Abstract
Background & Aims
Risk for colorectal cancer (CRC) can be greatly reduced through screening. To aid in development of screening strategies, we refined models designed to determine risk of CRC by incorporating information from common genetic susceptibility loci.
Methods
Using data collected from more than 12,000 participants in 6 studies performed from 1990 through 2011 in the United States (US) and Germany, we developed risk determination models based on sex, age, family history, genetic risk score (number of risk alleles carried at 27 validated common CRC susceptibility loci), and history of endoscopic examinations. The model was validated using data collected from approximately 1800 participants in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial, conducted from 1993 through 2001 in US.
Results
We identified a CRC genetic risk score that independently predicted which patients in the training set would develop CRC. Compared with determination of risk based only on family history, adding the genetic risk score increased discriminatory accuracy from 0.51 to 0.59 (P=.0028) for men and from 0.52 to 0.56 (P=.14) for women. We calculated age- and sex-specific 10 y CRC absolute risk estimates based on the number of risk alleles, family history, and history of endoscopic examinations. A model that included a genetic risk score better determined the recommended starting age for screening in subjects with and without family histories of CRC. The starting age for high-risk men (family history of CRC and genetic risk score=90%) was 42 y, and for low-risk men (no family history of CRC and genetic risk score=10%) was 52 years. For men with no family history and a high genetic risk score (90%), the starting age would be 47 years; this is an intermediate value that is 5 years earlier than it would be for men with a genetic risk score of 10%. Similar trends were observed in women.
Conclusions
By incorporating information on CRC risk alleles, we created a model to more accurately determine risk for CRC. This model might be used to develop screening and prevention strategies.
Keywords: Risk determination, genome-wide association study, colorectal cancer screening, risk stratification
Introduction
Worldwide, colorectal cancer (CRC) is the third most commonly diagnosed cancer in men and second in women. With about 1.36 million new cancer cases and 694,000 deaths estimated to have occurred in 2012, it is the second leading cause of cancer death.6 Substantial evidence shows that the risk of CRC can be greatly reduced through screening allowing early detection and removal of precancerous lesions.7–11 About 5% of the Westernized population will develop CRC over their lifetime. Individuals at high risk may benefit from earlier or more frequent screening whereas others at lower risk could delay, or reduce frequency of, screening. An improved estimation of the risk of developing CRC would assist both physicians and patients in making more informed screening decisions, and can identify a high-risk subgroup of the population that could benefit from preventive interventions, utilizing various modifiable risk factors of CRC.
Current guidelines for CRC screening generally recommend men and women begin at age 50 years with earlier and more frequent screening for those who have a positive family history of CRC, inflammatory bowel disease, or suspected hereditary CRC syndromes.12, 13 The recent progress in identifying CRC-associated common single nucleotide polymorphisms (SNPs) has offered new opportunities to refine risk determination beyond age and family history. Dunlop et al.14 examined 10 CRC susceptibility loci and found that these loci, along with sex, age, and family history, showed a slight improvement in discriminatory accuracy over a model that did not include these loci. However, the model did not consider prior history of endoscopy, which is a major predictor of future CRC risk as well as a strong correlate with family history.
In this study, we included 27 validated common CRC susceptibility loci identified from genome-wide association studies (GWAS). We built a risk determination model incorporating a genetic risk score computed as the number of risk alleles carried at these loci together with age, sex, family history, and history of endoscopy in more than 12,000 cases and controls and validated the results in approximately 1,800 additional cases and controls.
Methods
Study Participants
The included studies are described in detail in Supplemental Note A. The number of cases and controls and the distributions of age and sex are listed in Supplemental Table S1. Additional details for each study can be found in Peters et al.15 Briefly, CRC cases were defined as colorectal adenocarcinoma and confirmed by medical records, pathology reports, or death certificate. Controls had no history of CRC at the time of ascertainment. All participants gave written informed consent and the studies were approved by their respective Institutional Review Boards.
We included two case-control studies and four nested case-control studies as a training data set for building risk determination models (5,811 cases and 6,302 controls). These include DACHS (Darmkrebs: Chancen der Verhütung durch Screening, 2,369 cases and 2,206 controls); DALS (Diet, Activity and Lifestyle Study, 1,081 cases and 1,174 controls); HPFS (Health Professionals Follow-up Study, 256 cases and 305 controls); NHS (Nurses’ Health Study, 417 cases and 798 controls); VITAL (Vitamins And Lifestyle Study, 278 cases and 288 controls); and WHI (Women’s Health Initiative, 1,410 cases and 1,531 controls). We used PLCO (the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial, 866 cases and 869 controls) for model validation. All analyses were restricted to subjects of European descent because of small numbers from other population groups.
Reference Time, Family History, and Endoscopy
Data on family history and endoscopy were harmonized across studies as part of our standardized data-harmonization procedure, described elsewhere.16 Data were ascertained through self-reported or interviewer-administered questionnaire. We used the risk factor data at reference time, which was defined differently depending on the study design. For the population-based case-control study DACHS, the reference time was the date of diagnosis among the cases and the date of interview among the controls, and for DALS the reference time was approximately two years prior to date of diagnosis (cases) or selection (controls). For the cohort-based studies VITAL, WHI and PLCO, the reference time was study entry. For the HPFS (initiated in 1986) and NHS (initiated in 1976) cohort studies, we used risk-factor information collected nearest the time of blood-sample collection; reference times were blood collection years: 1994 and 1990, respectively.
A positive family history (yes/no) was defined as having one or more first-degree relatives with CRC. Endoscopy (yes/no) was defined as having a sigmoidoscopy or colonoscopy before the reference time to accommodate the limited endoscopy history data available from the studies. For DALS and the cohort-based studies, the reference time was prior to CRC diagnosis; hence, it was unlikely that endoscopy was performed for diagnostic purpose. For DACHS, detailed questionnaire data allowed exclusion of diagnostic endoscopy. For PLCO, we coded the endoscopy variable as “yes” if the participant had had an endoscopy prior to entering the study or had been assigned to the intervention arm and actually had screening; other participants were coded “no” to indicate not having had an endoscopy. Participants diagnosed with CRC within one year of follow-up were excluded. Details on the questions on endoscopy and family history for all studies can be found in Supplemental Note B.
Genotyping and SNP Selection
Genotype data, directly genotyped or imputed, for the known CRC loci were obtained from existing GWAS data. Details on genotyping, quality assurance/quality control, and imputation have been previously published.15
We extracted 31 CRC-risk-associated SNPs that were identified through GWAS as of December 2013 that reached or nearly reached the genome-wide significance 5×10−8 in discovery studies. For regions where multiple SNPs have been reported, we included only the statistically most significant SNP in the Genetics and Epidemiology of Colorectal Cancer Consortium if the D’ value17 or linkage disequilibrium was greater than 0.5. This resulted in a total of 27 SNPs: rs16892766,18 rs6983267,19 rs10795668,18 rs3802842,18 rs4444235,18, 20 rs4779584,20, 21 rs9929218,18 rs4939827,18, 19 rs10411210,18 rs961253,18, 20 rs6687758,22 rs10936599,22 rs1321311,23 rs719725,24, 25 rs3824999,23 rs7136702,22 rs1957636,20 rs4813802,20, 26 rs4925386,22 rs10911251,15 rs11903757,15 rs3217810,15 rs3217901,15 rs59336,15 rs647161,27 rs10774214,27 rs2423279.27 The call rate of genotyped SNPs was >98% and the average R2 for imputed SNPs was 0.93 (Supplemental Table S2).
Statistical Analysis
We estimated the odds ratio (OR) and 95% confidence intervals (CI) of the association of CRC risk with endoscopy, family history of CRC, and SNPs by logistic regression in the training data set. We stratified the model by sex (men and women) and anatomic tumor site (proximal colon, distal colon, and rectum) to allow for potential differential effects of risk factors on different tumor site for men and women. We adjusted for study and age in all models. For model evaluation and absolute risk estimation, we summed the risks for all three-tumor sites as the overall risk for colorectal cancer.
Each SNP was coded as 0, 1, or 2 copies of the risk allele if it was directly genotyped or the expected number of copies of the risk allele if it was imputed. We defined the genetic risk score as the count of risk alleles across all 27 SNPs, which ranged from 11 to 38. Additionally, we calculated the weighted genetic risk score with weights determined by the log-odds ratios estimated from prior studies (Table S2) to avoid potential bias. The performance of the model using the weighted genetic risk score was similar to the model using simple count of risk alleles (Table S7, Supplemental Note A). Therefore, we present the results based on the count score for its simplicity and easy interpretation.
For cohort-based nested case-control studies, we estimated the effect of endoscopy within 10 years of the reference time to reflect the diminished protective effect of endoscopy beyond 10 years.28 To achieve this, we excluded cases whose follow-up times were more than 10 years. We investigated potential population substructure by adjusting for the first three principal components of genetic ancestry obtained from GWAS genotyping data. We found little evidence for the substructure; and therefore, did not include them in the subsequent analyses. This is because the QC protocol for our previous GWAS analysis excluded any individuals who did not cluster well with the Utah residents with Northern and Western European ancestry in HapMap II. We assessed pairwise interactions among age, family history, endoscopy, and genetic risk score by including the product of paired variables, but no interaction was significant at α = 0.05, and hence, we did not include any interaction term in the final model.
We evaluated the discriminatory accuracy of the risk-prediction models by the area under the receiver-operating characteristic curve (AUC) in the validation data set. The AUC value is the probability that the risk score ranks a randomly selected case higher than a randomly selected control. We calculated the risk score for each tumor site based on the site-specific model and took the weighted average of the risk scores with weights equal to probabilities of developing rectal, proximal-colon, and distal-colon cancers obtained from the Surveillance, Epidemiology and End Results (SEER) registry.29 We then estimated the AUC for the risk scores that included family history alone, genetic risk score alone, and both while adjusting for study, age and endoscopy, separately for men and women.30 We obtained the 95% CIs of AUC estimates with 1,000 bootstrap samples. To compare the AUC of two models we calculated the Wald statistic by dividing the observed difference of AUC estimates of the two models by the standard error of the difference obtained from bootstrap.
We estimated the absolute risk and 95% confidence intervals for developing CRC for men and women separately based on the overall hazard rate for CRC, which is a sum of the hazard rates for proximal colon, distal colon, and rectal cancers given family history, genetic risk score and endoscopy.31 As CRC tends to occur at older ages, we accounted for the competing risks of death from non-CRC causes in the absolute risk estimation. The details of the calculation are provided in Supplemental Note A.
We considered a two-sided p-value<0.05 statistically significant. We performed all analyses using the R statistical software package.
Results
In both men and women, cases were more likely to have a positive family history of CRC and were less likely to have an endoscopy compared to controls (Table 1). In addition, cases tended to carry higher numbers of genetic risk alleles than controls (Supplemental Figure S1).
Table 1.
Study characteristics of the training and validation data sets for men and women
| Study | Men
|
Women
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Endoscopy | Family history | Genetic risk score | Total | Endoscopy | Family history | Genetic - risk score | |
|
|
|
|||||||
| N (yes %) | N (yes %) | Mean (SD) | N (yes %) | N (yes %) | Mean (SD) | |||
| Training data | ||||||||
| DACHS | ||||||||
| Proximal | 395 | 135 (34.3) | 60 (15.3) | 24.23 (3.0) | 363 | 119 (32.9) | 52 (14.4) | 24.44 (3.3) |
| Distal | 396 | 76 (19.2) | 53 (13.4) | 24.40 (3.3) | 266 | 56 (21.2) | 45 (16.9) | 24.60 (3.1) |
| Rectal | 615 | 101 (16.4) | 86 (14.0) | 24.42 (3.2) | 334 | 34 (10.2) | 44 (13.2) | 24.74 (3.1) |
| Controls | 1,344 | 748 (55.7) | 126 (9.4) | 23.75 (3.2) | 862 | 464 (54.0) | 106 (12.3) | 23.64 (3.1) |
| DALS | ||||||||
| Proximal | 288 | 97 (37.9) | 54 (18.8) | 24.68 (3.3) | 250 | 72 (33.5) | 46 (18.4) | 24.87 (3.3) |
| Distal | 306 | 65 (24.2) | 47 (15.4) | 24.69 (3.3) | 237 | 50 (24.6) | 33 (13.9) | 24.41 (3.2) |
| Rectal | - | - | - | - | - | - | - | - |
| Controls | 643 | 276 (46.9) | 58 (9.0) | 23.45 (3.3) | 531 | 177 (39.2) | 56 (10.5) | 23.57 (3.3) |
| HPFS | ||||||||
| Proximal | 97 | 21 (23.1) | 24 (24.7) | 24.24 (3.3) | - | - | - | - |
| Distal | 90 | 14 (17.3) | 13 (14.4) | 24.50 (3.4) | - | - | - | - |
| Rectal | 69 | 9 (14.5) | 11 (15.9) | 23.70 (3.0) | - | - | - | - |
| Controls | 305 | 83 (29.1) | 46 (15.1) | 23.46 (3.2) | - | - | - | - |
| NHS | ||||||||
| Proximal | - | - | - | - | 201 | 73 (37.8) | 33 (16.4) | 24.32 (3.4) |
| Distal | - | - | - | - | 123 | 36 (31.3) | 24 (19.5) | 24.47 (3.2) |
| Rectal | - | - | - | - | 93 | 22 (26.2) | 16 (17.2) | 24.43 (3.3) |
| Controls | - | - | - | - | 798 | 267 (36.6) | 109 (13.7) | 23.68 (3.4) |
| VITAL | ||||||||
| Proximal | 70 | 38 (55.9) | 8 (11.8) | 23.98 (3.3) | 73 | 43 (58.9) | 13 (18.1) | 24.41 (3.2) |
| Distal | 35 | 19 (54.3) | 3 (8.8) | 23.90 (3.3) | 34 | 14 (41.2) | 6 (18.2) | 24.24 (2.9) |
| Rectal | 41 | 14 (34.1) | 4 (10.3) | 24.53 (3.5) | 25 | 9 (36.0) | 5 (20.0) | 25.11 (3.1) |
| Controls | 150 | 104 (70.3) | 19 (12.9) | 23.52 (3.1) | 138 | 89 (65.9) | 18 (13.6) | 23.74 (3.3) |
| WHI | ||||||||
| Proximal | - | - | - | - | 783 | 443 (58.8) | 162 (22.2) | 24.19 (3.3) |
| Distal | - | - | - | - | 366 | 149 (43.8) | 55 (16.6) | 24.29 (3.4) |
| Rectal | - | - | - | - | 261 | 96 (38.6) | 44 (18.7) | 24.99 (3.2) |
| Controls | - | - | - | - | 1,531 | 846 (58.0) | 247 (17.6) | 23.92 (3.3) |
| Total | ||||||||
| Proximal | 850 | 291 (36.0) | 146 (17.3) | 24.36 (3.2) | 1,670 | 750 (47.0) | 306 (18.9) | 24.37 (3.3) |
| Distal | 827 | 174 (22.3) | 116 (14.0) | 24.50 (3.3) | 1,026 | 305 (31.9) | 163 (16.5) | 24.42 (3.2) |
| Rectal | 725 | 124 (17.3) | 101 (14.0) | 24.36 (3.2) | 713 | 161 (23.3) | 109 (15.9) | 24.80 (3.2) |
| Controls | 2,442 | 1,211 (51.2) | 249 (10.2) | 23.62 (3.3) | 3,860 | 1,843 (50.7) | 536 (14.4) | 23.75 (3.3) |
| Validation data | ||||||||
| PLCO | ||||||||
| Proximal | 254 | 135 (54.2) | 31 (12.3) | 24.38 (3.3) | 243 | 98 (40.3) | 40 (16.5) | 24.49 (3.3) |
| Distal | 139 | 44 (31.9) | 17 (12.5) | 24.41 (3.1) | 90 | 35 (38.9) | 19 (21.3) | 24.66 (3.0) |
| Rectal | 93 | 37 (40.7) | 13 (14.0) | 24.24 (3.4) | 47 | 18 (38.3) | 3 (6.5) | 24.30 (3.1) |
| Controls | 516 | 384 (75.1) | 56 (11.0) | 23.40 (3.2) | 353 | 216 (61.4) | 49 (14.0) | 23.85 (3.4) |
Association of Endoscopy, Family History, and Genetic risk score with CRC Risk by Tumor Sites in Men and Women
Endoscopy had a strong negative association with CRC risk, particularly with risk of distal colon and rectal cancers (Table 2). Additionally, within controls we found that both men and women with a positive family history of CRC were more likely to have had an endoscopy than those without (Men: OR=1.52, 95% CI, 1.15 to 2.02; Women: OR=1.97, 95% CI, 1.63 to 2.40; Supplementary Table S5). We adjusted for endoscopy in addition to study and age when assessing CRC risk in relation to family history and genetic risk score.
Table 2.
Odds ratio (95% CI) of risk factors associated with CRC risk in the training dataset†
| Variable | Men | Women |
|---|---|---|
| OR (95% CI) | OR (95% CI) | |
| Proximal colon cancer | ||
| Endoscopy | ||
| No | 1.00 | 1.00 |
| Yes | 0.49 (0.41 to 0.58) | 0.71 (0.62 to 0.81) |
| Family history | ||
| No | 1.00 | 1.00 |
| Yes | 2.03 (1.60 to 2.57) | 1.38 (1.16 to 1.63) |
| Genetic risk score | ||
| Allele | 1.07 (1.04 to 1.10) | 1.06 (1.03 to 1.08) |
| Distal colon cancer | ||
| Endoscopy | ||
| No | 1.00 | 1.00 |
| Yes | 0.26 (0.21 to 0.32) | 0.43 (0.36 to 0.50) |
| Family history | ||
| No | 1.00 | 1.00 |
| Yes | 1.68 (1.29 to 2.17) | 1.38 (1.11 to 1.71) |
| Genetic risk score | ||
| Allele | 1.08 (1.06 to 1.11) | 1.08 (1.05 to 1.10) |
| Rectal cancer | ||
| Endoscopy | ||
| No | 1.00 | 1.00 |
| Yes | 0.16 (0.13 to 0.21) | 0.28 (0.22 to 0.34) |
| Family history | ||
| No | 1.00 | 1.00 |
| Yes | 1.67 (1.25 to 2.23) | 1.41 (1.09 to 1.81) |
| Genetic risk score | ||
| Allele | 1.06 (1.03 to 1.09) | 1.12 (1.08 to 1.15) |
The logistic regression model includes study, age, endoscopy, family history and genetic risk score.
Both family history and genetic risk score were independent risk factors for CRC after adjusting for study, age, and endoscopy (Table 2). A positive family history of CRC significantly increased risk for all tumor sites. Similarly, the genetic risk score was statistically significantly associated with increased risk of CRC at all tumor sites in both men and women.
We examined the association by type of study (population-based case-control studies; cohort-based nested case-control studies). The effects of family history and endoscopy were stronger in the population-based case-control studies than in the nested case-control studies; however, patterns were largely consistent (Supplementary Table S6). We also performed a sensitivity analysis excluding the two SNPs, rs59336 (p-value = 3.7×10−7) and rs3217901 (p-value = 5.9×10−8), that did not reach the genome-wide significance and the results were essentially unchanged (Supplemental Table S8 and S9).
Assessment of Risk Determination Models
We assessed the discriminatory accuracy of the models obtained from the training data set in the validation data adjusted for study, age, and endoscopy (Table 3). For men, the AUC estimate for the family history alone was 0.51 (95% CI, 0.48 to 0.53) and for the genetic risk score alone it was 0.60 (95% CI, 0.56 to 0.65). Including both family history and genetic risk score in the model yielded an AUC = 0.59 (95% CI, 0.54 to 0.64), which showed no difference from the genetic-risk-score-only model (p-value = 0.57), but significantly improved prediction over the family-history-only model (p-value = 0.0028).
Table 3.
AUC comparisons between risk determination models†
| Men (N=890) AUC (95% CI) |
Women (N=666) AUC (95% CI) |
|||
|---|---|---|---|---|
| Model I | ||||
| Family history | 0.51 (0.48 to 0.53) | PI vs. III = 0.0028 | 0.52 (0.50 to 0.55) | PI vs. III = 0.14 |
| Model II | ||||
| Genetic risk score | 0.60 (0.56 to 0.65) | PII vs. III = 0.57 | 0.55 (0.50 to 0.60) | PII vs. III = 0.57 |
| Model III | ||||
| Family history & Genetic risk score | 0.59 (0.54 to 0.64) | 0.56 (0.51 to 0.61) | ||
The analyses were adjusted for study, age and endoscopy.
The AUC estimates for women were 0.52 (95% CI, 0.50 to 0.55) and 0.55 (95% CI, 0.50 to 0.60) for family history alone and genetic risk score alone, respectively. The AUC estimate for the model that included both family history and genetic risk score was 0.56 (95% CI, 0.51 to 0.61), which showed improvement but not statistically significant over the model with family history alone (p = 0.14). It did not show improvement over the model with the genetic risk score only (p-value = 0.57).
Probability of Developing CRC Based on Endoscopy, Family History, and Genetic risk score
The absolute risk estimates vary greatly for men depending on risk profiles (Table 4). To compare the absolute-risk estimates based on our model, we calculated two reference risks: the projected average risk based on SEER CRC incidence rates that included subjects with and without an endoscopy, and the projected average risk of subjects who did not have an endoscopy. To illustrate, for a 60-year-old man, the average risk of developing CRC in the next 10 years was 1.68% regardless of endoscopy status, and 2.70% in the absence of endoscopy. If he had not had an endoscopy, had a positive family history of CRC, and carried 28 risk alleles (the 90th percentile), his risk of developing CRC in the next 10 years was 5.79%. However, if he had had an endoscopy, despite his positive family history, his risk was only 1.75%, very similar to the average risk of a 60-year-old man in the general population (1.68%), and below the average risk of a 60-year-old without an endoscopy (2.70%). For a man with low-risk profile, e.g., no positive family history and carrying 20 risk alleles (the 10th percentile), the risk was 1.93%, even in the absence of endoscopy and only 0.55% if he had had an endoscopy. This is substantially lower than the average risk of a 60-year-old man in the general population (1.68%).
Table 4.
Examples of 10-year absolute risk estimates for CRC with different risk factor profiles.
| Endoscopy | Family history | Genetic-risk score | Men
|
Women
|
|||
|---|---|---|---|---|---|---|---|
| Risk(%) | 95% CI | Risk(%) | 95% CI | ||||
| Age=50 | |||||||
| Reference | Average risk1 | 0.69 | 0.49 | ||||
| Average risk (no endoscopy)2 | 1.13 | 1.07 to 1.19 | 0.68 | 0.65 to 0.71 | |||
| No | No | 20 | 0.81 | 0.73 to 0.89 | 0.47 | 0.42 to 0.51 | |
| No | No | 24 | 1.06 | 1.00 to 1.12 | 0.64 | 0.61 to 0.68 | |
| No | No | 28 | 1.39 | 1.26 to 1.53 | 0.90 | 0.82 to 0.97 | |
| No | Yes | 20 | 1.41 | 1.13 to 1.70 | 0.65 | 0.54 to 0.75 | |
| No | Yes | 24 | 1.85 | 1.52 to 2.19 | 0.89 | 0.76 to 1.03 | |
| No | Yes | 28 | 2.43 | 1.96 to 2.90 | 1.24 | 1.04 to 1.45 | |
| Yes | No | 20 | 0.22 | 0.19 to 0.24 | 0.22 | 0.20 to 0.24 | |
| Yes | No | 24 | 0.29 | 0.26 to 0.31 | 0.29 | 0.27 to 0.31 | |
| Yes | No | 28 | 0.38 | 0.34 to 0.43 | 0.39 | 0.35 to 0.43 | |
| Yes | Yes | 20 | 0.40 | 0.32 to 0.47 | 0.30 | 0.26 to 0.34 | |
| Yes | Yes | 24 | 0.52 | 0.43 to 0.62 | 0.40 | 0.35 to 0.45 | |
| Yes | Yes | 28 | 0.69 | 0.56 to 0.83 | 0.54 | 0.47 to 0.62 | |
| Age=60 | |||||||
| Reference | Average risk | 1.68 | 1.16 | ||||
| Average risk (no endoscopy) | 2.70 | 2.56 to 2.85 | 1.57 | 1.49 to 1.64 | |||
| No | No | 20 | 1.93 | 1.74 to 2.12 | 1.09 | 1.00 to 1.19 | |
| No | No | 24 | 2.53 | 2.38 to 2.68 | 1.48 | 1.41 to 1.56 | |
| No | No | 28 | 3.32 | 3.01 to 3.64 | 2.03 | 1.87 to 2.19 | |
| No | Yes | 20 | 3.38 | 2.72 to 4.04 | 1.51 | 1.27 to 1.75 | |
| No | Yes | 24 | 4.43 | 3.65 to 5.20 | 2.05 | 1.76 to 2.34 | |
| No | Yes | 28 | 5.79 | 4.70 to 6.88 | 2.81 | 2.37 to 3.24 | |
| Yes | No | 20 | 0.55 | 0.49 to 0.61 | 0.54 | 0.49 to 0.60 | |
| Yes | No | 24 | 0.72 | 0.66 to 0.79 | 0.72 | 0.67 to 0.77 | |
| Yes | No | 28 | 0.96 | 0.84 to 1.07 | 0.95 | 0.86 to 1.04 | |
| Yes | Yes | 20 | 1.00 | 0.81 to 1.20 | 0.75 | 0.65 to 0.86 | |
| Yes | Yes | 24 | 1.33 | 1.09 to 1.56 | 0.99 | 0.87 to 1.11 | |
| Yes | Yes | 28 | 1.75 | 1.41 to 2.09 | 1.31 | 1.13 to 1.49 | |
Average risks were calculated based on SEER incidence rates for men and women separately.
Average risks (no endoscopy) were calculated based on a model including endoscopy only and the SEER incidence rates for men and women separately.
The absolute risk of developing CRC among women exhibited a similar pattern to men, but with overall lower risk (Table 4). The population average risk of developing CRC for a 60-year-old woman was 1.16% regardless of endoscopy status and 1.57% in the absence of endoscopy. With a positive family history and carrying 28 risk alleles (the 90th percentile), her risk for developing CRC was 2.81% without endoscopy, but 1.31% with endoscopy, below the average risk of a woman who had not had an endoscopy (1.57%). In contrast, if the woman has low-risk profile (e.g., no positive family history of CRC and the genetic risk score at 10%), her 10-year risk for developing CRC was only 1.09% even without endoscopy, and the risk would be further reduced to 0.54% if she had had an endoscopy.
Utilizing Genetic risk score for Risk Stratification
We assessed the potential of our risk determination models to improve risk stratification by calculating age for screening if the subject’s calculated risk exceeded a pre-specified threshold. Since our models include history of endoscopy (yes/no), we can estimate absolute risk for subjects who have and have not had screening. Hence, for those who have not had screening, we can use the models to calculate the starting age for screening; for those who have had screening, it would be the age for the next screening. Here for illustration we focus on the former, i.e., the starting age for screening. To our knowledge, there is no agreed upon threshold of CRC risk to initiate screening. As 50 years of age is the recommended starting age for screening in the general population, we used as the threshold the average of 10-year risk of 50-year-old men (1.13%) and women (0.68%) who did not have endoscopy, i.e., 0.91% = (1.13% + 0.68%)/2.
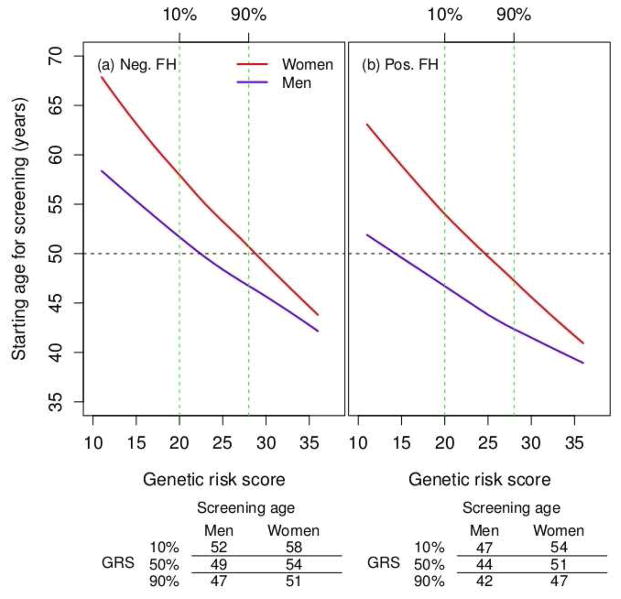
Based on this average risk threshold, the starting age for screening for men with and without a positive family history was 44 and 49 years, respectively based on the family-history-alone model. For women, the corresponding age was 50 and 54 years, respectively. Including genetic-variant information led to a wider age range for recommending the start of screening (Figure 1). The starting age for screening for high-risk men (a positive family history of CRC and a genetic risk score at the 90th percentile) was 42 years old. In contrast, for men at low-risk of CRC (no family history of CRC and a genetic risk score at the 10th percentile), the starting age was 52 years. For a man with no family history of CRC and a genetic risk score at the 90th percentile, the starting age would be 47 years, an intermediate value, 5 years earlier than it would be with a genetic risk score at the 10th percentile. The starting age for screening for high- and low-risk women was 47 and 58 years, respectively.
Figure 1.
Starting age for screening vs. genetic risk score by family history. (a) negative family history of CRC (Neg. FH); (b) positive family history of CRC (Pos. FH). The risk threshold is set to be 0.91%, the average 10-year risk for 50 years old subjects. The red and blue solid lines are for women and men, respectively. The horizontal line corresponds to reference age 50 years old. The two vertical lines correspond to the 10% and 90% of genetic risk score in the controls.
Discussion
Using data from six epidemiologic studies, we have shown that a genetic risk score based on common CRC susceptibility loci is an independent predictor of CRC risk in the presence of family history. Compared to the model including only family history, which the current guidelines use to define increased risk groups for screening,12 a risk-prediction model that incorporates both a genetic risk score and family history improves the discriminatory accuracy between CRC cases and controls in men and could lead to more individually tailored screening recommendations. It is important to note that while each risk locus by itself has moderate association, there is a noteworthy cumulative effect; particularly these are common variants and impact a sizeable fraction of the population. The differences in age to start screening for the top 90% and the bottom 10% of genetic risk score are 5 years for men and 7 years for women. This could mean that subjects at very low risk require fewer endoscopies in their lifetime, which will help reduce unnecessary procedures; those at very high risk may need to start endoscopy at an earlier age, resulting in more endoscopies over their lifetime to reduce risk for developing CRC.
Our study adds to previously reported risk-prediction models, see Win et al. 32 (and references therein) for a comprehensive review. To our knowledge, only two papers have used CRC-associated common genetic variants for risk determination. Jo et al.33 reported that the AUCs for genetic risk scores of top SNPs along with age and family history were 0.729 and 0.650 for Korean men and women, respectively. However, these top SNPs were chosen from the same data set that was used for calculating the AUCs. As a result, these AUC estimates are likely to be inflated. Dunlop et al.14 included 10 common genetic variants and estimated the AUC along with age, sex, and family history in independent validation studies of Swedish (n=3,067, AUC=0.56) and Finnish (n=1,120, AUC=0.57) populations. We included 27 validated CRC susceptibility loci that had been identified in the literature as of December 2013 (more than the previously published studies) in addition to family history, age, and sex. It is important to note that we included history of endoscopy in the model because it is strongly protective as well as strongly associated with a family history. By adjusting for endoscopy, we provide a more accurate assessment of the performance of risk-prediction models.
There are several strengths to our study. We have a large sample size with more than 12,000 participants in the training data set. Most CRC susceptibility loci were identified previously, not using our study samples, which avoids the bias from winner’s curse, where the effects are likely to be stronger in the discovery samples than in the general population.34 Our training data set included both population-based and cohort-based case-control studies. There are inherited differences such as recall and sample selection bias between these two study designs.35 Another notable difference is the timing of the risk factor information collected. For the population-based case-control studies risk factors are usually collected around the time of case and control ascertainment. In contrast, for case-control studies nested in prospective cohorts they are collected at study entry prior to the development of cancer in cases and ascertainment of matched controls. Because the protective effect of endoscopy diminishes over time and family history and endoscopy can change, it would be expected that the effects be attenuated towards the null for nested case-control studies. Despite these potential differences, we find the association patterns are consistent between these two types of studies (Supplemental Table S6), suggesting our risk estimates are generally robust. Our validation data set is based on the large PLCO screening trial with standardized protocol, implementation, and monitoring of outcomes, which allows us to evaluate risk-prediction models rigorously.
There are limitations in our study. First, information on hereditary nonpolyposis colorectal cancer was not collected across studies, hence, we were not able to examine directly if added common genetic risk loci would improve risk prediction for subjects with rare-high-penetrance mutations. Previous works have found the modifying effects of common SNPs in carriers of mismatch repair gene mutations;36 however these results were not replicated in other studies.37, 38 Using family history as a surrogate, we found that the genetic risk score of common loci remains to be significantly associated with increased risk of CRC in both men and women with a positive family history of CRC (results not shown), suggesting common genetic loci improve risk prediction in this group. Second, although our independent validation study includes approximately 1,800 subjects, we validated the models for men (~1,100) and women (~700) separately; therefore, the sample size is somewhat limited. Nevertheless, even with the current sample size, we were able to detect statistically significant AUC improvement for men although not for women. In addition, the validation study is a case-control study, which limits our ability to assess model calibration and fully evaluate the public health impact of our models. Third, our study samples are limited to subjects of European descent because the studies have small numbers from other populations. Previous work shows that, if the sample size is large enough, GWAS loci tend to replicate across ethnicities if tagging SNPs are included, and thus, the AUC estimates are likely to be generalizable. However, CRC incidence rates differ across populations and the distribution of risk factors probably differs. Our absolute risk estimates will need to be re-calibrated in non-European populations.
There are also limitations in predictors. We only included a variable for family history of CRC in first-degree relatives (yes/no). Although there was no association of the number of first-degree relatives with CRC risk after adjusting for this yes/no family history variable (data not shown), more detailed information on family history such as ages of affected relatives, history of non-CRC cancers and adenomas in first-degree relatives and family history beyond first-degree relatives could provide additional predictive power. Nonetheless, first-sgree family history is reported with more accuracy than second-degree family history and the risk conferred by second-degree relatives is comparably small.39 In addition, the level of family history data that we have in GECCO seems reflective of what would actually be collected for an average person. For determining risk in the general population, it is important to use the information that is commonly available. In this case, it is interesting to note that the genetic risk score based on common variants is adding independent information beyond the yes/no first-degree family history. We combined colonoscopy and sigmoidoscopy into one variable, endoscopy, due to lack of more detailed information in some studies, nor did we include data on frequency of screening and the results of endoscopy, nor can we account for endoscopy that was not reported or tracked. Accordingly, our model could be further refined if this additional information were available.
Our risk models can be further improved. Many environmental and lifestyle factors such as diet, obesity, non-steroidal anti-inflammatory drugs, post-menopausal hormone use, smoking and alcohol have been shown associated with CRC risk.40 In our study we also identified lower risks in women; this translated into an upward shift in the age to start screening of approximately 4–7 years across categories in women compared to men. This may in part be explained by differences in the risk profile of environmental factors such as red meat intake, hormone replacement therapy use, smoking and alcohol use. Accordingly, it will be important to investigate in future models if adding these environmental factors will further improve the risk determination and possibly explain the difference between men and women. Our genetic risk score is calculated based on tagging variants. There is ongoing work to identify the underlying functional or causal variants with potentially stronger effects, and updated analysis using these may further improve the risk determination. Furthermore, CRC has a sizable heritable component and there remains more information in the GWAS data that has not been fully utilized. Indeed, it has been shown that incorporating a larger number of variants, for example, the 500 or 1,000 most statistically significant variants, not just those reaching genome-wide significance, improves risk determination.41–43 Besides common variants, rare variants with stronger effects may also help identify individuals, though few in number, at markedly increased risk for CRC. A comprehensive risk determination model based on genome-wide genetic variants including rare variants, environmental factors, and gene-gene and gene-environment interactions will likely further improve risk determination as well as help understand the risk differences between men and women.
In summary, we demonstrate that the genetic risk score comprised of 27 known CRC loci is an independent risk factor and improves discriminatory accuracy compared to a model based on family history. We also show that genetic risk score could potentially be used to refine screening decision such as age to start screening. However, an important factor for determining whether the genetic risk score should be incorporated into screening strategies is the costs and comparative gain of targeted or genome-wide genetic testing in relation to current guidelines. A formal evaluation of the cost-effectiveness is beyond the scope of this paper. However, our findings show that risk-prediction model development using genetic information permits more accurate, individually tailored screening and prevention strategies, warranting comparative effectiveness studies.
Supplementary Material
Acknowledgments
We thank all study participants and investigators for making this project possible. The detailed acknowledgements are provided in Supplementary Note A.
Grant Support
This work was supported by the National Institutes of Health (P01 CA53996 and R01AG14358, R25CA094880). The grant support for individual studies is provided below.
GECCO: National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088; R01 CA059045).
CORECT: National Cancer Institute, National Institutes of Health under RFA # CA-09-002 (U19 CA148107). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in CORECT, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or CORECT.
DACHS: German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1), and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814).
DALS: National Institutes of Health (R01 CA48998 to M.L.S).
HPFS is supported by the National Institutes of Health (P01 CA 055075, UM1 CA167552, R01 137178, and P50 CA 127003), and NHS by the National Institutes of Health (R01 CA137178, P01 CA 087969 and P50 CA 127003).
PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. Additionally, a subset of control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS)1 Prostate Cancer GWAS,2 Colon CGEMS pancreatic cancer scan (PanScan),3, 4 and the Lung Cancer and Smoking study.5 The prostate and PanScan study datasets were accessed with appropriate approval through the dbGaP online resource (http://cgems.cancer.gov/data/) accession numbers phs000207.v1.p1 and phs000206.v3.p2, respectively, and the lung datasets were accessed from the dbGaP website (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000093.v2.p2. Funding for the Lung Cancer and Smoking study was provided by National Institutes of Health (NIH), Genes, Environment and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446, and NIH GEI U01 HG 004438. For the lung study, the GENEVA Coordinating Center provided assistance with genotype cleaning and general study coordination, and the Johns Hopkins University Center for Inherited Disease Research conducted genotyping.
VITAL: National Institutes of Health (K05 CA154337).
WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
Abbreviations
- AUC
area under the receiver-operating characteristic curve
- AR
attributable risk
- CI
confidence intervals
- CORECT
Colorectal Transdisciplinary Study
- CRC
colorectal cancer
- D′
normalized linkage disequilibrium
- DACHS
Darmkrebs: Chancen der Verhütung durch Screening
- DALS
Diet, Activity and Lifestyle Study
- GECCO
Genetics and Epidemiology of Colorectal Cancer Consortium
- GWAS
genome-wide association studies
- HPFS
Health Professionals Follow-up Study
- NHS
Nurses’ Health Study
- OR
odds ratio
- PLCO
the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial
- SEER
Surveillance, Epidemiology and End Results registry
- SNPs
single nucleotide polymorphisms
- VITAL
Vitamins And Lifestyle Study
- WHI
Women’s Health Initiative
Footnotes
Disclosures: There are no conflicts of interest to disclose
Part of this study was presented as an oral presentation at 2014 AACR in San Diego.
Author Contributions
Conceived and designed the experiments: SB, HB, AC, JCC, SG, TH, RH, MH, LH, DT, CH, JJ, PN, UP, JP, RP, RS, FS, DS, MS, MT, YZ. Collected phenotype data and biological samples and contributed these as investigators for their respective study: SB, HB, JCC, RH, MH, PN, JP, RP, RS, FS, MS, EW, JG, MD. Analyzed and interpreted the data: LH, JJ, RP, UP, YZ. Contributed reagents/materials/analysis tools: LH, JJ, YZ. Wrote the manuscript: LH, JJ, UP, EW. Critically reviewed the manuscript drafts and approved the final manuscript: All authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Institute NC. Cancer Genetic Markers of Susceptibility (CGEMS) data website. 2009;2013 [Google Scholar]
- 2.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 3.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–90. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15. 33. Nat Genet. 2010;42:224–8. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlay JSI, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] Lyon, France: International Agency for Research on Cancer; 2013. [Google Scholar]
- 7.Brenner H, Stock C, Hoffmeister M. Reduction of colorectal cancer incidence and mortality by screening sigmoidoscopy and screening colonoscopy: Systematic review and meta-analysis of randomized controlled trials and observational studies. BMJ. 2014 doi: 10.1136/bmj.g2467. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–60. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 9.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoen RE, Pinsky PF, Weissfeld JL, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012;366:2345–57. doi: 10.1056/NEJMoa1114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RL, Ward EM, Jemal A. Trends in colorectal cancer incidence rates in the United States by tumor location and stage, 1992–2008. Cancer Epidemiol Biomarkers Prev. 2012;21:411–6. doi: 10.1158/1055-9965.EPI-11-1020. [DOI] [PubMed] [Google Scholar]
- 12.USPSTF. Screening for colorectal cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2008;149:627–37. doi: 10.7326/0003-4819-149-9-200811040-00243. [DOI] [PubMed] [Google Scholar]
- 13.Rex DK, Johnson DA, Anderson JC, et al. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected] Am J Gastroenterol. 2009;104:739–50. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- 14.Dunlop MG, Tenesa A, Farrington SM, et al. Cumulative impact of common genetic variants and other risk factors on colorectal cancer risk in 42,103 individuals. Gut. 2013;62:871–81. doi: 10.1136/gutjnl-2011-300537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters U, Jiao S, Schumacher FR, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutter CM, Chang-Claude J, Slattery ML, et al. Characterization of geneenvironment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72:2036–44. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23. 3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 19.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24. 21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 20.Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genomewide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13. 33. Nat Genet. 2010;42:973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunlop MG, Dobbins SE, Farrington SM, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–6. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 25.Kocarnik JD, Hutter CM, Slattery ML, et al. Characterization of 9p24 risk locus and colorectal adenoma and cancer: gene-environment interaction and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19:3131–9. doi: 10.1158/1055-9965.EPI-10-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters U, Hutter CM, Hsu L, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131:217–34. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia WH, Zhang B, Matsuo K, et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet. 2013;45:191–6. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb PA, Storer BE, Morimoto LM, et al. Long-term efficacy of sigmoidoscopy in the reduction of colorectal cancer incidence. J Natl Cancer Inst. 2003;95:622–5. doi: 10.1093/jnci/95.8.622. [DOI] [PubMed] [Google Scholar]
- 29.SEER. Surveillance, Epidemiology, and End Results (SEER) Program. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch; 2012. ( www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 9 Regs Research Data, Nov 2011 Sub (1973–2010) <Katrina/Rita Population Adjustment> - Linked To County Attributes - Total U.S., 1969–2010 Counties. released April 2013, based on the November 2012 submission. [Google Scholar]
- 30.Janes H, Longton G, Pepe M. Accommodating Covariates in ROC Analysis. Stata J. 2009;9:17–39. [PMC free article] [PubMed] [Google Scholar]
- 31.Freedman AN, Slattery ML, Ballard-Barbash R, et al. Colorectal cancer risk prediction tool for white men and women without known susceptibility. J Clin Oncol. 2009;27:686–93. doi: 10.1200/JCO.2008.17.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Win AK, Macinnis RJ, Hopper JL, et al. Risk prediction models for colorectal cancer: a review. Cancer Epidemiol Biomarkers Prev. 2012;21:398–410. doi: 10.1158/1055-9965.EPI-11-0771. [DOI] [PubMed] [Google Scholar]
- 33.Jo J, Nam CM, Sull JW, et al. Prediction of Colorectal Cancer Risk Using a Genetic Risk Score: The Korean Cancer Prevention Study-II (KCPS-II) Genomics Inform. 2012;10:175–83. doi: 10.5808/GI.2012.10.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong H, Prentice RL. Correcting “winner’s curse” in odds ratios from genomewide association findings for major complex human diseases. Genet Epidemiol. 2010;34:78–91. doi: 10.1002/gepi.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koepsell TD, Weiss NS. Epidemiologic methods : studying the occurrence of illness. Oxford ; New York: Oxford University Press; 2003. [Google Scholar]
- 36.Talseth-Palmer BA, Wijnen JT, Brenne IS, et al. Combined analysis of three Lynch syndrome cohorts confirms the modifying effects of 8q23.3 and 11q23. 1 in MLH1 mutation carriers. Int J Cancer. 2013;132:1556–64. doi: 10.1002/ijc.27843. [DOI] [PubMed] [Google Scholar]
- 37.Win AK, Hopper JL, Buchanan DD, et al. Are the common genetic variants associated with colorectal cancer risk for DNA mismatch repair gene mutation carriers? Eur J Cancer. 2013;49:1578–87. doi: 10.1016/j.ejca.2013.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houlle S, Charbonnier F, Houivet E, et al. Evaluation of Lynch syndrome modifier genes in 748 MMR mutation carriers. Eur J Hum Genet. 2011;19:887–92. doi: 10.1038/ejhg.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor DP, Burt RW, Williams MS, et al. Population-Based Family History-Specific Risks for Colorectal Cancer: A Constellation Approach. Gastroenterology. 2010;138:877–885. doi: 10.1053/j.gastro.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–7. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatterjee N, Wheeler B, Sampson J, et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45:400–5. 405e1–3. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Z, Wang W, Bradfield J, et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92:1008–12. doi: 10.1016/j.ajhg.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kooperberg C, LeBlanc M, Obenchain V. Risk prediction using genome-wide association studies. Genet Epidemiol. 2010;34:643–52. doi: 10.1002/gepi.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.