN neurocircuit is satiating and appetitive.
a, Rabies terminal-mapping revealed that PVHMC4R→LPBN neurons do not collateralize to any other efferent field (representative images from one of two animals). b–g, In vivo optogenetic photostimulation of PVHMC4R→LPBN terminals (b) significantly reduced food consumption during the dark-cycle (c; n=11, Repeated measures ANOVA, main effect of treatment (F(1,40)=185.4, p<0.0001), main effect of time (F(3,40)=74.88, p<0.0001) and interaction (F(3,40)=32.92, p<0.0001); post-hoc: 1h, ***p=0.0002; 2h ****p<0.0001; 3h, ****p<0.0001) and re-feeding following an overnight fast (d; n=7, Repeated measures ANOVA, main effect of treatment (F(1,24)=117.4, p<0.0001), main effect of time (F(3,24)=174.6, p<0.0001) and interaction (F(3,24)=13.21, p<0.0001; post-hoc: 1h, ****p<0.0001; 2h ****p<0.0001; 3h, ****p<0.0001), due to increased latency to feed and (e; n=6, Paired two-tailed t-test, t(5)=1.82, *p=0.05.) decreased total time feeding (f; n=6, Paired two-tailed t-test, p, **p=0.003), number of feeding bouts (g; n=6, Paired two-tailed t-test, t(5)=2.98, *p=0.03) and the average bout duration (h; n=6, Paired two-tailed t-test, t(5)=3.63, *p=0.02), compared to the same mice without photostimulation. i, The PVHMC4R→LPBN terminal field (red) is spatially distinct from that of LPBNCGRP soma (green). j, CRACM from PVHMC4R neurons (red) onto putative post-synaptic LPBNCGRP neurons (green) demonstrated the absence of monosynaptic excitatory connections. k, Mc4r-t2a-Cre::ChR2-mCherry mice (n=6) exhibited a significant place preference for the photostimulation-paired chamber during a real-time place preference assay, compared to Mc4r-t2a-Cre::mCherry mice (n=7, Repeated measures ANOVA, main effect of state (F(1,11)=9.47, p=0.01) and interaction (F(1,11)=12.52, p=0.005), no effect of genotype (F(1,11)=2.95, p=0.11); post-hoc: ***p=0.001). Abbreviations, 3v, third ventricle; Aq, aqueduct; c, central LPBN; cc; central canal; e, external LPBN; scp, superior cerebellar peduncle. Scale bar in panel a, 100 μm and relates to all images. All values presented as mean±SEM.
Next, PVHMC4R neurons were virally-transduced with ChR2-mCherry and optic fibres bilaterally implanted over the LPBN (Fig 6b; Fig S9). Consistent with the chemogenetic activation of the MC4RPVH population as a whole, and the ability of PVHMC4R→LPBN projections to modulate post-synaptic LPBN neuron activity, we observed a significant attenuation of food intake during PVHMC4R→LPBN terminal photostimulation in two states of physiological hunger (Fig 6c-d), compared to the same mice without photostimulation or an independent cohort receiving photostimulation in the absence of ChR2-expression (Fig S10). Micro-analysis of feeding parameters found that activation of PVHMC4R→LPBN neurons increased latency to feed (Fig 6e) and decreased total time spent feeding (Fig 6f), as well as number (Fig 6g) and duration of individual feeding bouts (Fig 6h), consistent with the promotion of satiety. Optogenetic stimulation of PVHMC4R projections to the vlPAG (Fig S9a) or NTS/DMV (Fig S9b) had no effect on feeding behaviour. Thus, an explicit PVHMC4R→LPBN circuit is sufficient to reduce the homeostatic drive to consume food even in the face of caloric insufficiency.
While the nature of PVHMC4R-engaged LPBN satiety neurons remains to be determined, recently published work has identified the presence of an illness-associated appetite suppressing axis arising from central amygdala-projecting calcitonin gene-related peptide (CGRP) expressing LPBN neurons.35 To determine whether these LPBNCGRP neurons lie downstream of PVHMC4R→LPBN neurons we assessed monosynaptic connectivity using CRACM. Consistent with the neuroanatomical segregation of the PVHMC4R→LPBN efferent and LPBNCGRP cell body fields, in the central and external LPBN respectively (Fig 6i), no light-evoked EPSCs were detected on any LPBNCGRP (Fig 6j) or LPBNMC4R (Fig S10d) neurons (which have been shown to be overlapping populations)40 following photostimulation of PVHMC4R→LPBN terminals in an Mc4r-t2a-Cre::CGRP-CreERT2::L10-GFP or Mc4r-t2a-Cre::L10-GFP mouse line. These data affirm the non-overlapping nature of these two physiologically distinct appetite-suppressing axes.
Finally, to determine whether satiety promoted by PVHMC4R→LPBN terminal stimulation is associated with the induction of an aversive physiological state we assessed the motivational valence of this circuit using a real-time place preference assay41. In the physiologically hungry state Mc4r-t2a-CrePVH→LPBN::ChR2-mCherry mice exhibited a significant place preference for the photostimulation-paired chamber, while Mc4r-t2a-CrePVH→LPBN::mCherry controls showed an absence of preference (Fig 6k). This finding establishes that, in hungry mice, the PVHMC4R→LPBN circuit is not only satiating but also appetitive (i.e. encodes positive emotional valence). Remarkably, we observed that caloric repletion abolished this preference for the photo-stimulated side of the arena, suggesting that in the sated state, in contrast to the hungry state, stimulation of the PVHMC4R→LPBN circuit loses saliency. Together these data suggest that the PVHMC4R→LPBN circuit encodes state-dependent valence and may contribute to the motivational drive underlying homeostatic feeding behaviour.
Discussion
Despite a well defined role in energy balance, regulation circuit-level analysis of the broader melanocortin network has hitherto been lacking. Here, using an Mc4r-t2a-Cre mouse line to facilitate direct genetic entry into the MC4R circuitry, we comprehensively interrogated MC4R-expressing neuronal populations downstream of orexigenic ARCAgRP neurons. Our data establish a fundamental role for PVHMC4R, but not aBNSTMC4R or LHMC4R, neurons in the real-time regulation of feeding behaviour and further identify the LPBN as the site of functional outflow for this satiety promoting population.
ARCAgRP neurons drive robust feeding behaviour via their inhibition of second-order satiety neurons within the PVH, aBNST, LH and PVT6,8,25, yet the identity of these post-synaptic targets remains to be fully clarified. The expression of cognate receptor isoforms within efferent sites of ligand producing neurons would seem a strong predictor of co-functionality. Thus, we systematically interrogated the contribution of PVH, aBNST and LH MC4R-expressing neurons to ARCAgRP driven hunger and overall feeding behaviour.
As evidenced by both our ex vivo electrophysiological (CRACM) and in vivo behavioural (ARCAgRP driven hunger occlusion) data we find that MC4R-expressing neurons do not form part of the orexigenic ARCAgRP→aBNST or ARCAgRP→LH axes. Furthermore, functional manipulation of aBNSTMC4R or LHMC4R neurons, independently of ARCAgRP neurons also failed to influence real-time feeding behaviour. Thus it is likely that ARCAgRP efferents to these sites drive food consumption through an as of yet unidentified population of non-MC4R expressing neurons.
This finding is perhaps not surprising in light of genetic evidence that strongly supports the notion of functional divergence between neuroanatomical populations of MC4Rs, with those in the PVH being the principle regulators of satiety11,20. Our CRACM analysis of ARCAgRP→PVH projections has established that PVHMC4R neurons are the preponderant second-order target for this orexigenic circuit. Specifically, PVHMC4R neurons are an enriched population of ARCAgRP→PVH inhibited neurons with a connectivity rate far higher than that observed of PVHSim1 neurons24, which represent the majority of PVH neurons. However, our behavioural finding that concomitant stimulation of ARCAgRP→PVH terminals and PVHMC4R soma did not fully occlude ARCAgRP→PVH feeding, together with the fact that 20% of non-MC4R PVH neurons receive synaptic input from ARCAgRP neurons, suggests that a non-MC4R expressing satiety-promoting population of PVH neurons remains to be identified.
Interestingly, our CRACM analysis revealed that 17% of PVHMC4R neurons do not receive inhibitory input from ARCAgRP neurons. It is possible that despite not being in fast synaptic contact with GABAergic ARCAgRP efferents these neurons are still responsive to neuropeptidergic transmission upon volume release of AgRP. Along similar lines, the absence of a consistent fast neurotransmitter identity in the majority of ARCPOMC neurons precluded our assessment of their engagement of PVHMC4R neurons, although we did observe that 100% of synaptically isolated PVHMC4R→LPBN neurons were responsive to a selective MC4R agonist. While this does not establish the neuroanatomical source of endogenous POMC neuropeptide, convergent synaptic connections of ARCAgRP and ARCPOMC neurons have been observed on unlabelled PVH neurons42. Thus it is likely that ARC derived POMC neuropeptide plays a significant role in activating satiety-promoting PVHMC4R neurons.
Consistent with this melancortinergic engagement, we observe that, much like upstream ARCAgRP neurons, PVHMC4R neurons exhibit functional reciprocity with regards to their artificial inhibition and activation, driving hunger and satiety, respectively. Chemogenetic suppression, mimicking ARCAgRP neuron mediated inhibition of PVHMC4R, increased food consumption during the light-cycle and was correlated with an increased motivation to attain a food reward in an instrumental nosepoke assay. Conversely, activation of PVHMC4R neurons, as may be expected from stimulation by endogenous POMC ligands, promoted satiety in two states of physiological hunger. This latter observation is supported by the activation of PVHMC4R neurons by post-fast refeeding, as evidenced by increased FOS-IR. As a population PVHMC4R neurons would thus seem bona fide regulators of homeostatic satiety.
A previous investigation of ARCAgRP neuron engaged PVH neurons identified PVHOXT neurons as the principle exponent of ARCAgRP→PVH stimulated food consumption7. Inconsistent with this, other studies which have used Oxt-ires-Cre knockin mice to gain access to PVHOXT neurons, found that these neurons, unlike ARCAgRP and PVHMC4R neurons, appear to lack the capacity to influence feeding behaviour when acutely ablated or chemogenetically manipulated27,33(also this study). We also find in the present study that PVHOXT neurons, as identified using Oxt-ires-Cre knockin mice, do not receive GABAergic input from ARCAgRP neurons, and importantly that optogenetic stimulation of PVHOXT neurons fails to occlude ARCAgRP→PVH feeding. In total, these data argue that PVHOXT neurons do not regulate feeding in general, and do not appear to be downstream targets of ARCAgRP→PVH driven feeding.
Having established PVHMC4R neurons as a critical satiety-promoting population we next sought to determine the third-order node through which the central melanocortin system regulates feeding. The LPBN has been repeatedly demonstrated to integrate viscerally derived energy balance signals and has the capacity to influence food consumption within a number of contexts35–38. Based on this evidence and the results of our neuroanatomical and CRACM analyses we hypothesised the LPBN to be a likely site of functional outflow for satiety-promoting PVHMC4R neurons. Indeed, PVHMC4R→LPBN neurons, a subset of non-collateralizing PVHMC4R cells, exhibited nutritional regulation and melanocortinergic responsiveness, consistent with their role in promoting satiety in reaction to caloric repletion. Furthermore, we found that optogenetic stimulation of PVHMC4R→LPBN terminals reduced food intake in calorically depleted mice, while the chemogenetic silencing of LPBNvGLUT2 neurons engendered reciprocal hyperphagia in calorically replete mice. Thus, PVHMC4R neurons promote satiety via their excitation of an as of yet unidentified population of anorectic glutamatergic LPBN neurons.
Although PVH projections to the caudal NTS have long been predicted to play a role in the regulation of energy balance43,44 we find that, despite robust axonal innervation, only a small proportion of NTS neurons receive glutamatergic input from PVHMC4R neurons. By contrast, pre-ganglionic parasympathetic neurons in the DMV exhibited a level of connectivity similar to that of the LPBN. However, optogenetic stimulation of PVHMC4R→NTS/DMV projections had no effect on food consumption, suggesting that these efferents likely contribute to other aspects of PVHMC4R regulated physiology. Similarly, and consistent with the very low CRACM connectivity, we find that PVHMC4R→vlPAG projections also have no affect on acute feeding behaviour. This finding contrasts a recent study that identified the vlPAG/dorsal raphe nucleus as the downstream target for satiety-promoting PVHSim1 neurons45, of which PVHMC4R neurons are a subpopulation. In reconciling these observations there are two potential explanations 1) that the PVHSim1→vlPAG circuit and subsequent feeding effect is defined by a population of Sim1-expressing non-MC4R expressing PVH satiety neurons and thus not represented in our PVHMC4R→vlPAG optogenetic study or 2) that the PVHSim1→vlPAG feeding effect may be explained by the spread of CNO to the LPBN, which is represented at the relevant neuroanatomical levels (Bregma: −4.8→5.35) identified in this study45 by Stachniak et al.
The role of the LPBN in appetite regulation has been further highlighted by the identification of an anorectic circuit arising from LPBNCGRP neurons that is associated with appetite-suppression under a variety of pathophysiological contexts35,46. Importantly, as a means of promoting satiety the engagement of such an illness-associated circuit would run contrary to the physiological manifestation of fullness following food consumption (see below). Indeed, although closely apposing we find that PVHMC4R→LPBN neurons do not directly engage LPBNCGRP neurons. Thus, the identification of two non-overlapping anorectic LPBN axes suggests that this nucleus integrates multiple appetite-regulating inputs and, via distinct downstream mechanisms, is able to suppress food consumption under multiple contexts.
Further distinguishing the nature of the PVHMC4R→LPBN versus LPBNCGRP→CeA circuit is the finding that they encode opposite emotional valence. Consistent with the aversive conditions under which they are activated, optogenetic LPBNCGRP neuron stimulation drives conditioned taste aversion47. By contrast however, we find that optogenetic stimulation of PVHMC4R→LPBN terminals promoted conditional place preference, indicating the appetitive nature of this axis. While this is an interesting finding that neatly highlights the interoceptive difference between the satiety- and illness-induced anorexia, it is in fact the state-dependent valence of the PVHMC4R→LPBN circuit that is most remarkable.
While satiety would be predicted to be reinforcing, so as to ensure the motivation required to meet episodes of energetic demand, genuine hunger induces a homeostatic disturbance that promotes recognizable physiological and psychological discomfort that is ultimately alleviated upon food consumption48. Thus the negative state of tension arising from unmet caloric need serves as a motivational incentive to fulfil the homeostatic disturbance by consuming food, and is subsequently reinforced by the appetitive nature of satiety. This notion is the principle tenet of the drive reduction hypothesis of motivation1,49 whereby the motivation to enact the behaviour (food consumption) that fulfils an unmet need (caloric insufficiency) is nullified in the absence of the initiating drive (hunger). Using RTPP in freely-behaving naive animals we were able to mimic this physiological construct independently of food consumption (i.e. food consumption substituted with photostimulation of PVHMC4R→LPBN terminals). Consistent with the conceptual imperative of drive reduction theory we find that the motivational incentive of satiating PVHMC4R→LPBN neurons is only salient within the context of physiological hunger and that in the sated state this axis is emotionally inert. This finding provides real-time circuit level evidence that reestablishes the drive-reduction hypothesis of motivation as a valid construct underlying homeostatic feeding. Contrasting our experimental support of drive reduction are recent optogenetic studies of the LH demonstrating that activation of glutamatergic neurons drives an aversive form of hypophagia, while stimulation of GABAergic neurons promotes appetitive feeding behaviour30,31, consistent with historical LH electrode stimulation studies50. It is possible however that this consummatory motor program, driven collectively by multiple neuronal subtypes, differs from genuine homeostatic feeding in the integration of hedonic facets of feeding and general arousal mechanisms. Thus, although these observations must be assimilated into our broader consideration of feeding behaviour, our experimental isolation of a de facto homeostatic axis, defined by PVHMC4R→LPBN neurons, has facilitated the disambiguation of mammalian feeding circuitry and clearly establishes a place for drive reduction in the biological basis of energy homeostasis.
In conclusion, we now extend the circuit-level wiring diagram for melanocortin-regulated appetite. Our studies establish PVHMC4R neurons as the principle downstream target for ARCAgRP→PVH driven food consumption and further identify the LPBN as the third-order node in this system. Moreover, we determine that PVHMC4R→LPBN neurons not only promote physiological satiety but encode positive emotional valence in a state-dependent manner. Thus, this circuit may serve as a promising target for appetite-suppressing pharmacotherapies.
Material and Methods
Animals
AgRP-ires-Cre51, Oxt-ires-Cre27, Slc17a6(vGLUT2)-ires-Cre39, Crh-ires-Cre24, Calca(Cgrp)-CreERT2,52 R26-loxSTOPlox-L10-GFP17 and Ai9-tdTomato53 mice were generated and maintained as previously described. All mice are on a mixed background. All animal care and experimental procedures were approved by the National Institute of Health and Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. Mice were housed at 22–24 °C with a 12 h light:12 h dark cycle with standard mouse chow (Teklad F6 Rodent Diet 8664; 4.05 kcal g−1, 3.3 kcal g−1 metabolizable energy, 12.5% kcal from fat; Harlan Teklad) and water provided ad libitum. All diets were provided as pellets. For all behavioral studies male mice between 6–10 weeks were used. For electrophysiological studies male and female mice between 4–10 weeks were used.
Generation of Mc4r-t2a-Cre mice
Mice were generated using recombineering techniques as previously described24. Briefly, a selection cassette containing the t2a peptide54 linked to Cre- recombinase and an Frt-flanked kanamycin resistance gene was targeted just downstream of the stop codon of the melanocortin-4 receptor gene, in a bacterial artificial chromosome, so that Cre-recombinase expression was driven by the endogenous regulatory elements. A targeting plasmid containing the Cre-containing selection cassette and 4 kb genomic sequence upstream and downstream of the melanocortin-4 receptor stop codon was isolated and used for embryonic stem cell targeting. Correctly targeted clones were identified by long range PCR and injected into blastocysts. Chimaeric animals generated from blastocyst implantation were then bred for germline transmission of the altered melanocortin-4 receptor allele. Flp-deletion mice were then used to remove the neomycin selection cassette.
Brain tissue preparation
Mice were terminally anesthetised with chloral hydrate (Sigma Aldrich) and transcardially perfused with phosphate-buffered saline (PBS) followed by 10% neutral buffered formalin (Fisher Scientific). Brains were extracted, cryoprotected in 20% sucrose, and sectioned coronally on a freezing sliding microtome (Leica Biosystems) at 30 μm and collected in four equal series.
Immunohistochemistry
Brain sections were washed in 0.1 M phosphate-buffered saline with Tween-20, pH 7.4 (PBST), blocked in 3% normal donkey serum/0.25% Triton X-100 in PBS for 1 hour at room temperature and then incubated overnight at room temperature in blocking solution containing primary antiserum (sheep anti-melanocyte-stimulating hormone, Millipore (#5087) 1:1000; rabbit anti-dsRed, Clonetech (#632496)1:1000; chicken anti-GFP, Life Technologies (#A10262) 1:1,000; rabbit anti-oxytocin, Peninsula Laboratories (#T4084), 1/1000; goat anti-AgRP, Neuromics (#GT15023) 1/1000 or rabbit anti-FOS, Calbiochem (#PC38) 1/25,000). The next morning sections were extensively washed in PBS and then incubated in Alexa fluorophore secondary antibody (Molecular Probes, 1:1000) for 2 h at room temperature. After several washes in PBS, sections were mounted onto gelatin-coated slides and fluorescent images were captured with Olympus VS120 slide scanner microscope.
Single cell RNA-sequencing
PVHMC4R neurons were labeled by a germline Cre-reporter transgene (R26-loxSTOPlox-L10-GFP) or injection of a Cre-dependent AAV-mCherry. Non-PVH neurons were labeled with different Cre/lox systems to be described elsewhere but otherwise sampled and processed the same. The PVH was micro-dissected from 1mm coronal sections of chilled, freshly extracted mouse brain and then dissociated according to a published protocol.55 Following other published protocols, fluorescently-labeled cells were manually picked and washed as per,56 frozen at −80degC then lysed for cDNA synthesis and amplification as per.57 To control for mRNA contamination during cell picking, an equivalent volume of cell-picking buffer was sampled and processed with each batch of single-cell samples. Amplified cDNA was analysed by quantitative PCR (qPCR) for housekeeping gene expression (Actb, Gapdh, Rplp0). Single-cell samples showing low or undetectable levels of gene expression were excluded from further use, as were batches of samples in which the cell-picking buffer showed evidence of mRNA contamination (i.e., housekeeping genes detected in cDNA amplified from cell-picking buffer). Samples passing this quality control were further amplified and size-selected to remove primer artifacts.57 Multiplex RNA-seq libraries were constructed with a commercially available kit (Ovation Rapid Library Kit NuGen, California, USA) and sequenced by Illumina HiSeq (50 base single-end reads). Pass-filter reads were aligned to the mouse genome (mm10) and quantified by TopHat/Bowtie2/Cufflinks software58–60 in units of FPKM (fragments per kilobase exon per million fragments mapped).
In situ hybridisation
In situ hybridisation experiments were performed in Mc4r-t2a-Cre::AAV-DIO-GFP injected mice employing a mouse-specific antisense 35S-labeled Mc4r cDNA probe11 generated using the following primers: F 5′-ATTACCTTGACCATCCTGAT-3′ and R 5′-ATGTCAATTCATAACGCCCA-3′. Linearised recombinant plasmids were subjected to in vitro transcription with a T7 RNA polymerase (Ambion Inc, Austin, Tx) in the presence of 35S-labeled UTP. cRNA riboprobes were diluted to 2x107 cpm/ml in hybridisation solution and the tissue treated as previously described.11 Tissue was then washed in PBS before commencement of the immunohistochemistry protocol. Sections were treated for 30 min in 0.3% H2O2 in PBS, rinsed in PBS and blocked in 0.5% BSA/ 0.5% Triton-X 100 in PBS for 1 hour. Sections were incubated in blocking buffer containing goat α-GFP antibody (1/1000; Abcam) overnight. Sections were washed in PBS and a biotinylated donkey anti-goat secondary antibody (Vector Laboratories) applied at 1/1000 in blocking buffer for 1 hour. Sections were then washed in PBS and incubated for 1 hour in VectaStain ABC reagent and chromogenic detection conducted using 3,3′-diaminobenzidine (DAB) reagent (Vector Laboratories). Sections were mounted onto microscope slides and air-dried. Slides were dipped in photographic emulsion (Kodak) and stored at 4°C for two weeks before being developed in D-19 developer and fixer (Kodak). Double labelled cells were recorded if GFP positive cell bodies contained overlying black grains that were in a quantity greater than 3X background and conformed to the shape of the GFP-immunoreactive cell bodies.
Viral injections
Stereotaxic injections were performed as previously described.8 Mice were anaesthetised with xylazine (5 mg per kg) and ketamine (75 mg per kg) diluted in saline (350 mg per kg) and placed into a stereotaxic apparatus (KOPF Model 963 or Stoelting). For postoperative care, mice were injected intraperitoneally with meloxicam (0.5 mg per kg). After exposing the skull via small incision, a small hole was drilled for injection. A pulled-glass pipette with 20–40 mm tip diameter was inserted into the brain and virus was injected by an air pressure system. A micromanipulator (Grass Technologies, Model S48 Stimulator) was used to control injection speed at 25 nl min 1 and the pipette was withdrawn 5 min after injection. For electrophysiology and in vivo optogenetic experiments, AAV8-DIO-ChR2(H134R)-mCherry (University of North Carolina Vector Core; titer 1.3 × 10 12 genome copies per ml) or AAV9-FLEX-ChR2(H134R)-tdTomato (University of Pennsylvania School of Medicine; titer 1.6 × 10 12 genome copies per ml) were injected into the ARC (50–100 nl, bregma: AP: −1.50 mm, DV: −5.80 mm, ML: +/−0.20 mm), PVH (15–40 nl, bregma: AP: −0.70 mm, DV: −4.70 mm, L: +/−0.20 mm), BNST (30–50 nl, bregma: AP: +0.50 mm, DV: −4.25mm, ML: +/−0.50 mm) and/or LH (50–100 nl, bregma: AP: −1.50 mm, DV: −5.00 mm, ML: +/−1.00 mm). For electrophysiology and in vivo chemogenetic experiments, AAV8-DIO-hM3Dq-mCherry (University of North Carolina Vector Core; titer 1.2 ×1012 genome copies per ml) or AAV8-DIO-hM4Di-mCherry (University of North Carolina Vector Core; titer 1.7 × 10 12 genome copies per ml) were bilaterally injected into the PVH (15–40 nl, coordinates as above), BNST (30–50 nl,coordinates as above), LH (50–100 nl, coordinates as above) or LPBN (30–40 nl, bregma: AP: −5.20 mm, DV: −3.35 mm, ML: +/−1.35 mm). For tracing experiments, AAV8-DIO-synaptophysin-mCherry34 (Virovek, Inc; titer 1.3 × 10 12 genomes copies per ml) was unilaterally injected into the PVH (15 nl, coordinates as above). For retrograde rabies tracing, AAV8-FLEX-TVA-mCherry, (University of North Carolina Vector Core; titer 1.1 × 10 12 genomes copies per ml) was injected unilaterally into the PVH (15 nl, coordinates as above) and 3 weeks later SADΔG–EGFP (EnvA) rabies (Massachusetts General Hospital Vector Core; titer 107 genomes copies per ml) was unilaterally injected into the LPBN (50 nl, coordinates as above). For electrophysiology experiments, red retrobeads (Lumafluor) were bilaterally injected into the LPBN (50 nl, coordinates as above).
Optic fibre implantation
For occlusion experiments, optical fibres (200 μm diameter core; BFH37-200 Multimode; NA 0.37; Thor Labs) were implanted unilaterally over the PVH (bregma: AP: −0.70 mm, DV: −4.20 mm, ML: −0.10 mm), bilaterally over the BNST (bregma: AP: +0.50 mm, DV: −4.00 mm, ML: +/−0.50 mm) orbilaterally over the LH (bregma: AP: −1.50 mm, DV: −4.80 mm, ML: +/−1.00 mm). For PVHMC4R in vivo terminal stimulation experiments, optical fibres were implanted bilaterally over the LPBN (bregma: AP: −5.20 mm, DV: −3.15 mm, ML: +/−1.35 mm) and vlPAG (bregma: AP: −4.60 mm, DV: (R) −2.30 mm and (L) −2.43 mm, ML: (R) +0.5 mm and (L at 20°) −1.43 mm). For PVHMC4R→NTS/DMV a unilateral optical fibre was implanted (bregma: AP: −7.5 mm, DV: −4.3 mm, ML: +0.25 mm). Fibres were fixed to the skull using dental acrylic and mice were allowed 2 weeks for recovery before acclimatisation to the recording chambers (12 h light/dark cycle starting at 6am) for 1 week.
Neuron counting
Neurons with red fluorescence were counted manually using Zen, image software. Abercrombie’s correction was applied to the neuron counts (slice thickness, 50 μm; AGRP neuron diameter, 14 ± 1 μm; correction factor, 0.78).6,61 In the text, neuron counts were rounded to the nearest hundred.
SADΔG–EGFP (EnvA) rabies terminal mapping
Three weeks after unilateral injection of AAV-FLEX-TVA-mCherryinto the PVH of Mc4r-t2a-Cre mice, SADΔG–EGFP (EnvA) rabies was unilaterally injected into the LPBN. Animals were allowed 6 days for retrograde transport of rabies virus and EGFP transgene expression before tissue collection. Comprehensive examination of SADΔG–EGFP (EnvA) axonal and retrograde transductions were obtained using 10–15 confocal images of PVHMC4R boutons along the neuraxis using an Zeiss LSM-510 confocal microscope.
Electrophysiology and channelrhodopsin-assisted circuit mapping (CRACM)
For brain slice preparation, 6–10-weeks-old mice were anaesthetised with isoflurane before decapitation and removal of the entire brain. Brains were immediately submerged in ice-cold, carbogen-saturated (95% O2, 5% CO2) high sucrose solution (238 mM sucrose, 26 mM NaHCO 3, 2.5 mM KCl, 1.0 mM NaH2PO4, 5.0 mM MgCl2, 10.0 mM CaCl2, 11 mM glucose). Then, 300-μM thick coronal sections were cut with a Leica VT1000S Vibratome and incubated in oxygenated aCSF (126 mM NaCl, 21.4 mM NaHCO 3, 2.5 mM KCl, 1.2 mM NaH2PO4, 1.2 mM MgCl2, 2.4 mM CaCl2, 10 mM glucose) at 34 °C for 30 min. Then, slices were maintained and recorded at room temperature (20–24 °C). The intracellular solution for voltage clamp recording contained the following (in mM): 140 CsCl, 1 BAPTA, 10 HEPES, 5 MgCl2, 5 Mg-ATP, 0.3 Na2GTP, and 10 lidocaine N-ethyl bromide (QX-314), pH 7.35 and 290 mOsm. The intracellular solution for current clamp recordings contained the following (in mM): 128 K gluconate, 10 KCl, 10 HEPES, 1 EGTA, 1 MgCl2, 0.3 CaCl2, 5 Na2ATP, 0.3 NaGTP, adjusted to pH 7.3 with KOH.
For CRACM, photostimulation-evoked EPSCs and IPSCs were recorded in the whole cell voltage clamp mode, with membrane potential clamped at −60 mV. Photostimulation-evoked IPSCs and EPSCs were recorded in presence of CNQX (20 μM) and picrotoxin (100 μM), respectively, to block inhibitory or excitatory postsynaptic currents, as appropriate. All recordings were made using a Multiclamp 700B amplifier, and data were filtered at 2 kHz and digitised at 10 kHz. To photostimulate channelrhodopsin2-positive fibres, a laser or LED light source (473 nm; Opto Engine; Thorlabs) was used. The blue light was focused onto the back aperture of the microscope objective, producing a wide-field exposure around the recorded cell of 10–15 mW mm2. The light power at the specimen was measured using an optical power meter PM100D (Thorlabs). The light output was controlled by a programmable pulse stimulator, Master-8 (A.M.P.I.) and pClamp 10.2 software (Axon Instruments). Photostimulation-evoked EPSCs/IPSCs detection protocol consisted of four blue light laser pulses administered 1 s apart during the first 4 s of a 8 s sweep, repeated for a total of 30 sweeps. Number of animals used per study: ARCAgRP→PVHMC4R n=4; ARCAgRP→PVHOXT n=3; ARCAgRP→PVHCRH n=2; ARCAgRP→BNSTMC4R n=2; ARCAgRP→LHMC4R n=3; PVHMC4R→LPBN/pLC n=3; PVHMC4R→LPBNMC4R n=2; PVHMC4R→LPBNCGRP n=2; PVHMC4R→ARCAgRP n=1; PVHMC4R→vlPAG n=3; PVHMC4R →NTS/DMV n=3.
To assess the effect of MC4R selective agonist (THIQ; 100 nM; Tocris) onto PVHMC4R→LPBN neurons, whole cell current clamp recordings were performed on double fluorescent-labeled PVHMC4R neurons in Mc4r-t2a-Cre mice injected with AAV8-DIO-GFP (PVH, green) and retrobeads (LPBN, red). Synaptic blockers (1 mM kynurenate and 100 μM picrotoxin) were added in aCSF to synaptically isolate PVHMC4R neurons. Number of animals n = 3.
To assess the effect of distinct states of appetite on PVHMC4R→LPBN neurons, whole cell current clamp recordings were performed on double fluorescent-labeled PVHMC4R neurons in Mc4r-t2a-Cre mice injected with AAV8-DIO-GFP (green) into the PVH and retrobeads (red) into the LPBN. Food was removed from home cages at the onset of the dark cycle. Fasted animals, n = 3 were euthanised the following morning in the hungry state. Fed animals, n = 3 were given food for 2 hours the following morning and euthanised in the sated state.
To assess the effect of CNO on PVHMC4R neurons, aCSF solution containing 5 μM CNO was perfused into the brain slice preparation after acquisition of stable whole-cell recordings for 2–5 min.8
FOS analysis
Mc4r-t2a-Cre:: Ai9-tdTomato mice were handled for 10 consecutive days before the assay to reduce stress response. Food was removed from home cages at the onset of the dark cycle. Fasted animals, n=3 were euthanised the following morning in the hungry state with 7% chloral hydrate for histological assay. Fed animals, n=3 were given food for 2 hours the following morning and euthanised in the sated state. Tissue was collected and processed as detailed above. Assessment of FOS induction was performed using a previously developed method62 modified for fluorescent co-localisation with tdTomato in PVHMC4R neurons, as described above. Data are expressed as the percentage of all PVHMC4R neurons that were double-positive for FOS. The experimenter was blinded to the conditions during the analysis stage.
Photostimulation protocol
Fibre optic cables (1.5 m long, 200 mm diameter; Doric Lenses) were firmly attached to the implanted fibre optic cannulae with zirconia sleeves (Doric Lenses). During occlusion and PVHMC4R→LPBN photostimulation experiments, light pulse trains (10-ms pulses of 20 Hz; 1 second on, 3 seconds off; 5 seconds on, 2 seconds off, respectively) were programmed using a waveform generator (PCGU100; Valleman Instruments or Arduino electronics platform) that provided input to a blue light laser (473 nm; Laserglow). We adjusted the light power of the laser such that the light power exiting the fibre optic cable was at least 10 mW using an online light transmission calculator for brain tissue http://web.stanford.edu/group/dlab/cgi-bin/graph/chart.php. We estimated the light power at the PVH, BNST, LH and LPBN to be 6.01, 4.99, 4.99 and 11.25 mW/mm2, respectively. Note that this is probably a high estimation because some light was probably lost at the interface between the fibre optic cable and the implanted fibre optic cannula. After the completion of photostimulation experiments, mice were perfused and the approximate locations of fibre tips were identified based on the coordinates of Franklin and Paxinos.63
Food intake studies
Food intake studies on chow were performed as previously described.8 All animals were singly housed for at least 2.5 weeks following surgery and handled for 10 consecutive days before the assay to reduce stress response. Studies were conducted in a home-cage environment with ad libitum food access. A full trial consisted of assessing food intake from the study subjects after they received injections of saline or pseudo-photostimulation on day 1 and CNO or photostimulation on day. Animals received a week ‘off’ between trials before another trial was initiated. The food intake data from all days were then averaged and combined for analysis. Mice with ‘missed’ injections, incomplete ‘hits’ or expression outside the area of interest were excluded from analysis after post hoc examination of mCherry/tdTomato expression. In this way, all food intake measurements were randomised and blind to the experimenter.
For chemogenetic studies, CNO was administered at 0.3 mg per kg of body weight (i.p). Saline was delivered at the same volume to maintain consistency in the studies. Previous publications suggest the duration of the drugs effect at the dosage used throughout the studies is approximately 8 h.29 For optogenetic studies, mice tethered to an optical patch cord 1 hour prior to the commencement of the study. Light-cycle and occlusion feeding studies were conducted between 9:00am to 12:00pm and intake was monitored 1 h, 2 h and 3 h after i.p. injection. Dark-cycle feeding studies were conducted between 6:00pm to 9:00pm and intake was monitored 1 h, 2 h and 3 h after i.p. injection. For post-fast refeed studies, animals were fasted overnight at 5:00pm and food returned the following morning at 9:00am. Food intake was monitored 1 h, 2 h and 3 h after i.p. injection. For micro-analysis offeeding parameters, behavior was recorded and scored using Noldus Tracking Software.
Progressive ratio task
Fully acclimated and handled mice were maintained at 85% of their respective body weights during the training protocol in the testing chamber (Noldus Phenotyper) where familiarity of the pellet delivery (active versus non-active nosepoke) and food retrieval systems (pellet dispenser into dish) were obtained. Training sessions took place over 5 consecutive days on a fixed ratio 1 (FR1) schedule. To continue training, each animal had to consume at least 5 pellets (20 mg grain pellets; TestDiet) during each two-hour session. After 5 training sessions, animals that were not nosepoking sufficiently to earn at least 5 pellets were eliminated from the study. For PVHMC4R neuron silencing experiments, breakpoint testing was performed on a progressive ratio 2 (PR2) schedule, such that each successive food pellet increased the nosepoke schedule by two additional responses. Breakpoint was defined as the maximum number of consecutive nosepokes an animal performed to procure a single food pellet. Mice received either a saline or CNO (0.3 mg per kg) i.p. injectionimmediately before being placed in the test chamber.
Real-time place preference
Mice were run in a real-time place preference paradigm,41 whereby one chamber was paired with constant 20 Hz photostimulation (10–15 mW, 473 nm) and in the other identical chamber the animals received no photostimulation. Percent time spent in the photostimulation chamber versus non-photostimulation chamber and velocity was recorded via a CCD camera interfaced with Ethovision software.
Energy expenditure measurements
Energy expenditure was assessed by measuring oxygen consumption as previously described8 using a comprehensive lab animal monitoring system (CLAMS; Columbus Instruments).
Statistical analysis
Statistical analyses were performed using Origin Pro 8.6 and Prism 6.0 (GraphPad) software. Power analyses were calculated to estimate sample size using statistical conventions for 80% power, assuming a standard deviation of change of 1.0, a difference between the means of 1.5-fold and alpha level of 0.05. In all statistical tests normal distribution and equal variance was established. The data presented met the assumptions of the statistical test employed. No randomisation of animals was conducted since all behavioural tests were within-subject comparisons. Exclusion criteria for experimental animals were a) sickness or death during the testing period or b) if histological validation of the injection site demonstrated an absence of reporter gene expression. These criteria were established prior to data collection. N-numbers represent final number of healthy/validated animals.
Supplementary Material
Acknowledgments
This work was supported by the University of Edinburgh Chancellor’s Fellowship (ASG) and National Institutes of Health to BBL: R01 DK096010, R01 DK089044, R01 DK071051, R01 DK075632, R37 DK053477, BNORC Transgenic Core P30 DK046200, BADERC Transgenic Core P30 DK057521); to MJK: F32 DK089710; to DPO K08 DK071561; to JKE R01 DK088423 and R37 DK0053301; to JNC, American Heart Association (AHA) Postdoctoral Fellowship 14POST20100011; to BAT viral vector production core P30 NS045776; and an ADA Mentor-Based Fellowship to BPS and BBL. This research was supported, in part, by the Intramural Research Program of the NIH, The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; DK075087, DK075088).
The authors gratefully acknowledge the NIDDK Mouse Metabolism Core for technical support. Dr. Paul Greer (Datta Lab, Harvard Medical School), Dr. Ben Okaty (Dymecki Lab, Harvard Medical School), Dr. Linus Tsai (Rosen Lab, Beth Israel Deaconess Medical Center/Harvard Medical School/Broad Institute) for advice on single-cell RNA-Seq and Dr. Victoria Petkova (Beth Israel Deaconess Medical Center Molecular Medicine Core Facility) for assistance with qPCR and sample preparation. Sequencing and initial data processing were performed at Massachusetts General Hospital’s Next-Gen Sequencing Core. Sequencing was supported in part by funding from the Boston Area Diabetes Endocrinology Research Center (BADERC P30 DK057521). Ms. Danielle Morse for the production of the rabies virus.
Footnotes
Disclosure summary: The authors have nothing to disclose
Author contribution
ASG, MJK, BPS and BBL conceived the studies. ASG, MJK, CL and JCM conducted the studies with assistance from BPS, EW, JSS and DPO. Single cell RNA-Seq profiling was conducted by JNC. Energy expenditure assays were conducted by OG. In situ validation of the MC4R-t2a-Cre mouse line was conducted by CEL and JKE. Rabies virus was provided by BAT. ASG, MJK and BBL wrote the manuscript.
References
- 1.Berridge KC. Motivation concepts in behavioral neuroscience. Physiology & behavior. 2004;81:179–209. doi: 10.1016/j.physbeh.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Cannon WB. The wisdom of the body. xv. W.W. Norton and Company; 1932. [Google Scholar]
- 3.Cowley MA, et al. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron. 1999;24:155–163. doi: 10.1016/s0896-6273(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 4.Sternson SM. Hypothalamic survival circuits: blueprints for purposive behaviors. Neuron. 2013;77:810–824. doi: 10.1016/j.neuron.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garfield AS, Lam DD, Marston OJ, Przydzial MJ, Heisler LK. Role of central melanocortin pathways in energy homeostasis. Trends in endocrinology and metabolism: TEM. 2009;20:203–215. doi: 10.1016/j.tem.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nature neuroscience. 2011;14 doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. The Journal of clinical investigation. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krashes MJ, Shah BP, Koda S, Lowell BB. Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell metabolism. 2013;18:588–595. doi: 10.1016/j.cmet.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan C, et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. The Journal of neuroscience. 2013;33:3624–3632. doi: 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 12.Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature. 1997;385:165–168. doi: 10.1038/385165a0. [DOI] [PubMed] [Google Scholar]
- 13.Huszar D, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 14.Rossi M, et al. A C-terminal fragment of Agouti-related protein increases feeding and antagonizes the effect of alpha-melanocyte stimulating hormone in vivo. Endocrinology. 1998;139:4428–4431. doi: 10.1210/endo.139.10.6332. [DOI] [PubMed] [Google Scholar]
- 15.Shah BP, et al. MC4R-expressing glutamatergic neurons in the paraventricular hypothalamus regulate feeding and are synaptically connected to the parabrachial nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13193–13198. doi: 10.1073/pnas.1407843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Small CJ, et al. Effects of chronic central nervous system administration of agouti-related protein in pair-fed animals. Diabetes. 2001;50:248–254. doi: 10.2337/diabetes.50.2.248. [DOI] [PubMed] [Google Scholar]
- 17.Yaswen L, Diehl N, Brennan MB, Hochgeschwender U. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nature medicine. 1999;5 doi: 10.1038/12506. [DOI] [PubMed] [Google Scholar]
- 18.Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nature genetics. 1998;20 doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 19.Yeo GS, et al. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nature genetics. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 20.Rossi J, et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell metabolism. 2011;13:195–204. doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leibowitz SF, Hammer NJ, Chang K. Hypothalamic paraventricular nucleus lesions produce overeating and obesity in the rat. Physiology & behavior. 1981;27:1031–1040. doi: 10.1016/0031-9384(81)90366-8. [DOI] [PubMed] [Google Scholar]
- 22.Xi D, Gandhi N, Lai M, Kublaoui BM. Ablation of Sim1 neurons causes obesity through hyperphagia and reduced energy expenditure. PloS one. 2012;7:e36453. doi: 10.1371/journal.pone.0036453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of neuroscience. 2008:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krashes MJ, et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 2014;507:238–242. doi: 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betley JN, Cao ZF, Ritola KD, Sternson SM. Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell. 2013;155:1337–1350. doi: 10.1016/j.cell.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nature neuroscience. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 27.Wu Z, et al. An obligate role of oxytocin neurons in diet induced energy expenditure. PloS one. 2012;7:e45167. doi: 10.1371/journal.pone.0045167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu XY, Barsh GS, Akil H, Watson SJ. Interaction between alpha-melanocyte-stimulating hormone and corticotropin-releasing hormone in the regulation of feeding and hypothalamo-pituitary-adrenal responses. The Journal of neuroscience. 2003;23:7863–7872. doi: 10.1523/JNEUROSCI.23-21-07863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jennings JH, Rizzi G, Stamatakis AM, Ung RL, Stuber GD. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science. 2013;341 doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennings JH, et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell. 2015;160:516–527. doi: 10.1016/j.cell.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proceedings of the National Academy of Sciences of the United States of America. 2007;104 doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton AK, et al. Control of food intake and energy expenditure by nos1 neurons of the paraventricular hypothalamus. The Journal of neuroscience. 2014;34 doi: 10.1523/JNEUROSCI.0226-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garfield AS, et al. A parabrachial-hypothalamic cholecystokinin neurocircuit controls counterregulatory responses to hypoglycemia. Cell metabolism. 2014;20:1030–1037. doi: 10.1016/j.cmet.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter ME, Soden ME, Zweifel LS, Palmiter RD. Genetic identification of a neural circuit that suppresses appetite. Nature. 2013;503:111–114. doi: 10.1038/nature12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai K, Ino H, Yamamoto H, Nakagawa H, Yamano M, Tohyama M, Shiosaka S, Shiotani Y, Inagaki S, Kitoh S. Lesions in the lateral part of the dorsal parabrachial nucleus caused hyperphagia and obesity. J Clin Biochem Nutr. 1987:103–112. [Google Scholar]
- 37.Wu Q, Boyle MP, Palmiter RD. Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell. 2009;137:1225–1234. doi: 10.1016/j.cell.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Q, Clark MS, Palmiter RD. Deciphering a neuronal circuit that mediates appetite. Nature. 2012;483:594–597. doi: 10.1038/nature10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paues J, Mackerlova L, Blomqvist A. Expression of melanocortin-4 receptor by rat parabrachial neurons responsive to immune and aversive stimuli. Neuroscience. 2006;141:287–297. doi: 10.1016/j.neuroscience.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis AM, Stuber GD. Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nature neuroscience. 2012;15 doi: 10.1038/nn.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atasoy D, et al. A genetically specified connectomics approach applied to long-range feeding regulatory circuits. Nature neuroscience. 2014;17:1830–1839. doi: 10.1038/nn.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirchgessner AL, Sclafani A. PVN-hindbrain pathway involved in the hypothalamic hyperphagia-obesity syndrome. Physiology & behavior. 1988;42:517–528. doi: 10.1016/0031-9384(88)90153-9. [DOI] [PubMed] [Google Scholar]
- 44.McCabe JT, DeBellis M, Leibowitz SF. Clonidine-induced feeding: analysis of central sites of action and fiber projections mediating this response. Brain research. 1984;309:85–104. doi: 10.1016/0006-8993(84)91013-8. [DOI] [PubMed] [Google Scholar]
- 45.Stachniak TJ, Ghosh A, Sternson SM. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus-->midbrain pathway for feeding behavior. Neuron. 2014;82:797–808. doi: 10.1016/j.neuron.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai H, Haubensak W, Anthony TE, Anderson DJ. Central amygdala PKC-delta(+) neurons mediate the influence of multiple anorexigenic signals. Nature neuroscience. 2014;17 doi: 10.1038/nn.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carter ME, Han S, Palmiter RD. Parabrachial calcitonin gene-related Peptide neurons mediate conditioned taste aversion. The Journal of neuroscience. 2015;35:4582–4586. doi: 10.1523/JNEUROSCI.3729-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keys A. Human starvation and its consequences. Journal of the American Dietetic Association. 1946;22:582–587. [PubMed] [Google Scholar]
- 49.Hull CL. Principles of behavior, an introduction to behavior theory. x. D Appleton Century; 1943. pp. 421–422. [Google Scholar]
- 50.Margules DL, Olds J. Identical “feeding” and “rewarding” systems in the lateral hypothalamus of rats. Science. 1962;135:374–375. doi: 10.1126/science.135.3501.374. [DOI] [PubMed] [Google Scholar]
- 51.Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nature neuroscience. 2008;11:998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song H, et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17531–17536. doi: 10.1073/pnas.1207238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saxena A, et al. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. BioTechniques. 2012;52:381–385. doi: 10.2144/0000113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nature protocols. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- 57.Tang F, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nature protocols. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abercrombie M. Estimation of nuclear population from microtome sections. The Anatomical record. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- 62.Liu T, et al. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron. 2012;73:511–522. doi: 10.1016/j.neuron.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franklin BJaPG. The Mouse Brain in Stereotaxic Coordinates. 3. Academic Press Elsevier; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.