Abstract
Profound insights regarding nucleic acid structure and function can be gleaned from very simple, direct, and chemistry-based strategies. Our approach strives to incorporate the elegant physical insights that Don Crothers instilled in those who trained in his laboratory. Don emphasized the advantages of focusing on direct and concise experiments, even when the final objective was to understand something complex – potentially including the large-scale architectures of the genomes of RNA viruses and the transcriptomes of cells. Here, I review the intellectual path, plus a few detours, that led to development of the SHAPE-MaP and RING-MaP technologies for interrogating RNA structure and function at large scales. I also argue that greater attention to creating direct, less inferential experiments will convert 'omics investigations into lasting and definitive contributions to our understanding of biological function.
The Crothers Lab signature of physical insight and simplicity
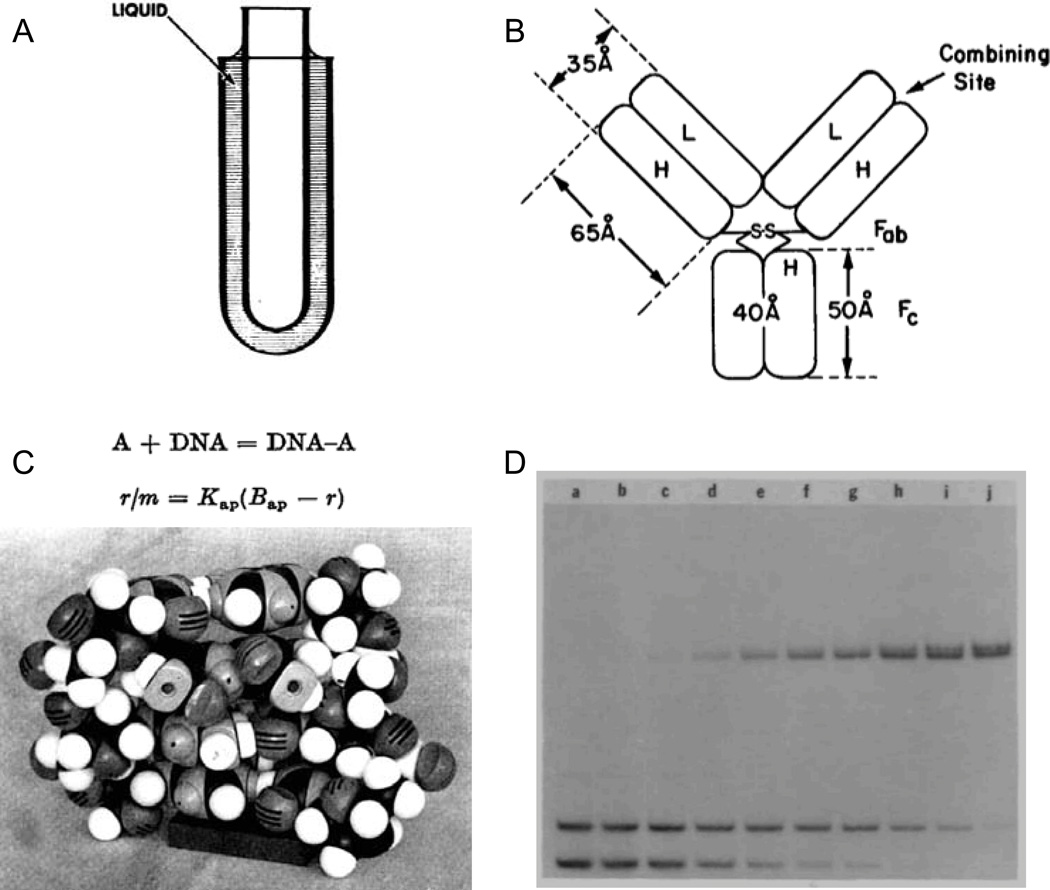
At one point during a (notably rare) cleanup day in the Crothers' lab, a dusty apparatus comprised of glass parts and tubing was discovered, and we wondered if it should be thrown out. When asked, Don responded with his funny laugh and said this was an example of the rotating viscometer he developed with Bruno Zimm and that we should keep it. Although it had the look of a discarded high school science project, the viscometer reflected deep and careful thinking and was capable of very accurate measurements of the effects of DNA on the properties of a solution. The critical insight in developing this instrument was to simplify it: The instrument measured the speed of rotation of an inner glass cylinder that was suspended in a solution containing DNA as an outer cylinder was rotated at a known speed (Fig. 1A). As noted in the abstract of the manuscript reporting this instrument1 "no mechanical devices [are] attached to the moving cylinder … This feature greatly simplifies the construction of the viscometer and permits the measurement of viscosities at very low shear rates with high precision."
Figure 1.
Representative Crothers’ laboratory work that emphasized simple and direct physical ideas and experimentation to understand biological macromolecules. (A) The Zimm-Crothers viscometer,1 (B) model for understanding the effect of polyvalency on antibody-ligand interactions,3 (C) model for drug-DNA intercalation based on measurements using equilibrium dialysis,4 and (D) the Fried-Crothers electrophoretic mobility shift assay.7
The Crothers lab applied this principle of simplicity and experimental concision in a remarkable variety of lasting work. Consider the following manuscripts and a few representative illustrations:
Simplified rotating cylinder viscometer for DNA. Zimm and Crothers, Proc. Natl. Acad. Sci. USA 1962.1 295 citations2 (Fig. 1A).
The influence of polyvalency on the binding properties of antibodies. Crothers and Metzger, Immunochem. 1972.3 410 citations (Fig. 1B).
Studies of the binding of actinomycin and related compounds to DNA. Müller and Crothers, J. Mol. Biol. 1968. and Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on the interaction of daunomycin with deoxyribonucleic acid. Chaires, Dattagupta and Crothers, Biochemistry 1982.4,5 1520 citations (Fig. 1C).
Bent helical structure in kinetoplast DNA. Marini, Levene, Crothers and Englund, Proc. Natl. Acad. Sci. USA 1982.6 630 citations.
Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Fried and Crothers, Nucl. Acids Res. 1981.7 2570 citations (Fig. 1D).
These papers, among Don's most influential, share key features. All represent lasting, non-trendy contributions to understanding macromolecular and nucleic acid structure and function; were published in thoughtfully edited, respected journals but not in the short list of science fashion journals; include extensive validation based on either new experiments or on the prior literature; use remarkably measured language in their Abstracts, different from the buzzword compliant prose in wide use today; and are distinguished by their experimental simplicity. Even though each describes or uses a relatively simple approach, the results and information obtained from the experiments described are superior to more complex alternatives. Finally, and perhaps most strikingly, you can explain the core idea any of these works to a bright undergraduate in about six minutes.
Challenges in RNA structure interrogation
I transition here to reviewing recent work from my laboratory in which we strive to quantitatively examine structural and functional features of nucleic acids, especially RNA, with an intense focus on using the simplest and most concise possible approaches. RNA continues to surprise us, in part, because of its special ability to encode information at many levels and scales. Representative specific roles of the RNA primary sequence, secondary structure, higher-order tertiary structure, and quaternary structures involving interactions with metabolites, other RNAs, and proteins are well-established. What is new and perhaps most exciting are the vast scales over which the distinct levels of RNA structural organization operate, often simultaneously. For example, long viral and mRNAs encode proteins at the level of individual nucleotide triplet codons and form long-range interactions often spanning hundreds or thousands of nucleotides that are essential for juxtaposition of critical regulatory elements. These RNAs, plus the diverse world of long non-coding RNAs, are recognized by proteins and transacting small and large RNAs whose molecular interactions are governed by short nucleotide sequences and are modulated by post-transcriptional modifications such as methylation of single functional groups. The accessibility of these interaction sites can be further enhanced or sequestered by secondary and tertiary structure motifs spanning hundreds of nucleotides. Recovering all this information, operating on such divergent chemical, length, and structural scales is a major contemporary challenge for understanding the genetic code as expressed through RNA.
SHAPE
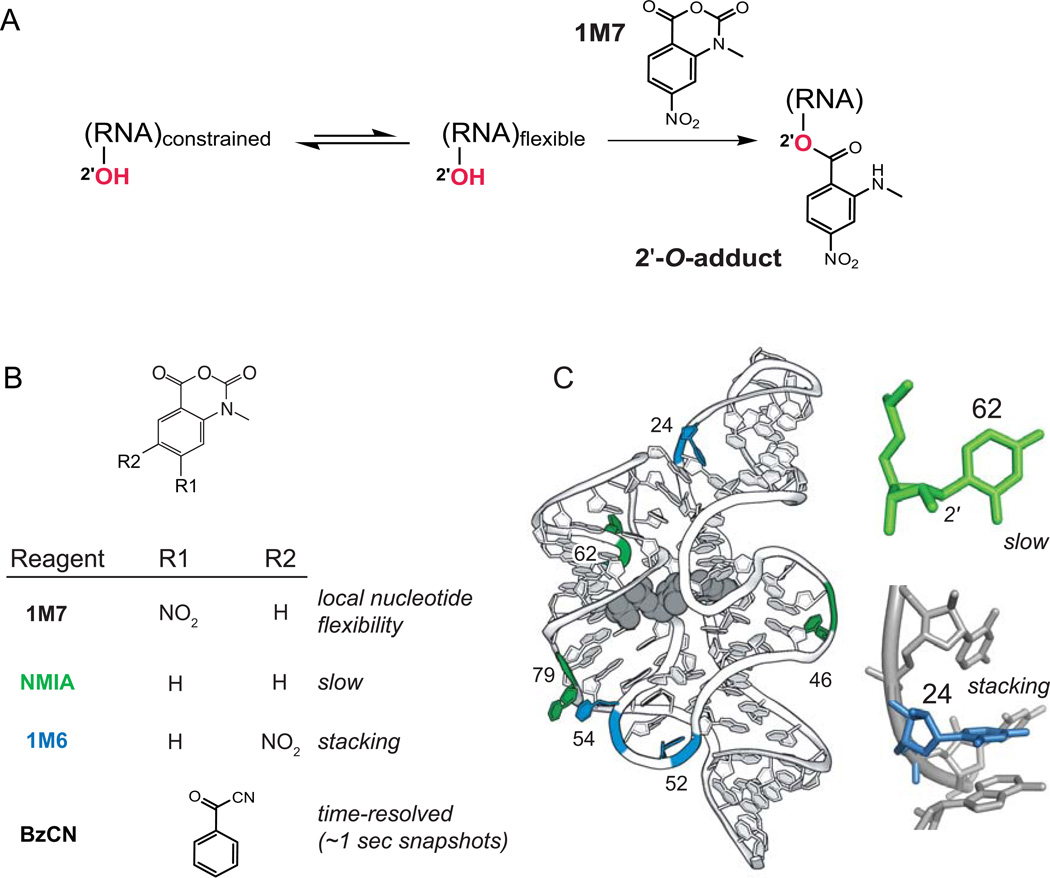
Thinking as students of Don Crothers, my colleagues and I sought to find a simple chemical strategy that might structurally interrogate a ubiquitous functional group in RNA. We focused on performing chemistry at the ribose 2'-hydroxyl group under biologically realistic conditions. Since almost every RNA nucleotide has a 2'-hydroxyl (except those with post-transcriptional modifications at this position), by probing this functional group, every nucleotide in an RNA could be examined in a single experiment. Ultimately, we found a family of anhydride agents that react with the 2'-hydroxyl group (Fig. 2A).8,9 Reactivity of these reagents is highly sensitive to the local nucleotide flexibility of a given nucleotide in way that correlates strongly with much more complex measurements including the model free order parameter.10 The reason for this dependence on local nucleotide flexibility is that 2'-hydroxyl groups are not very reactive, especially in water. The reaction with SHAPE reagents is facilitated by general base catalysis by neighboring groups in the RNA.11 Conformationally dynamic nucleotides more readily achieve the unusual states that juxtapose 2'-hydroxyl groups proximal to a catalytic group.
Figure 2.
SHAPE chemistry monitors local nucleotide dynamics and non-canonical interactions. (A) Mechanism of RNA SHAPE chemistry to form 2'-O-adducts at flexible nucleotides. (B) Core set of useful SHAPE reagents. (C) Structural contexts for nucleotides exhibiting NMIA (green) and 1M6 (blue) enhancements superimposed on a model for the TPP riboswitch aptamer domain; the bound ligand shown in gray. Structure illustrations for the TPP riboswitch are based on 2gdi.30
Initially, the sites of structure-selective 2'-hydroxyl acylation were detected as stops to reverse transcriptase-mediated primer extension, an approach now called SHAPE. Extensive biophysical investigations showed that SHAPE reactivity is independent of nucleotide identity and faithfully measures local nucleotide disorder and dynamics.9 SHAPE gives a high-resolution view of the structural dynamics of complex RNAs and can be used to visualize perturbations to RNA structure including those induced by small molecule ligand binding, solution conditions and ion environment, mutation, interaction with proteins, and the complex effects of the cellular environment (Fig. 3).9,12,13
Figure 3.
SHAPE reactivities used to understand the structure and ligand-induced folding of the TPP riboswitch aptamer domain. (A) Superposition of absolute SHAPE reactivities. (B) Changes in local nucleotide dynamics induced upon binding by the TPP ligand.
Because SHAPE reagents work by such straightforward chemistry and involve simple molecular frameworks, they can be varied in numerous useful ways. One advance was to identify reagents that react very rapidly with RNA. Benzoyl cyanide, BzCN (Fig. 2B), is an example. BzCN reacts with RNA in a period of 1 sec, allowing RNA folding and RNA-protein assembly reactions to be readily monitored over short snapshots.14–16 A second development has been to create reagents whose physical properties cause them to react in distinct ways with RNA nucleotides that are in unusual or special conformations. By varying the substituents on the simple two-ring isatoic anhydride framework, it was possible to create reagents that (i) react on relatively slow timescales and (ii) react selectively at sites where it is possible to stack on the exposed face of a nucleobase (Fig. 2B,C). It turns out that nucleotides that react slowly with a SHAPE reagent tend to exist in the relatively unusual C2'-endo conformation (most nucleotides in RNA are in the C3'-endo conformation). Thus, using these "slow" and "stacking" reagents – NMIA and 1M6, respectively – non-canonical and tertiary interactions can be detected using very simple chemistry (Fig. 2C).17,18
SHAPE and RNA secondary structure modeling
One compelling application of the SHAPE RNA structure probing framework has been to develop robust experimentally directed models of RNA secondary structures, at large scales and at nucleotide resolution. The starting point for SHAPE-directed RNA structure modeling is the nearest-neighbor thermodynamic model, to which the Crothers lab made important contributions, in collaboration with the laboratories of Nacho Tinoco and Olke Uhlenbeck.19 The thermodynamic parameters that enable this model have been broadly extended by Doug Turner and colleagues and, together with RNA structure modeling algorithms, do an outstanding job of predicting the relative stability of helices and of simple RNA structures.20 Critical features of RNA structure – higher-order and non-canonical interactions – are not included in the nearest-neighbor model, however. In addition, the kinetic history of RNA folding cannot be predicted.
SHAPE reactivities are roughly inversely proportional to the probability that a nucleotide is based paired and simple considerations of the ideas that underlie statistical mechanics emphasize that the logarithm of a quantity related to a probability yields an energy. In collaboration with Dave Mathew's lab, we therefore devised a very simple pseudo-free energy change potential, based on these ideas and making use of the workhorse 1M7 reagent (Fig. 2B). By adding a "∆GSHAPE" term to the nearest-neighbor model used in most RNA folding algorithms, we were able to recover RNA secondary structures at high accuracies.21,22 This very simple approach does a good job of modeling a large variety of RNA structures including many that had previously been considered among the most difficult to model. However, there were still a few structures, especially those with a high proportion of stable tertiary and non-canonical interactions, for which this simple one-reagent approach, using 1M7, did not recover the accepted secondary structure at high accuracy.
We hypothesized that it might be possible to increase the accuracy of SHAPE-directed secondary structure modeling further by specifically detecting higher-order interactions using the NMIA and 1M6, reagents sensitive to non-canonical interactions (Fig. 2B). Again, we created a pseudo-free energy change potential and added this term into the algorithm. This "three-reagent" experiment (1M7, NMIA, and 1M6) achieves highly accurate RNA structure models for diverse RNAs including most currently known challenging RNAs (Fig. 4). Intriguingly, there is a near-monotonic improvement in RNA secondary structure modeling going from the Turner nearest-neighbor model, to one-reagent 1M7 modeling, to the three-reagent experiment.18 One interpretation of the increase in accuracy in RNA structure modeling is that these three types of data reflect increasingly complex levels of structural information: local base pairing (Turner's rules), local loop and non-canonical interactions (1M7 reactivity), and a subset of tertiary interactions (three-reagent SHAPE including NMIA and 1M6) (Fig. 4).
Figure 4.
Accuracy of SHAPE-directed modeling of secondary structure. Secondary structure modeling accuracies reported as a function of sensitivity (sens) and positive predictive value (ppv) for calculations performed without experimental constraints (no data), with SHAPE data obtained with the 1M7 reagent, or with three-reagent data26. Results are colored on a scale to reflect low (red) to high (green) modeling accuracy.
In our view, this three-reagent SHAPE experiment nicely balances simplicity and experimental concision with accurate recovery of RNA structure. Although there remain important regimes for improvement, the three-reagent SHAPE experiment, coupled with the nearest-neighbor model, appears to resolve the RNA secondary structure modeling problem for many classes of RNA.
Massively parallel Rube Goldberg machines
Massively parallel sequencing has been applied to interrogate many levels of RNA structure. Most approaches for performing massively parallel sequencing – whether detected in two-dimensional arrays, visualized using single-molecule polymerization in real time, or analyzed using nanopores23 – are direct and elegant and continue to benefit from advances in the magnitude of nucleotides examined, increases in accuracy, and reduction in costs. Moving beyond the strict analysis of sequence, RNA-seq experiments are now widely used to count the numbers of each kind of transcript in a cellular state.24 In principle, this experiment seems straightforward: sequence representative fragments of RNA in a given cell or state and quantify each type of transcript detected. In practice, however, apparently similar RNA-seq experiments performed in different laboratories or on different platforms often yield disparate results.25
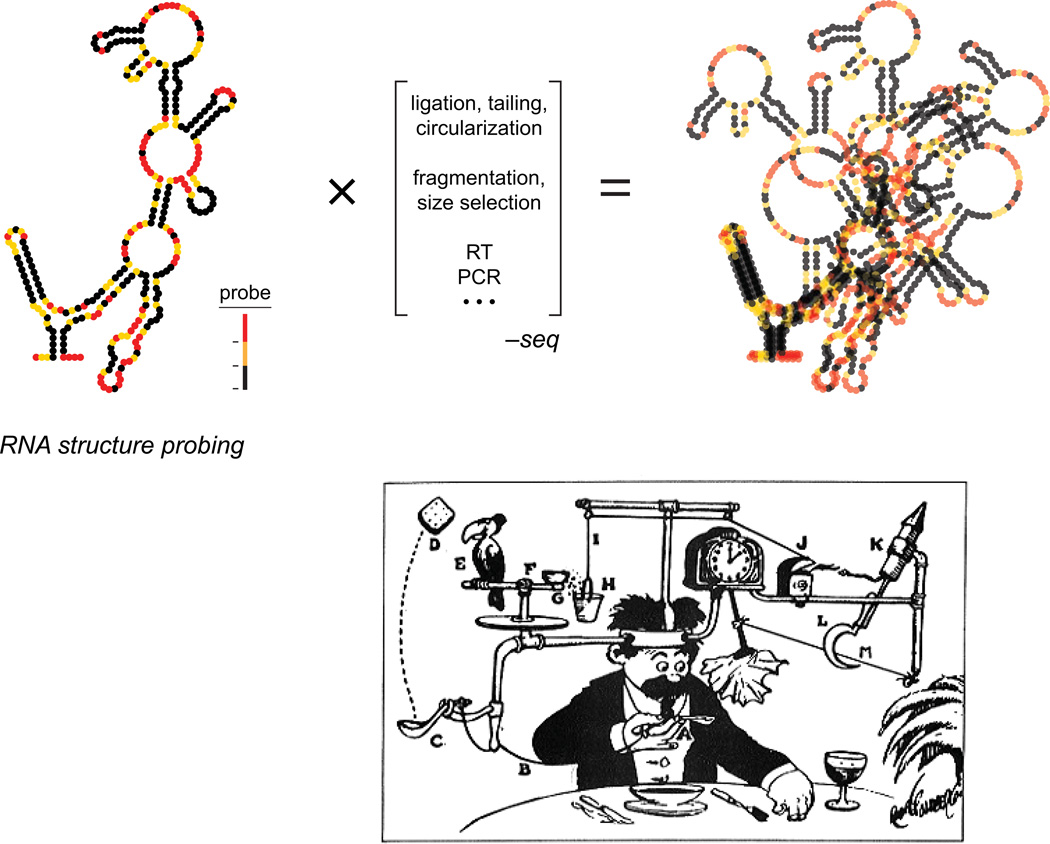
The problem becomes much harder when the goal is not just to count RNAs but to measure their structure. Sequencing does not report structural information directly; instead, an RNA first must be probed using a chemical or protein reagent that modifies or cleaves the RNA via a mechanism that is sensitive to the underlying RNA structure. This general approach, termed footprinting, was developed over 20 years ago; SHAPE falls into this class of approaches. We and others reasoned that a transformative advance would be to link RNA structure probing with readout by massively parallel sequencing. Many groups, including mine, pursued strategies that involved gluing together current molecular biology methods to convert the initial result of a structure probing experiment into a form readable by massively parallel sequencing. In most approaches, the probing information is recovered by counting the number of sequences that terminate at a given position in the RNA target. For example, one method my laboratory initially explored involved treating an RNA with SHAPE reagent, performing primer extension such that the reaction terminated at the site of the probing event, ligating on pieces of an adapter DNA, circularizing, amplifying, and ultimately sequencing. This approach completely violates the principles of simplicity emphasized in Don Crothers’ lab! The results were predictable: in nearly every step there was some selection bias or blurring of the information obtained in the original probing step (Fig. 5). If we include references to all of the steps that are likely to influence the final probing experiment, some of the early experiments developed in our lab, and in other laboratories, might be called structure probe-fragment-size select-3' ligate-RT-ligate again-circularize-PCR-sequence; another experiment we tried could reasonably have been called structure probe-fragment-exonuclease-reverse chemistry-ligate-RT-ligate again-PCR-sequence. Whew.
Figure 5.
Illustration of the challenges in melding a structure probing experiment with readout by massively parallel sequencing. The many intervening steps (in brackets) impose selection on the final data that, in essence, blur and conflate the results of the original experiment. Lower panel shows a comparative example of a complex process used to complete a task. Illustration is "Professor Butts and the Self-Operating Napkin" by Rube Goldberg.32
Given the complexity of these experiments, it should have been no surprise that the final results were unsatisfactory. The RNA structure probing profiles using these methods look a bit like profiles read out by more conventional methods like capillary electrophoresis but fail to capture the fine details of RNA structure that are required to, for example, model RNA structure accurately or characterize regulatory elements in individual RNAs. In essence, these were massively parallel Rube Goldberg machines that were quite complex to implement and, critically, disrupted the relationship between the initial probing information and the final read out (Fig. 5). For several years, our laboratory has since focused intensively on trying to create a no-compromises approach for interrogating RNA structure such that all nucleotides are examined simultaneously and that the probing data can be read out by massively parallel sequencing with no loss of the fundamental RNA structure information.
Mutational profiling (MaP)
The problem with many approaches for reading out RNA structure probing information by massively parallel sequencing is that there are just too many steps between the initial physical probing step and the final sequencing data. After having worked for many years to create the generic, quantitative, and physically accurate SHAPE strategy for probing RNA structure at nucleotide resolution, my laboratory was frustrated by efforts that converted SHAPE probing to a qualitative approach and thought this was an ill-advised step backwards for the current era of 'omics and massively parallel sequencing.
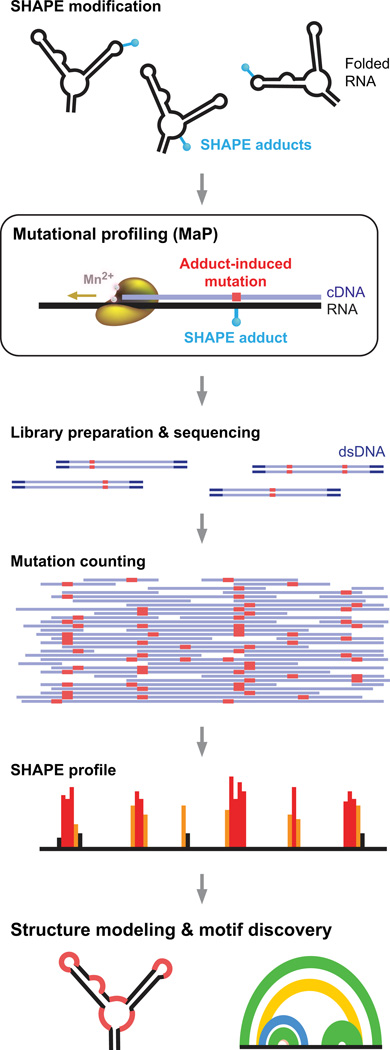
To simplify analysis of RNA structure probing, we needed a method that records the presence of a chemical adduct in an RNA in as direct and concise way possible. We thought that chemical adduct detection might be fully accomplished in the first analysis step, during reverse transcription. Most reverse transcriptase enzymes are derived from enzymes encoded by retroviruses that have likely evolved to be specifically mutagenic when they pause during cDNA synthesis. There was also a precedent that reverse transcriptase polymerase enzymes could read through certain kinds of chemical adducts with low efficiency. Based on these insights, we developed conditions under which reverse transcriptase reads through diverse kinds of chemical modifications in RNA and, in doing so, permanently records these sites by incorporating a nucleotide non-complementary to the original templated nucleotides.26 Detection efficiencies for individual chemical adducts are high, in the neighborhood of 50% or better. We call this process mutational profiling, or MaP (Fig. 6).
Figure 6.
SHAPE-MaP overview. RNA is treated with a SHAPE reagent that reacts at conformationally dynamic nucleotides. Specialized reverse transcription conditions allow the polymerase to read through chemical adducts in the RNA and to record the site as a nucleotide non-complementary to the original sequence (red) in the cDNA. The resulting cDNA is subjected to massively parallel sequencing to create a mutational profile. Sequencing reads are then converted to a SHAPE reactivity profile. SHAPE reactivities can be used to model secondary structures, visualize competing and alternative structures, discover functional RNA motifs, and quantify any process that modulates local nucleotide RNA dynamics. Figure reprinted from Ref. 26.
With mutational profiling, nucleic acid structure information is "stored" during reverse transcription and is thus rendered nearly impervious to downstream manipulations required to prepare the cDNA for massively parallel sequencing. Comparison of the accuracy of SHAPE-MaP with the prior gold standard using capillary electrophoresis showed that the new approach is fully comparable to the prior standard.26 Current evidence indicates that SHAPE-MaP represents a no-compromises strategy for nucleotide-resolution structure analysis of chemical probing experiments (Fig. 4). If named strictly by the steps that impact recovery of the underlying structure probing information, it really is appropriate to summarize this new experiment simply, as SHAPE-MaP-sequence.
By making the experiment simpler, structure probing was improved in decisive ways. First, every read is included in the analysis, which reduces or eliminates the possibility that researchers inadvertently over-interpret rare events in their experiments by either physical purification or computational filtering. Second, because a chemical modification is only detected when the reverse transcriptase enzyme fully reads through the site of modification, the MaP approach is relatively insensitive to pre-existing RNA degradation and to complications related to the efficiency and processivity of reverse transcription. Third, because the sequencing data are digital, interpretation can be fully automated. Moreover, because the chemical modification information is encoded internally in the DNA and can be compared to an experiment in which the reagent is omitted, cDNAs can be heavily amplified by PCR. Thus, SHAPE-MaP can be used to obtain nucleotide-resolution RNA structural information on very small amounts of RNA, even if this RNA is in a mixture with large excesses of other RNAs. The MaP approach is so robust that it has been used successfully to probe the structures of synthetic genetic polymers (XNAs), molecules with backbone chemistries not found in nature.27
RNA motif discovery
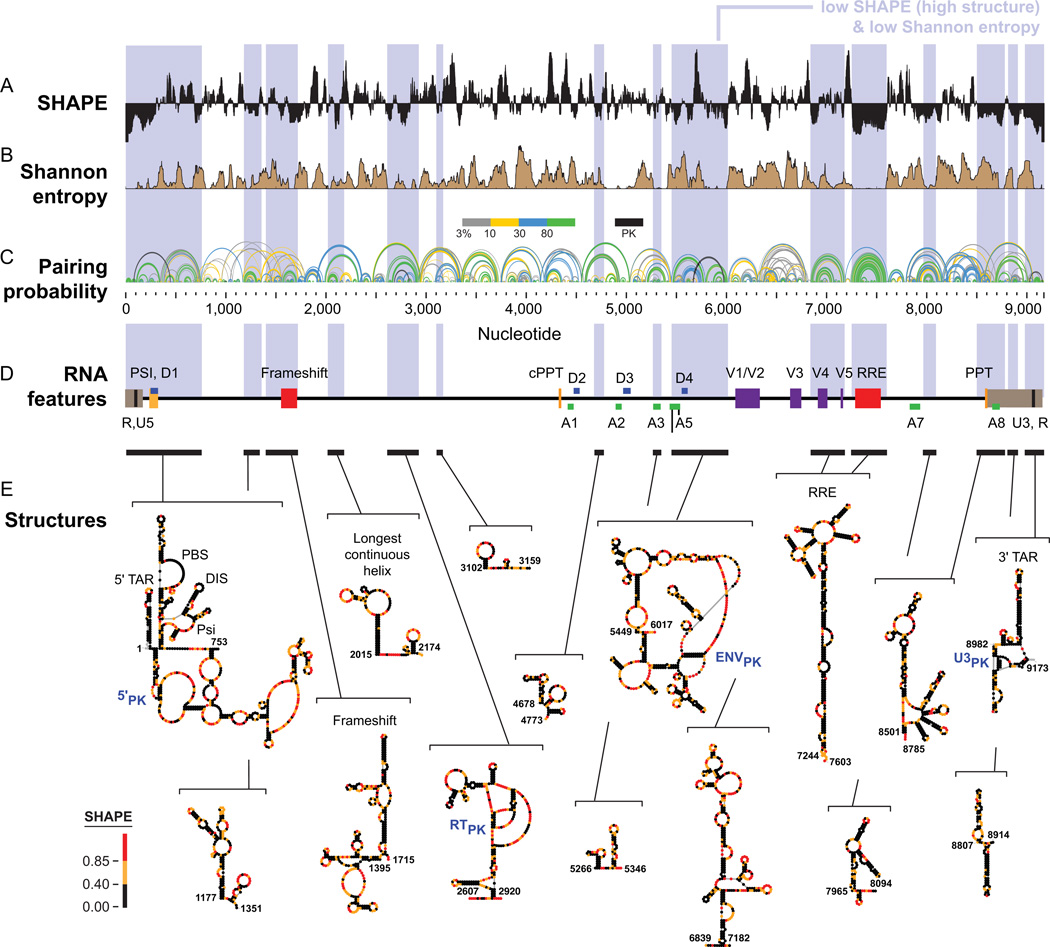
One intriguing way to use large-scale SHAPE-MaP analysis is to identify RNA regions that are both highly structured, as evidenced by low SHAPE reactivity, and simultaneously have low structural entropy, indicating that a structural model is well-determined. In an analysis of an entire HIV-1 RNA genome, we found that such regions are highly likely to correspond to known RNA elements with important functional roles in viral replication (Fig. 7). We also identified many new regions with no previously defined function. We are betting that many or most of these regions will turn out to be functionally important, and initial viral replication studies support this view. Thus, using very simple chemical principles, de novo structure-first RNA motif discovery seems to be tractable.26 As SHAPE-MaP makes it possible to examine the structures of long RNAs in a matter of days or a few weeks, accurate and quantitative transcriptome-wide analyses will be coming soon.
Figure 7.
SHAPE-MaP analysis of the HIV-1 RNA genome (NL4-3 strain). (A) SHAPE reactivities. Reactivities are shown as the centered 55-nt median window, relative to the global median. (B) Shannon entropies. (C) Pairing probabilities. Arcs representing base pairs are colored by their pairing probabilities, with green arcs indicating highly probable helices. Areas with overlapping arcs have multiple potential structures. Black arcs indicate pseudoknots (PK). (D) RNA regions with known biological functions. Bars and blue shading enclose low SHAPE (highly structured) and low Shannon entropy regions; these regions overlap with known RNA functional motifs much more frequently than expected by chance. (E) Secondary structure models for regions identified de novo. Names of known structures are given. Figure adapted from Ref. 26.
Keep on MaP-ing: tertiary structure
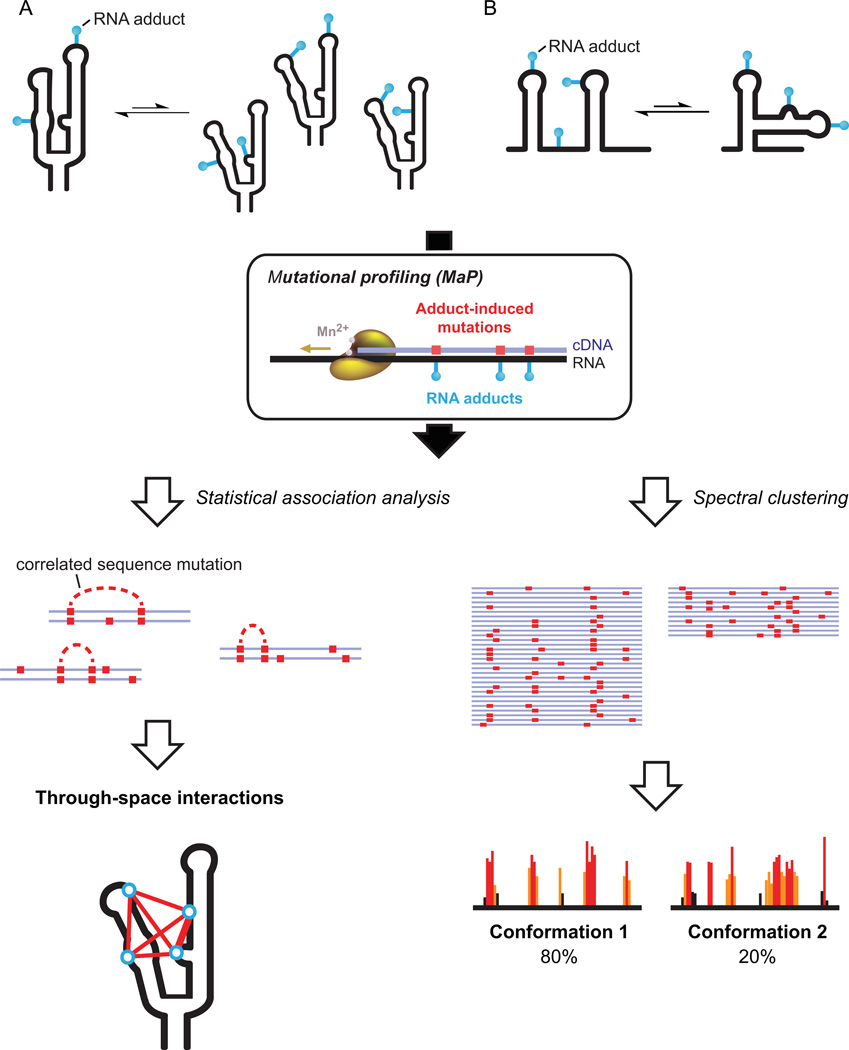
The utility of the extremely simple MaP approach extends far beyond merely creating a high-throughput technology for analyzing SHAPE probing information. Critically, if an experiment reads through one chemical adduct, then it is possible to read through two adducts or ten! By using the MaP strategy to detect multiple chemical modification events in a single RNA strand, we reasoned that might be possible to convert chemical probing, coupled with massively parallel sequencing, into a single-molecule experiment specifically sensitive to higher-order and tertiary interactions in RNA. We call this strategy for detecting correlated modifications RING-MaP, for RNA interaction groups by mutational profiling.28 Conceptually, there are at least three broad ways in which RING-MaP can be used.
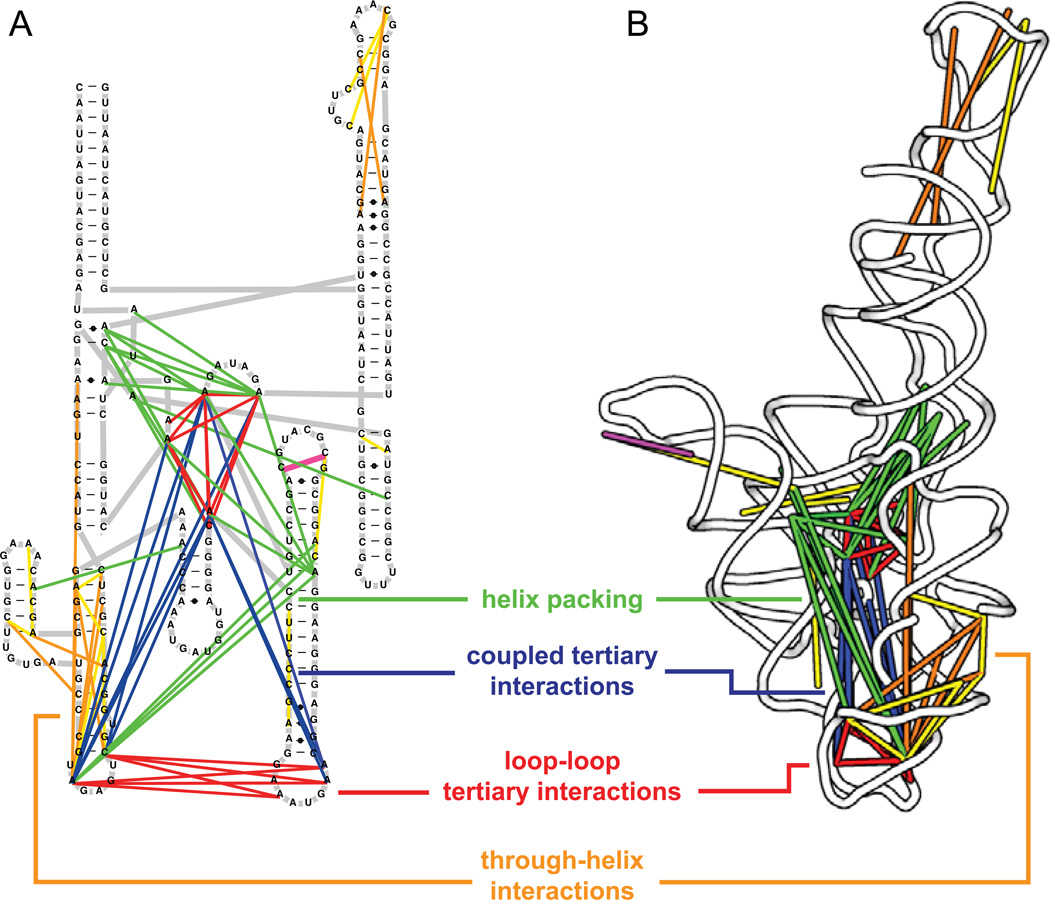
The first application recognizes that RNA molecules that form stable structures undergo "breathing" transitions that will transiently expose RNA nucleotides in a coordinated way. By detecting the correlated chemical reactivities that occur at a rate that exceeds that expected based on independent modification of the individual nucleotides in the form of easily measured mutation rates, the RING-MaP experiment identifies nucleotides participating in through-space communication (Fig. 8A). When we plot these correlated interactions on the secondary and tertiary structures of RNAs with complex folds, almost exclusively tertiary and higher-order interactions, rather than simple base pairing, are detected (Fig. 9).28 Roughly a third of through-space interactions detected by the RING approach correspond to the kinds of higher-order tertiary interactions that are now well understood to stabilize complex RNA folds (Fig. 9, in red and magenta).
Figure 8.
RING-MaP overview. (A) Detection of through-space structural communication. RNA molecules experience local structural variations in which nucleotides become reactive to a chemical probe in a correlated way. Statistical association analysis detects these interdependencies. (B) Analysis of multiple coexisting RNA conformations based on clustering analysis. Initial RING-MaP experiments were performed with the RNA-modifying reagent dimethyl sulfate (DMS).28
Figure 9.
Through-space RNA structural relationships revealed by single-molecule correlated chemical probing. Direct, through-helix, and global internucleotide interactions are illustrated on both (A) secondary structure and (B) three-dimensional models of the RNase P catalytic domain.28 Each class of interaction is indicated by a line of a distinct color. Conventional tertiary interactions are red; non-canonical base pairs are magenta; through-helix interactions are yellow and orange; coupled tertiary interactions are blue; and helix packing is green. The RNase P structure is from model 3dhs.33
Intriguingly, another third of all the correlated interactions that we observed in our initial study of three RNAs with complex folds involve single-stranded or loop nucleotides located on the opposite ends of a single helix (Fig. 9, in orange and yellow).28 These interactions suggest that structural communication can be propagated very long distances through individual helices. In addition, the RING-MaP analysis indicated that almost all tertiary interactions are strongly connected to other tertiary interactions (Fig. 9, in blue and green). Perhaps it comes as no surprise that tertiary interactions operate as complex and tightly interrelated networks that are in effect brittle.29 Disruption of or "breathing" at one interaction often means that many tertiary interactions are weakened in a correlated way.
RNA molecules are distinctive among large biological macromolecules as they are able to form multiple stable structures that often coexist. A second broad application of RING-MaP is based on our finding that, when two populations exist simultaneously in solution, each population has distinctive patterns of chemical reactivity (Fig. 8B). We analyzed RING-MaP data on the riboswitch aptamer domain that recognizes the metabolite thiamine pyrophosphate (TPP) using a clustering approach.28 In both the absence of ligand and in the presence of saturating TPP, the RNA exists in two distinct conformations. In the presence of TTP, the major cluster (at approximately 80%) has correlated SHAPE reactivities consistent the three-dimensional structure of the ligand-bound state characterized in crystallographic studies.30 The other 20% of molecules are in a conformation that resembles the no-ligand structure. In the absence of ligand, the correlated SHAPE reactivities are consistent with the no-ligand structure, but ~20% of molecules had reactivity patterns consistent with formation of a highly structured ligand binding pocket (Fig. 10).28 This emphasizes that, even in the absence of ligand, the riboswitch RNA samples a state in which the part of the ligand-binding pocket that binds thiamine is pre-organized to bind ligand. Identification of this hidden state also immediately supports a multi-state mechanism for specific ligand recognition by this RNA.
Figure 10.
Multiple in-solution conformations for TPP riboswitch in the absence of TPP ligand. (A) Spectral clustering analysis. There are two clusters in the no-ligand state. The major cluster (red) reflects an unstructured state with few internucleotide interactions. The minor cluster (blue) is more highly structured than the major cluster specifically in the region of the thiamine binding pocket (blue closed circles). (B, C) Nucleotides that are more structured in the minor cluster – which flank the thiamine-binding pocket – are emphasized in blue on RNA secondary and tertiary structure models. The thiamine moiety is gray in panel C.
Finally, given the very dense array of through-space interactions that can be identified using the RING-MaP approach, we should be able to use it to discover and refine three-dimensional models for complexly folded RNAs.28 In exploratory work on large RNAs, the models generated using RING-MaP compare favorably with, and may be superior to, those generated using biophysical approaches including FRET, SAXS, and NMR. These models do not yet yield the resolution achievable by X-ray crystallography; however, the RNA modeling field is advancing rapidly31 and RING-MaP constraints will likely prove useful when melded with emerging approaches.
Sequencing toward all RNA structures, concisely
Massively parallel sequencing is a powerful and transformative technology.23 Sequencing in real time – enabled by two-dimensional arrays, by visualization of polymerization of single molecules, or in nanopores – is direct and elegant. So, too, is the idea that massively parallel sequencing can be used to move far beyond simply sequencing nucleic acids to a wide variety of RNA counting (as in RNA-seq), nucleic acid structure-measuring, and RNA-protein proximity-determining experiments (of which there are now an extremely wide variety). Many of the current generation experiments are extremely complex and involve numerous incompletely characterized steps, most of which directly affect the final measured outcome (Fig. 5). Because these experiments are often first employed on very complex large-scale systems, research teams are often unable to or unenthusiastic about thoroughly validating new methods. It is commonplace to employ experimental and algorithmic filters such that only small fractions of the total data are actually examined.
Widespread assumptions that underlie many 'omics and massively parallel sequencing-based experiments might be roughly summarized as (i) "Hey, I just obtained 10 million (or 100 million or 1 billion) sequences, I must have measured something!" and (ii) "I know these data are really noisy, but I have so much of it, I will use statistics to work things out!" There is no question that statistical analysis of the enormous data sets generated by massively parallel sequencing can reveal important features; however, data quantity and statistics are often overemphasized. Instead, focusing on making experiments themselves more direct (and less inferential) measurements of diverse 'omes will make each reported experiment a more definitive contribution to our understanding of biological function.
The MaP approach represents one new way to use massively parallel sequencing to directly, concisely, and accurately measure features involving nucleic acid structure and function in a no-compromises way. Essentially, the active site of a polymerase enzyme becomes a direct, physical device for measuring nucleic acid structure (Figs. 6 & 8). To use massively parallel sequencing to make accurate and lasting measurements of nucleic acid structure and, likely, to measure interactions with protein and other ligands requires that the field take a Crothers laboratory approach (Fig. 1). It is almost always superior to use concise chemical principles and to make the physical measurement in as few steps as possible. That one can generate millions of pieces of sequencing information, or any other type of data, cannot replace concise and careful experimental design.
Many extensions to the MaP approach can be imagined. For example, the MaP approach could be used to read out and score post-transcriptional and epigenetic modifications in RNA and DNA, perhaps by selecting specific polymerases for these purposes. The single-molecule vision, as encapsulated in the RING-MaP correlated chemical probing approach, can be extended to measure RNA-RNA interactions. Direct detection of RNA-protein interactions (via cross-linking), RNA-DNA interactions (perhaps to reveal nuclear organization) seem feasible. SHAPE-MaP and RING-MaP were validated by identification of previously well-determined elements of secondary and tertiary structure, respectively. Given that regions with well-defined higher-order RNA structures tend to be highly overrepresented among functional elements, variations on the MaP approach are likely to make it possible to identify de novo functionally critical regions across viral RNA genomes and cellular transcriptomes.
Acknowledgements
This manuscript is dedicated to the memory and mentorship of Don Crothers. It has been a privilege to work with the many student and postdoctoral colleagues in my own laboratory who made numerous decisive and innovative contributions to the ideas reviewed here. The National Science Foundation and National Institutes of Health support our work on creating high-content chemical microscopes for understanding structure-function interrelationships in RNA. The NSF has consistently supported many projects at early high-risk stages, as did the NIH via the EUREKA program. Steve Busan provided critical assistance in preparing figures.
References
- 1.Zimm BH, Crothers DM. Simplified rotating cylinder viscometer for DNA. Proc Natl Acad Sci U S A. 1962;48:905–911. doi: 10.1073/pnas.48.6.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Citation numbers were retrieved from Google Scholar on 1 Oct 2014.
- 3.Crothers DM, Metzger H. The influence of polyvalency on the binding properties of antibodies. Immunochemistry. 1972;9:341–357. doi: 10.1016/0019-2791(72)90097-3. [DOI] [PubMed] [Google Scholar]
- 4.Müller W, Crothers DM. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968;35:251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- 5.Chaires JB, Dattagupta N, Crothers DM. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry. 1982;21:3933–3940. doi: 10.1021/bi00260a005. [DOI] [PubMed] [Google Scholar]
- 6.Marini JC, Levene SD, Crothers DM, Englund PT. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982;79:7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 9.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGinnis JL, Dunkle JA, Cate JHD, Weeks KM. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGinnis JL, Weeks KM. Ribosome RNA assembly intermediates visualized in living cells. Biochemistry. 2014;53:3237–3247. doi: 10.1021/bi500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J Am Chem Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 15.Mortimer SA, Weeks KM. C2'-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci U S A. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grohman JK, et al. A guanosine-centric mechanism for RNA chaperone function. Science. 2013;340:190–195. doi: 10.1126/science.1230715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steen K-A, Rice GM, Weeks KM. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J Am Chem Soc. 2012;134:13160–13163. doi: 10.1021/ja304027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20:846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tinoco I, et al. Improved estimation of secondary structure in ribonucleic acids. Nature New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 20.Turner DH, Sugimoto N, Freier SM. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- 21.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 24.Wold B, Myers RM. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 25.SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor AI, et al. Catalysts from synthetic genetic polymers. Nature. 2014 doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homan PJ, et al. Single-molecule correlated chemical probing of RNA. Proc Natl Acad Sci U S A. 2014;111:13858–13863. doi: 10.1073/pnas.1407306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritz J, Martin JS, Laederach A. Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics. 2012;13(Suppl 4):S6. doi: 10.1186/1471-2164-13-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruz JA, et al. RNA-Puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. RNA. 2012;18:610–625. doi: 10.1261/rna.031054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikimedia Commons, in the public domain.
- 33.Kazantsev AV, Krivenko AA, Pace NR. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA. 2009;15:266–276. doi: 10.1261/rna.1331809. [DOI] [PMC free article] [PubMed] [Google Scholar]