Abstract
Height is the result of many growth and development processes. Most of the genes associated with height are known to play a role in skeletal development. Single-nucleotide polymorphisms in the SPAG17 gene have been associated with human height. However, it is not clear how this gene influences linear growth. Here we show that a targeted mutation in Spag17 leads to skeletal malformations. Hind limb length in mutants was significantly shorter than in wild-type mice. Studies revealed differences in maturation of femur and tibia suggesting alterations in limb patterning. Morphometric studies showed increased bone formation evidenced by increased trabecular bone area and the ratio of bone area to total area, leading to reductions in the ratio of marrow area/total area in the femur. Micro-CTs and von Kossa staining demonstrated increased mineral in the femur. Moreover, osteocalcin and osterix were more highly expressed in mutant mice than in wild-type mice femurs. These data suggest that femur bone shortening may be due to premature ossification. On the other hand, tibias appear to be shorter due to a delay in cartilage and bone development. Morphometric studies showed reduction in growth plate and bone formation. These defects did not affect bone mineralization, although the volume of primary bone and levels of osteocalcin and osterix were higher. Other skeletal malformations were observed including fused sternebrae, reduced mineralization in the skull, medial and metacarpal phalanges. Primary cilia from chondrocytes, osteoblasts, and embryonic fibroblasts (MEFs) isolated from knockout mice were shorter and fewer cells had primary cilia in comparison to cells from wild-type mice. In addition, Spag17 knockdown in wild-type MEFs by Spag17 siRNA duplex reproduced the shorter primary cilia phenotype. Our findings disclosed unexpected functions for Spag17 in the regulation of skeletal growth and mineralization, perhaps because of its role in primary cilia of chondrocytes and osteoblasts.
Introduction
Human height is a highly heritable, classic polygenic trait [1]. Genetic alterations affecting height variation provide insight into human growth and development, as well as the genetic architecture of a number of diseases [2]. Prenatal longitudinal bone development and growth is a determinant of an individual’s postnatal height [3]. After formation of the cartilage anlage, longitudinal growth occurs at the epiphyseal growth plate, a layer of cartilage cells that are arranged in columns between epiphyseal and metaphyseal bone present in long bones and scapulae. In normal fetal growth plates, immature chondrocytes are located in a reserve cell zone (resting zone) that is oriented toward the epiphysis. Under hormonal, growth factor and mechanical regulation, these cells proliferate (proliferating cell zone), which increases length of the bone. After a set number of divisions, the cells undergo hypertrophy, remodeling their extracellular matrix to accommodate the change in cell shape (hypertrophic cell zone). Finally, the chondrocytes mineralize their extracellular matrix (calcifying cell zone), which is then vascularized, leading to bone formation [4]. In addition to long bone growth in the fetus, the diaphyseal bone also undergoes development with cortical bone forming around the cartilage anlage as the core is replaced with marrow [5,6].
Several genes have been shown to be involved in height [1,2,7–12], including those encoding proteins that are involved in signaling pathways known to be important in skeletal growth and development. Interestingly, single-nucleotide polymorphisms (SNPs) in the sperm associated antigen 17 protein gene (SPAG17) have been associated with human height [1,7,9–13] and idiopathic short stature in a Korean population [2]. However, it is not clear whether this gene directly influences height and/or skeletal growth.
Spag17 encodes a protein present in cilia and flagella with a “9+2” axoneme structure. SPAG17 protein is present in the central pair complex (CPC). It is the orthologue of Chlamydomonas reinhardtii PF6 [14], a protein located on a projection from the C1 CPC microtubule in green algae [15]. Domains near the C-terminus of PF6 are essential for flagellar motility and assembly of the C1a projection. The N-terminal half is not required for the assembly, but it is important for stability of C1a complex members [16].
PF6 protein interacts with a number of other proteins, including calmodulin [17]. Consequently, PF6 is thought to be a proximal effector of CPC action on the sliding activity of the outer doublets that control cilia or flagellar beat. Chlamydomonas pf6 mutants have paralyzed flagella, and lack the CPC 1a projection [15].
The murine Spag17 gene encodes a full-length 250 kDa protein found in testis and tissues with motile cilia, but other splice variants and proteolytically generated fragments have been described [14,18]. Targeting of Spag17 resulted in a severe phenotype characterized by motile cilia dysfunction in the nares and trachea, reduced clearance of nasal mucus, profound respiratory distress associated with lung fluid accumulation and disruption of the alveolar epithelium, cerebral ventricular expansion consistent with emerging hydrocephalus, failure to suckle, and neonatal demise within 12 h of birth. Ultrastructural analysis revealed loss of one CPC microtubule in approximately one quarter of tracheal cilia axonemes, absence of a C1 microtubule projection, as well as less frequent CPC structural abnormalities [19].
In the present study, the effect of a targeted mutation in Spag17 on the skeleton during differentiation, maturation and growth was examined. The effect of silencing of Spag17 -/- on osteoblasts and chondrocytes was examined to evaluate the role of Spag17 in cells involved in osteochondral development. Our findings suggest that SPAG17 protein plays a role in endochondral bone formation, most likely because of a function in primary cilia.
Results
Spag17 plays a role in skeletal development
Spag17 -/- mice have a severe phenotype characterized by motile cilia dysfunction [19]. Here we show that Spag17 -/- mice also have skeletal malformations. This finding was verified using a variety of experimental methods and provides evidence for a functional role for SPAG17 in height, consistent with multiple genetic association studies.
Spag17-deficient mice exhibit defects in development of the femur
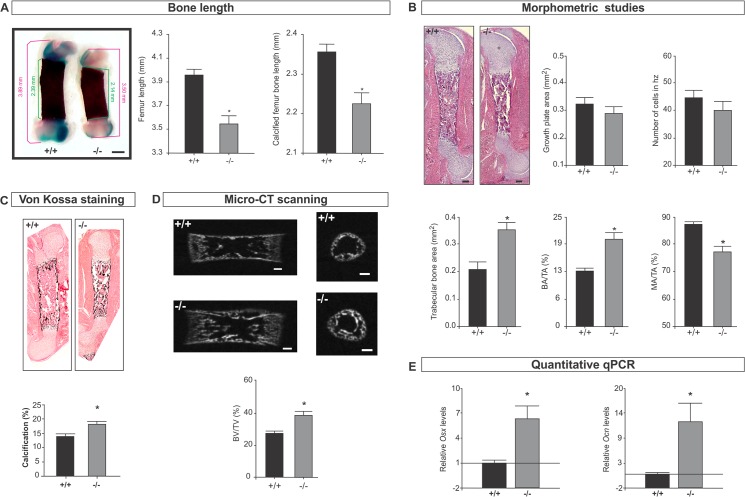
Because SPAG17 has been associated with human height [1,7,9–13], we investigated hind limb length. Femurs and tibias from wild-type and mutant mice were isolated, stained with Alcian blue and Alizarin red for cartilage and bone detection, respectively, and then length was measured as described in Materials and Methods. Fig 1A shows that femurs from mutant mice are significantly shorter than wild-type mice. However, no differences in bone width were observed. Morphometric studies showed normal cartilage development without significant differences in the growth plate area and number of cells in the hypertrophic zone. Trabecular bone area and bone area/total area were increased, leading to a reduction in marrow area/total area in the mutant mice (Fig 1B and S1 Fig). Because bone formation is related to bone mineralization, we examined the mineral deposits by von Kossa staining. This analysis revealed significant differences indicating that the Spag17 mutation has a significant effect on mineral deposition within the bone matrix (Fig 1C and S1 Fig). Increases in the ratio of BV/TV(%) were also observed by micro-CT scanning (Fig 1D). In addition, the expression of two genes involved in osteogenesis, osterix (Osx) and osteocalcin (Ocn), was higher in the mutant mice in comparison to wild-type mice (Fig 1E). Osteoclasts are cells that degrade bone to initiate normal bone remodeling and mediate bone loss in pathologic conditions and by increasing their resorptive activity. Although osteoclast numbers were slightly reduced in femurs of knockout mice, the reduction was not statistically significant, S3 Fig Together, the data collected from the femurs suggest that the Spag17-mutant mice have enhanced bone formation, which may lead to bone shortening due to premature ossification.
Fig 1. Femur bone development is affected in Spag17-deficient mice.
(A) Alcian Blue/ Alizarin Red staining of femur from wild-type (+/+) and Spag17-mutant (-/-) mice. Measurement of femur length and calcified femur bone length shows Spag17-mutant limbs are shorter than wild-type mice. Scale bars, 0.5 mm. (B) Histological and morphometric studies on femur shows increase bone formation in mutant mice. Scale bars, 200 μm. (C) von Kossa staining reveals increased calcification in mutant mice. (D) Micro-CT structural analysis on femurs; mid-sagittal sections (left) and cross-sectional trans-axial views at the mid-diaphysis (right). Images were acquired in 600 projections in 180 degrees of rotation with 500 miliseconds exposure per projection. Scale bars, 200 μm. (E) mRNA expression of osterix (Osx) and osteocalcin (Ocn) measured by qPCR. (+/+), wild-type; (-/-), Spag17-mutant mice. Data are presented as means ± SEM from ≥7 mice. * Indicates statistically significant differences, p< 0.05. hz: hypertrophic zone; BA/TA: bone area/total area; MA/TA: marrow area/total area; BV/TV: bone volume/total volume; Osx: osterix; Ocn: osteocalcin.
Spag17-deficient mice exhibit defects in tibia development
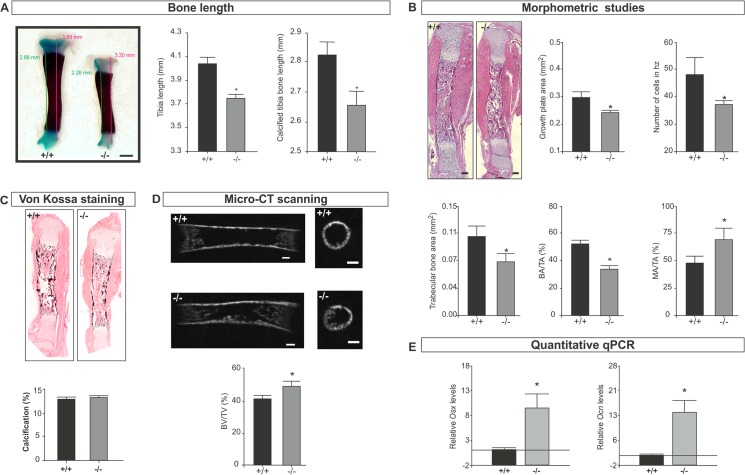
Measurement of tibia and calcified bone length revealed a shorter distal limb on Spag17-mutant mice (Fig 2A). Morphometric studies showed reduced growth plate area, characterized by a decrease in the number of hypertrophic cells (terminally differentiated chondrocytes) (Fig 2B and S2 Fig). Also bone morphology was affected by Spag17 disruption. Tibias had significantly less trabecular bone area, a reduction in the relation of bone area/total area (BA/TA%) and an increased ratio of marrow area/total area (MA/TA%) (Fig 2B). Calcification evaluation from von Kossa staining did not show statistically significant differences between wild-type and Spag17-mutant mice (Fig 2C and S2 Fig). However, micro-CT detected increased bone volume/total volume (BV/TV%) (Fig 2D). In addition, expression of Ocn and Osx was higher in the mutant mice in comparison to wild-type mice (Fig 2E). No statistically significant differences in the osteoclast number were observed in tibias comparing wild-type and knockout mice, S3 Fig These data suggest that the effect of Spag17 in hind limb development depends on the maturation state of the individual bones. Reduced Spag17 results in delayed tibial development. The shorter growth plate and the presence of mainly primary bone might explain the shorter bone length compared to wild type animals.
Fig 2. Tibia bone development is affected in Spag17-deficient mice.
(A) Alcian Blue/ Alizarin Red staining on tibia from wild-type (+/+) and Spag17-mutant (-/-) mice. Measurement of tibia length and calcified tibia bone length shows Spag17-mutant limbs are shorter than wild-type mice. Scale bars, 0.5 mm. (B) Histological and morphometric studies on tibia shows cartilage and bone defects. Scale bars, 200 μm. (C) von Kossa staining. (D) Micro-CT structural analysis on tibias; mid-sagittal sections (left) and cross-sectional trans-axial views at the mid-diaphysis (right). Images were acquired in 600 projections in 180 degrees of rotation with 500 miliseconds exposure per projection. Scale bars, 200 μm. (E) mRNA expression of osterix (Osx) and osteocalcin (Ocn) measured by qPCR. (+/+), wild-type; (-/-), Spag17-mutant mice. Data are presented as means ± SEM from ≥7 mice. * Indicates statistically significant differences, p< 0.05. hz: hypertrophic zone; BA/TA: bone area/total area; MA/TA: marrow area/total area; BV/TV: bone volume/total volume; Osx: osterix; Ocn: osteocalcin.
Spag17-deficient mice present fused sternebrae
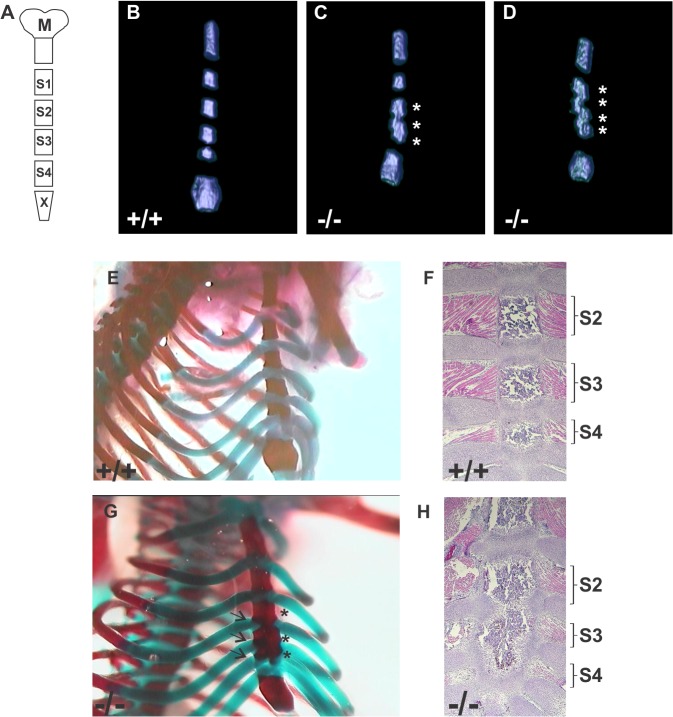
Spag17 -/- mice had additional skeletal malformations. The sternum is a segmented structure (Fig 3A) consisting of the manubrium, four sternebrae, and a xiphoid [20]. Micro-CT showed Spag17 -/- mice had fused sternebrae frequently (70%) between S3 and S4 and occasionally (20%) between S2, S3 and S4 (Fig 3C and S2 Video), but rarely (10%) between all sternebrae (Fig 3D and S3 Video). No fused sternebrae were observed in wild-type mice. Sternebrae malformations were also observed by Alcian blue/ Alizarin red staining (Fig 3G). The failure of ribs 4, 5 and 6 to complete their attachment to the sternum and promote sternebrae separation was apparent in the stained preparations in the Spag17 -/- mice. Similarly, histological examination showed failure of the last ribs to complete their attachment to the sternum, leading fused sternebrae (Fig 3H).
Fig 3. Disrupted development of the sternum in Spag17 -/- mice.
(A) Schematic representation of normal sternal bones (M, manubrium; S1 to S4, sternebrae 1 to 4; X, xiphoid). (B,C and D) Analysis of the sternum by micro-CT scanning shows fused sternebrae in the mutant mice. Pictures are representative images from respective supporting videos. (Fig 3B is representative of S1 Video; Fig 3C is representative of S2 Video; and Fig 3D is representative of S3 Video.) (E and G) Alcian blue/ Alizarin red staining. Failure of ribs 4, 5 and 6 to complete their attachment to the sternum and promote sternebrae separation is apparent in the stained preparations in the Spag17 -/- mice (arrows). (F and H) Representative H&E staining shows the presence of normal cartilage development that prevents fusion of the vertebrae in wild type animals. (+/+), wild-type; (-/-), Spag17-mutant mice. * Fused sternebrae.
Mineralization defects in skull and phalanges in Spag17-deficient mice
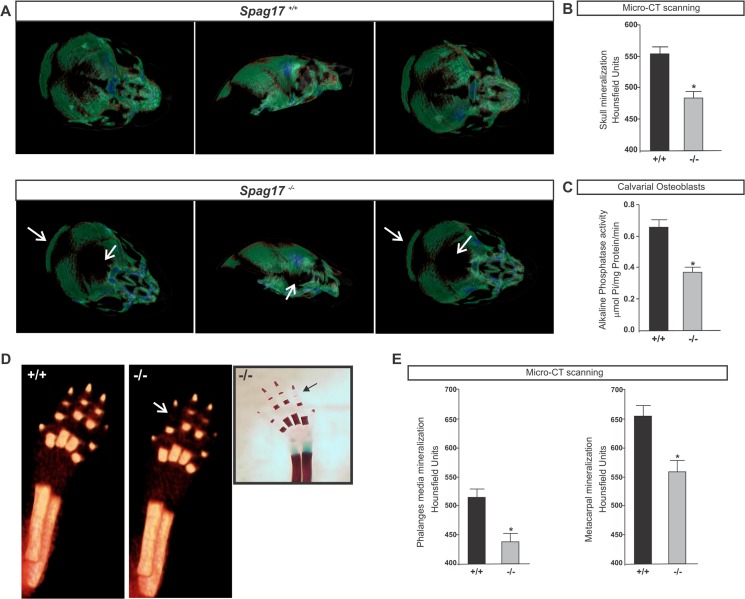
Micro-CT studies in Spag17-mutants showed separation in the bone from skull, which may be related to the prominent hydrocephalous and the intracranial pressure (Fig 4A) [19]. In addition, there was reduced mineralization in the skull (Fig 4B), suggesting membranous ossification defects. Medial and metacarpal phalanges in Spag17 -/- mice also showed reduced mineralization (Fig 4D and 4E). To investigate defects in mineralization, osteoblasts were isolated from calvaria and cultured using a standard protocol (see Materials and Methods), and alkaline phosphatase (ALP) activity, an early marker of osteoblast activity, was measured. Fig 4C shows a reduction in ALP activity in Spag17-mutant mice.
Fig 4. Cranial and phalanges defects in Spag17 -/-.
(A) Micro-CT imaging of mineralized skull. Note the demineralization of the skull in the mutant mouse (white arrows). (B and E) Quantitative mineralization measurement by micro-CT scanning. (C) Alkaline phosphatase activity in cultured calvarial osteoblast. (D) Micro-CT imaging of mineralized forelimb. Insert shows reduced metacarpal mineralization by Alcian Blue/ Alizarin Red staining. Micro-CT images were acquired in 360 projections by 360 degree rotation, with 680 millisecond exposure per projection. White and black arrows indicate less metacarpal mineralization in the mutant mouse. (+/+), wild-type; (-/-), Spag17-mutant mice. Data are presented as means ± SEM. * Indicates statistically significant differences, p< 0.05.
Primary cilia are disrupted in Spag17-mutant mice
Cilia play important roles in proliferation, differentiation and functions of many cells. Ciliary defects also results in skeletal development abnormalities [21]. Spag17 is expressed in tissues with “9+2” motile cilia like testis, brain, oviduct, lung and trachea [14,19]. However, Spag17 expression in cartilage and bone has not been reported in mice. RT-PCR analysis on RNA isolated from articular cartilage and bone showed that this gene is indeed expressed in wild-type mice, S4 Fig
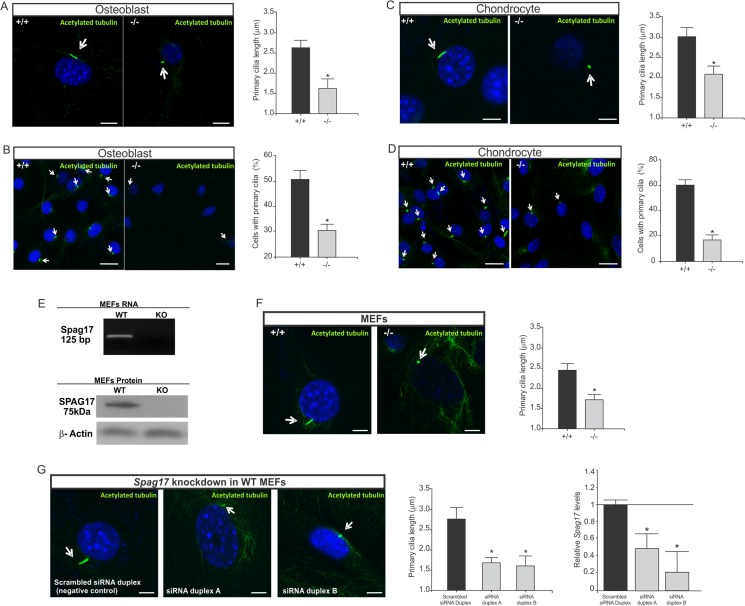
In order to determine if primary cilia in bone and cartilage are affected in Spag17-mutant mice, cultures of osteoblasts that were isolated from calvaria and cultures of chondrocytes that were isolated from the costochondral cartilages from wild-type and Spag17-mutant mice were stained with anti-acetylated α-tubulin antibody (a primary cilia marker). Fig 5 shows that primary cilia from Spag17 -/- mice are shorter than those from Spag17 +/+ mice, and there is a reduction in the percentage of cells expressing primary cilia in the knockout cells.
Fig 5. Primary cilia are altered in Spag17-deficient osteoblast, chondrocyte and MEFs cells.
Cells were stained with anti-acetylated α-tubulin to visualize primary cilia and DAPI as a nuclei marker. (A) Primary cilia length from osteoblast cells. Scale bars, 5 μm. (B) Percentage of osteoblast cells expressing primary cilia. Scale bars, 20 μm (C) Primary cilia length from chondrocyte cells. Scale bars, 5 μm. (D) Percentage of chondrocyte cells expressing primary cilia. Scale bars, 20 μm. Mouse embryonic fibroblasts (MEFs) were isolated from embryos at E12.5. (E) Detection of Spag17 mRNA and protein in MEFs. RT-PCR products were generated by primers from exon 4 and 5. The knockout mice have a deletion of the entire exon 5. Detection of SPAG17 protein by western blot using an antibody against C-terminal domain. (F) After serum-starvation to promote cilia growth, primary cilia were visualized in the MEFs with acetylated tubulin antibody, and DAPI as a nuclei marker. Primary cilia from Spag17-mutant MEFs were significantly shorter than wild-type. Scale bars, 5 μm. (G) Spag17 knockdown in WT MEFs cells after treatment with Spag17 siRNA duplex reproduced the shorter primary cilia phenotype. Scale bars, 5 μm. Arrows indicate primary cilia. (+/+), wild-type, (-/-); Spag17-mutant mice. Data are presented as means ± SEM. * Indicates statistically significant differences, p< 0.05.
To further evaluate an effect of Spag17 in primary cilia, mouse embryonic fibroblasts (MEFs) were isolated from wild-type and Spag17-mutant embryos at E12.5. Expression of SPAG17 in wild-type MEFs was shown by Western blot analysis (Fig 5E) using an anti-SPAG17 antibody against the C-terminal domain. This antibody detected a protein ~ 75 kDa molecular weight, suggesting that MEFs express a SPAG17 variant instead of the 250 kDa full length protein [14,18]. This ~75 kDa protein may be derived from alternative splicing or post-translational processing, processes [14]. SPAG17 mRNA was also detected in wild-type MEFs by RT-PCR (Fig 5E). After serum-starvation to promote cilia growth, primary cilia were visualized in the MEFs with anti-acetylated α-tubulin antibody. Primary cilia from Spag17-mutant MEFs were significantly shorter than wild-type (Fig 5F). The same primary cilia phenotype was reproduced by infecting wild-type MEFs with two different Spag17 siRNA duplex molecules (Fig 5G).
Discussion
The embryonic limb is first visible as a small bud that protrudes from the body and contains mesenchymal cells that are rounded by a layer of ectoderm. The limb skeleton is laid down progressively, starting from the proximal end to the distal end. The mesenchymal cells condense, differentiate into cartilage, and later into bone. Many genes are known to regulate growth and patterning of the limb in three dimensions; however, is not to clear how these genes interact [22]. The results showed here revealed an unexpected function of Spag17 gene in skeletal development. It appears that mutation in Spag17 differentially altered femurs and tibias. Femurs had mature Haversian bone formation, potentially leading to bone shortening due to premature ossification. Tibia bone shortening may be due to a delay in cartilage and primary bone development. These results suggest defects in limb patterning and also chondrocyte and osteoblast differentiation and maturation.
It is not clear yet how SPAG17 influences bone development and mineralization, and how its mutation can reduce and enhance this process in different bone locations as observed in skull and phalanges and limbs from mutant mice, respectively. The site-specific differences in the bone defects in Spag17 -/- mice may be related to a location-specific cell signaling process. Whether these are a consequence of inherent differences in development or are the cause of differences in development is not clear. We have observed differential effects of gene deficiency in the femur and tibia in other mice models due to the rate at which different bones in the embryo develop [23]. The phenotypic changes in the femur of the Spag17 -/- mice suggest increased bone formation. In contrast, changes in the tibia, which matures at a later time than the femur, indicate that the gene defect altered features of the growth plate. Similar developmentally related changes were observed in sternum, where mineral was observed in the intersternebral spaces at early times, when the presence of cartilage prevents fusion of the vertebrae in wild type animals. The observation that osteoclast number was not affected by Spag17 knockout may suggest that the effect of Spag17 in bone and cartilage was on the anabolic phase (bone formation and differentiation) and not on the remodeling phase. The effect of Spag17 on bone development differentiation and calcification depends on the stage of bone maturation and the type of bone. One potential reason why calvaria and long bones respond differentially to the lack of this important cilia protein are their distinct functions, which lead to differences in the mechanical forces they undergo and to which the cilia are exposed.
Normal sternum development requires that all the ribs form a continuous bar of cartilage between sternebrae by embryonic day 16 [20], suggesting that the defects in the Spag17 -/- mice may be due to alterations in early skeletal development. Fgfr1-mutant mice develop a similar fused sternum phenotype as observed in Spag17 -/- mice [24]. Additionally, Runx1 has been shown to play an essential role in the development of the sternum [25], and Runx2 cooperates with Runx1 during sternal morphogenesis [26]. However, the link between these genes and their possible regulation by Spag17 remains to be clarified.
Mechanotransduction is a process that helps bone cells to convert mechanical forces into biochemical signals. It is important for maintaining bone health and homeostasis [27]. Primary cilia extend from the surface of cells and function mainly as sensory antenna [28]. This organelle is believed to play a role in bone mechanotransduction [27]. In this context, calcium deposition by osteoblasts has been shown to be mediated by primary cilia [29]. Cultures of MLO-A5 cells (murine long osteocyte cell line A5), which display shorter and fewer cilia per cell, show reduced mineralization [29]. The same phenotype was observed in calvarial osteoblasts from Spag17-mutant mice suggesting this may be one of the mechanisms where Spag17 plays a role and has an effect in bone mineralization. In addition, several cell signaling pathways take place in primary cilia development and function. For instances, vesicular trafficking, intraflagellar transport (IFT), Hedgehog (Hh) and Wnt signaling converge in the organization and functional maintenance of the primary cilium [30,31]. Similar pathways influence cartilage and bone development and growth [32]. Ciliary chondrodysplasias are believed to result from imbalances in the signaling pathways that normally occur in functional cilia. Moreover, several knockout mouse models for genes related to primary cilia made important contribution to the knowledge of skeletal development [33].
Our understanding of how disruption of the Spag17 gene leads to alterations in primary cilia function and subsequent cellular pathology is limited. It is unknown, if this CPC protein is related to a structural function, as primary cilium lack the CPC, or it may plays a role in any of the signaling (vesicular trafficking, intraflagellar transport (IFT), Hedgehog (Hh), Wnt signaling, etc). Our results show that primary cilia are shorter in knockout mice. We hypothesize that Spag17 may be involved in ciliogenesis like Spag6 [34], although its role and/or mechanisms related to ciliogenesis may be different from Spag6. In addition, a ~75 kDa SPAG17 protein is expressed in MEFs suggesting that it is a variant instead of the 250 kDa full length protein. This ~75 kDa protein may be derived from alternative splicing or post-translational processing, processes. Further studies are required to determine how this protein functions in primary cilia.
Several studies have reported an association of SPAG17 gene variants with height [1,7,9–13]. However, until now there has been no known link between this gene, whose function was thought to be restricted to motile cilia, and height. Our findings have revealed functions for Spag17 that extend the role of this gene to regulation of skeletal differentiation, growth and mineralization, perhaps because of a function in primary cilia. This newly defined role could explain the influence of SPAG17 on linear growth and bone development.
Materials and Methods
Mice
All animal procedures were performed in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals. Animals were euthanized in accordance with protocol AD10000167 approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee. Target disruption of Spag17 was made by deletion of exon 5 using Cre-lox and FLP-FRT recombination system. Briefly, embryonic stem (ES) cells were obtained from the KOMP Repository (Davis, CA). The chimera males were crossed to C57BL/6J wild-type females, the resulting heterozygous offspring were crossed to the 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J to remove the Neo cassette. The Spag17 +/flox mice were used for mating with CMV-Cre (B6.C-Tg(CMV-cre)1Cgn/J) mice to generate Spag17 +/- ;CMV-Cre mice. Male and female Spag17 +/- ;CMV-Cre mice were mated resulting in the expecting Mendelian inheritance for Spag17 -/- ;CMV-Cre mice. After these mattings the mice had a mixed background of C57BL/6J−129S4/SvJ. Deletion of this gene was published in detail in Teves et al., 2013 [19].
Histology studies
Bones were removed from newborn mice and fixed overnight in 4% formalin/PBS, dehydrated in ethanol and embedded in paraffin. Slides were stained with hematoxylin and eosin. Histology preparations were performed at the VCU Massey Cancer Center Macromolecule Core Facility, Virginia Commonwealth University.
Morphometric studies
Histological sections were imaged (Zeiss AxioObserver, Germany) using a 20x objective. Cartilage (growth plate area, percent hypertrophic cells in the growth plate, percent resting zone cells, height of hypertrophic region, number of cells in hypertrophic region) and bone (total bone area, cortical bone area, trabecular bone area, bone area/total area, and marrow area/total area) were measured in the tibia and femur using Zen Black software (Zeiss). Data presented are mean ± standard error of n = 7 mice. Osteoclast number was counted in 8 knockout and Wild-type mice. Statistical comparisons were made using unpaired t-test; p < 0.05 was considered significant.
High resolution micro-CT
Newly born mouse pups were euthanized using isoflurane, and the hindlimb and sternum were removed and fixed in 10% neutral buffered formalin. High resolution microcomputed tomography (micro-CT) was performed using the Brϋker Skyscan 1173 micro-CT scanner (Skyscan NV, Kontich, Belgium). Sternal and hind limb images were acquired in 600 projections in 180 degrees of rotation, with 500 millisecond exposure per projection, at 50 kilovolts and 160 microamps, an image pixel size of 7.91 micrometers, and a rotation step of 0.40 degrees. Images were acquired at a resolution of 2240 x 2240 pixels. Sample images were reconstructed standard Feldkamp reconstruction using NRecon Software (Kontich, Belgium) and a Gaussian smoothing kernel of 2. Region of Interest (ROI) analysis was conducted using CT-analysis software (CTAn, version 1.13.11.0, Skyscan NV) to assess the amount of attenuated tissue present as a percentage of the total three dimensional volume in the ROI (% Bone Volume/Total Volume, %BV/TV). Data are mean ± standard error of n = 7 mice and statistical comparisons were made using unpaired t-test; p < 0.05 was considered significant.
Measurement of bone length and width
Alcian blue/ Alizarin red stained hind limbs were scanned with Life Technologies EVOS Fl Auto Cell Imagining System. Then images were analyzed, and bone measured by ImageJ software (NIH). Data are mean ± standard error of n = 9 mice and statistical comparisons were made using t-test; p < 0.05 was considered significant.
RT-PCR and qPCR
Total RNA was isolated with TRIzol (Life Technologies, Inc., Grand Island, N.Y.) from newborn mice sternum, cartilage, and bone tissue. The RNA was reversed transcribed with RETROscript kit (Ambion, Austin, TX) according to the manufacturer’s instructions, and the cDNA were used for RT-PCR or qPCR. Amplification products were resolved on 1% agarose gels stained with ethidium bromide. The results presented are representative of seven different animals from each genotype shown.
Micro-CT scanning
Mouse pups were euthanized with excess of isoflurane and were then imaged in the micro-CT scanner. Micro-CT was performed using an Inveon micro-CT system (Siemens, USA). Three dimensional images were acquired in 360 projections by 360 degree rotation, with 680 millisecond exposure per projection, around a 31 x 31 mm field of view (FOV), with 75 kV voltage, 500 μA of current and 2 × 2 binning were used to yield an effective pixel size of 19.24 μm and a scan time of 6 minutes. A thin walled plastic tube containing water was scanned using the same parameters and was used for Hounsfield unit (HU) normalization. HU normalized micro-CT images were reconstructed using a modified Feldkamp algorithm using manufacturer provided software (COBRA Exiim). ROI analyses for bone density and bone length measurements were done in the Inveon Research Workplace 4.1 (Siemens Healthcare, PA). Statistical comparisons were made using Student’s t-test; p < 0.05 was considered significant.
Alcian blue/ Alizarin red staining
Skeletons from newborn mice were stained with Alcian Blue/ Alizarin Red as described by Ovchinnikov D [35]. Skeletal preparations were analyzed and photographed in glycerol/ethanol solution.
von Kossa staining
Bones were removed from newborn mice and fixed overnight in 4% formalin/PBS, dehydrated in ethanol and embedded in paraffin. Slides were stained for phosphate via the method of von Kossa at the VCU Massey Cancer Center Macromolecule Core Facility, Virginia Commonwealth University using a kit and following manufacturer’s instructions (American MasterTech Scientific Inc., CA, USA).
Osteoblast and chondrocyte isolation and culture
Newborn mice were euthanized by an overdose of isoflurane. Chondrocytes were isolated from the costochondral cartilage following methods previously described for chondrocyte isolation for rats [36]. Briefly, rib cages were removed and resting zone cartilage dissected. Cartilage pieces were washed in Hank's Balanced Salt Solution (HBSS), incubated in 0.25% trypsin-EDTA (Life Technologies) for 15 minutes, then in 0.02% collagenase type II (Worthington Biochemicals, Freehold, NJ) for 3 hours. Cells were separated from tissue debris using 40 μm cell strainers (BD, Franklin Lakes, NJ) and collected by centrifugation. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Life Technologies), 1% penicillin-streptomycin, and 50 ug/ml ascorbic acid (Sigma Aldrich). Osteoblasts were isolated from frontal and parietal bones as previously described [37]. Briefly, minced bone pieces were digested in trypsin followed by 0.01% collagenase type I (Sigma Aldrich) and 0.01% dispase (Life Technologies). Cells were separated from debris using cell strainers and collected by centrifugation. Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Passage one chondrocytes and osteoblasts were used for subsequent experiments.
MEFs isolation and culture
Mouse embryonic fibroblasts were isolated from E12.5 embryos. Head and heart tissues were removed, and the remaining embryos were digested with 0.25% trypsin/EDTA at 37°C for 15 min. After digestion and removal of undigested tissues, the cells were plated into a 100 mm dish, and allowed to grow in DMEM + 10% FBS. Early passages cells were used for all the experiments. Spag17 was knocked down in WT MEFs by treatment with Spag17 (mouse) -3 unique 27mer siRNA duplex (OriGene Technologies, Inc. Rockville, MD).
Alkaline phosphatase activity
The assay was performed following standard techniques. Briefly, confluent cultures of mouse osteoblasts were lysed in 0.05% Triton X-100 (Sigma Aldrich) and homogenized by sonication for 10 seconds. Alkaline phosphatase specific activity was measured in the cell lysates as the release of p-nitrophenol from p-nitrophenylphosphate at pH 10.2. Enzyme activity was normalized to total protein content of the lysates (BCA Assay, ThermoFisher).
Immunofluorescence studies
Osteoblasts and chondrocytes in passage one were fixed in 4% formalin for 1 hour at room temperature and blocked with 10% goat serum diluted in PBS containing 3% BSA and 0.2% Triton X-100. Then, cells were incubated with anti-acetylated α-tubulin mouse antibody overnight at 4°C. After several washes in PBS, cells were stained with anti-mouse IgG alexa-488 labeled antibody (dilution 1: 3,000). Finally, slides were sealed using VectaMount with DAPI (Vector Laboratories, Burlingame, CA). Images were taken by confocal laser-scanning microscopy (Leica TCS-SP2 AOBS). The same immunofluorescence staining protocol was used for MEFs cells. The results presented are representative of mean ± standard error obtained from four different animals of each genotype. Statistical comparisons were made using t-test; p < 0.05 was considered significant.
Western Blot
Equal amounts of protein (100 μg/lane) were heated to 95°C for 10 min in sample buffer, loaded onto 12% SDS-PAGE gels, electrophoretically separated, and transferred to PVDF membranes (Millipore, Billerica, MA) by semi-dry transference. A dual-color precision plus protein standard (BIO-RAD, Hercules, CA) was used. Membranes were blocked for 1 h in 5% milk-TTBS (BIO-RAD, Hercules, CA) and then incubated overnight with Anti- SPAG17 C-terminal antibody (1:5,000) at 4°C, After several washes in TTBS, the membranes were incubated with an anti-rabbit IgG horseradish-peroxidase labeled antibody (1:2,000 dilution) for 1 h at room temperature. Protein was detected with Super Signal Chemiluminescent Substrate (Pierce). β-actin antibody was used as loading control to assure relatively equivalent amounts of protein were present amongst sample types.
Statistical Analysis
InfoStat software was used for statistical analyses (Di Rienzo J.A., Casanoves F., Balzarini M.G., Gonzalez L., Tablada M., Robledo C.W. InfoStat ver 2009. Group InfoStat, FCA, Universidad Nacional de Córdoba, Argentina). Data are presented as means ± standard error. Normality of data was assessed by D'Agostino & Pearson omnibus normality test. Each set of data was analyzed comparing the means from two groups (wild-type and knockout) by Student’s t-test or one-way analysis of variance (ANOVA) followed by Duncan’s test for three group comparisons. Samples were considered significantly different when the p-value was < 0.05.
Ethics statements
All animal procedures were performed in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals. Experiments were in accordance with protocol AD10000167 approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee.
Supporting Information
(A) Representative high magnification pictures from H&E staining on femur from wild-type (+/+) and knockout (-/-) mice. (B) Representative high magnification pictures from von Kossa staining on femur from wild-type (+/+) and knockout (-/-) mice.
(TIF)
(A) Representative high magnification pictures from H&E staining on tibia from wild-type (+/+) and knockout (-/-) mice. (B) Representative high magnification pictures from von Kossa staining on tibia from wild-type (+/+) and knockout (-/-) mice.
(TIFF)
(A) Osteoclasts number in femur from wild-type (+/+) and knockout (-/-) mice. (B) Osteoclasts number in tibia from wild-type (+/+) and knockout (-/-) mice. Data are presented as means ± SEM from 8 mice for each group. No statistically significant differences were found between the two groups, p> 0.05.
(TIFF)
RT-PCR products generated by primers from exon 4 and 5. Notice the knockout mice have a deletion in the entire exon 5 [19]. RNA was isolated from wild-type and Spag17-mutant sternum (S), limb bone (B) and cartilage (C).
(TIF)
(MOV)
(MOV)
(MOV)
Acknowledgments
Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility. We thank Pamela J Gigliotti and Kimberly R Stratton for histological preparation and bone scanning at the VCU Massey Cancer Center Macromolecule Core Facility.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institutes of Health [grant number R01 HD-37416 to J.F.S., R01 HD-076257 to Z.Z.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Weedon MN, Frayling TM. Reaching new heights: insights into the genetics of human stature. Trends in Genetics. 2008; 24: 595–603. 10.1016/j.tig.2008.09.006 [DOI] [PubMed] [Google Scholar]
- 2. Kim J, Lee H, Park T, Kim K, Lee J, Cho N, et al. Identification of 15 loci influencing height in a Korean population. J Hum Genet. 2010; 55: 27–31. 10.1038/jhg.2009.116 [DOI] [PubMed] [Google Scholar]
- 3. Michigami T. Regulatory mechanisms for the development of growth plate cartilage. Cellular and Molecular Life Sciences. 2013; 70: 4213–4221. 10.1007/s00018-013-1346-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003; 423: 332–336. [DOI] [PubMed] [Google Scholar]
- 5. Bruder SP, Caplan AI. Cellular and Molecular Events During Embryonic Bone Development. Connective Tissue Research. 1989; 20: 65–71. [DOI] [PubMed] [Google Scholar]
- 6. Boyan BD, Schwartz Z, Swain LD, Khare A. Role of lipids in calcification of cartilage. The Anatomical Record. 1989; 224: 211–219. [DOI] [PubMed] [Google Scholar]
- 7. Zhao J, Li M, Bradfield J, Zhang H, Mentch F, Wang K, et al. The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature. BMC Med Genet. 2010; 11: 96 10.1186/1471-2350-11-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lango Allen H, Estrada K, Lettre G, Berndt S, Weedon M, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010; 467: 832–838. 10.1038/nature09410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008; 40: 575–583. 10.1038/ng.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takeuchi F, Nabika T, Isono M, Katsuya T, Sugiyama T, Yamaguchi S, et al. Evaluation of genetic loci influencing adult height in the Japanese population. J Hum Genet. 2009; 54: 749–752. 10.1038/jhg.2009.99 [DOI] [PubMed] [Google Scholar]
- 11. N'Diaye A, Chen G, Palmer C, Ge B, Tayo B, Mathias R, et al. Identification, replication, and fine-mapping of Loci associated with adult height in individuals of african ancestry. PLoS Genet. 2011; 7: e1002298 10.1371/journal.pgen.1002298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014; 46: 1173–1186. 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der Valk RJP, Kreiner-Møller E, Kooijman MN, Guxens M, Stergiakouli E, Sääf A, et al. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Human Molecular Genetics. 2014; 24: 1155–1168. 10.1093/hmg/ddu510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z, Jones BH, Tang W, Moss SB, Wei Z, Ho C, et al. Dissecting the Axoneme Interactome: The Mammalian Orthologue of Chlamydomonas PF6 Interacts with Sperm-Associated Antigen 6, The Mammalian Orthologue of Chlamydomonas PF16. Mol Cell Proteomics. 2005; 4: 914–923. [DOI] [PubMed] [Google Scholar]
- 15. Rupp G, O'Toole E, Porter ME. The Chlamydomonas PF6 Locus Encodes a Large Alanine/Proline-Rich Polypeptide That Is Required for Assembly of a Central Pair Projection and Regulates Flagellar Motility. Mol Biol Cell. 2001; 12: 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goduti D, Smith E. Analyses of functional domains within the PF6 protein of the central apparatus reveal a role for PF6 sub-complex members in regulating flagellar beat frequency. Cytoskeleton (Hoboken). 2012; 69: 179–194. 10.1002/cm.21010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wargo MJ, Dymek EE, Smith EF. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J Cell Sci. 2005; 118: 4655–4665. [DOI] [PubMed] [Google Scholar]
- 18. Silina K, Zayakin P, Kalnina Z, Ivanova L, Meistere I, Endzelinš E, et al. Sperm-associated Antigens as Targets for Cancer Immunotherapy: Expression Pattern and Humoral Immune Response in Cancer Patients. Journal of Immunotherapy. 2011; 34: 28–44 10.1097/CJI.1090b1013e3181fb1064fa [DOI] [PubMed] [Google Scholar]
- 19. Teves ME, Zhang Z, Costanzo RM, Henderson SC, Corwin FD, Zweit J, et al. Sperm-associated antigen-17 gene is essential for motile cilia function and neonatal survival. Am J Respir Cell Mol Biol. 2013; 48: 765–772. 10.1165/rcmb.2012-0362OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J. Studies on the morphogenesis of the mouse sternum. I. Normal embryonic development. J Anat. 1952; 86: 373–386. [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan X, Serra RA, Yang S. Function and regulation of primary cilia and intraflagellar transport proteins in the skeleton. Ann N Y Acad Sci. 2014; 1335: 78–99. 10.1111/nyas.12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niswander L. Pattern formation: old models out on a limb. Nat Rev Genet. 2003; 4: 133–143. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Nizkorodov A, Riemenschneider K, Lee CSD, Olivares-Navarrete R, Schwartz Z, et al. Impaired bone formation in Pdia3 deficient mice. PLoS One. 2014; 9: e112708 10.1371/journal.pone.0112708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hajihosseini MK, Lalioti MD, Arthaud S, Burgar HR, Brown JM, Twigg SRF, et al. Skeletal development is regulated by fibroblast growth factor receptor 1 signalling dynamics. Development. 2004; 131: 325–335. [DOI] [PubMed] [Google Scholar]
- 25. Liakhovitskaia A, Lana-Elola E, Stamateris E, Rice D, van 't Hof R, Medvinsky A. The essential requirement for Runx1 in the development of the sternum. Dev Biol. 2010; 340: 539–546. 10.1016/j.ydbio.2010.02.005 [DOI] [PubMed] [Google Scholar]
- 26. Kimura A, Inose H, Yano F, Fujita K, Ikeda T, Sato S, et al. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010; 137: 1159–1167. 10.1242/dev.045005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Temiyasathit S, Jacobs CR. The osteocyte primary cilium and its role in bone mechanotransduction. Annals of the New York Academy of Sciences. 2010; 1192: 422–428. 10.1111/j.1749-6632.2009.05243.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoey DA, Downs ME, Jacobs CR. The mechanics of the primary cilium: An intricate structure with complex function. Journal of Biomechanics. 2012; 45: 17–26. 10.1016/j.jbiomech.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Delaine-Smith RM, Sittichokechaiwut A, Reilly GC. Primary cilia respond to fluid shear stress and mediate flow-induced calcium deposition in osteoblasts. The FASEB Journal. 2014; 28: 430–439. 10.1096/fj.13-231894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gerdes J, Davis E, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009; 137: 32–45. 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goetz S, Anderson K. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010; 11: 331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Day TF, Yang Y. Wnt and Hedgehog Signaling Pathways in Bone Development. J Bone Joint Surg Am. 2008; 90: 19–24. 10.2106/JBJS.G.01174 [DOI] [PubMed] [Google Scholar]
- 33. Schmidts M. Clinical genetics and pathobiology of ciliary chondrodysplasias. Journal of pediatric genetics. 2014; 3: 46–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teves ME, Sears PR, Li W, Zhang Z, Tang W, van Reesema L, et al. Sperm-Associated Antigen 6 (SPAG6) Deficiency and Defects in Ciliogenesis and Cilia Function: Polarity, Density, and Beat. PLoS One. 2014; 9: e107271 10.1371/journal.pone.0107271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ovchinnikov D. Alcian Blue/Alizarin Red Staining of Cartilage and Bone in Mouse. Cold Spring Harb Protoc. 2009; 2009: pdb.prot5170-. [DOI] [PubMed] [Google Scholar]
- 36. Boyan BD, Schwartz Z, Carnes David L., Ramirez V. The Effects of Vitamin D Metabolites on the Plasma and Matrix Vesicle Membranes of Growth and Resting Cartilage Cells in Vitro. Endocrinology. 1988; 122: 2851–2860. [DOI] [PubMed] [Google Scholar]
- 37. Lohmann CH, Bonewald LF, Sisk MA, Sylvia VL, Cochran DL, Dean DD, et al. Maturation State Determines the Response of Osteogenic Cells to Surface Roughness and 1,25-Dihydroxyvitamin D3. Journal of Bone and Mineral Research. 2000; 15: 1169–1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representative high magnification pictures from H&E staining on femur from wild-type (+/+) and knockout (-/-) mice. (B) Representative high magnification pictures from von Kossa staining on femur from wild-type (+/+) and knockout (-/-) mice.
(TIF)
(A) Representative high magnification pictures from H&E staining on tibia from wild-type (+/+) and knockout (-/-) mice. (B) Representative high magnification pictures from von Kossa staining on tibia from wild-type (+/+) and knockout (-/-) mice.
(TIFF)
(A) Osteoclasts number in femur from wild-type (+/+) and knockout (-/-) mice. (B) Osteoclasts number in tibia from wild-type (+/+) and knockout (-/-) mice. Data are presented as means ± SEM from 8 mice for each group. No statistically significant differences were found between the two groups, p> 0.05.
(TIFF)
RT-PCR products generated by primers from exon 4 and 5. Notice the knockout mice have a deletion in the entire exon 5 [19]. RNA was isolated from wild-type and Spag17-mutant sternum (S), limb bone (B) and cartilage (C).
(TIF)
(MOV)
(MOV)
(MOV)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.