Abstract
Glaucoma is a multifactorial, neurodegenerative disease. The molecular mechanisms that underlie the pathophysiological changes in glaucomatous eyes, especially at the early stage of the disease, are poorly understood. Here, we report the findings from a quantitative proteomic analysis of retinas from experimental glaucoma (EG) eyes. An early stage of EG was modeled on unilateral eyes of five nonhuman primates (NHP) by laser treatment-induced elevation of intraocular pressure (IOP). Retinal proteins were extracted from individual EG eyes and their contralateral control eyes of the same animals, respectively, and analyzed by quantitative mass spectrometry (MS). As a result, a total, 475 retinal proteins were confidently identified and quantified. Results of bioinformatic analysis of proteins that showed an increase in the EG eyes suggested changes in apoptosis, DNA damage, immune response, cytoskeleton rearrangement and cell adhesion processes. Interestingly, hemoglobin subunit alpha (HBA) and Ras related C3 botulinum toxin substrate 1 (Rac1) were among the increased proteins. Results of molecular modeling of HBA- and Rac1-associated signaling network implicated the involvement of Mitogen-Activated Protein Kinase (MAPK) pathway in the EG, through which Rac1 may exert a regulatory role on HBA. This is the first observation of this potentially novel signaling network in the NHP retina and in EG. Results of Western blot analyses for Rac1, HBA and a selected MAPK pathway protein indicated synergistic changes in all three proteins in the EG eyes. Further, results of hierarchical cluster analysis of proteomes of control eyes revealed a clear age-proteome relationship, and such relationship appeared disrupted in the EG eyes. In conclusion, our results suggested an increased presence of a potentially novel signaling network at the early stage of glaucoma, and age might be one of the determinant factors in retinal proteomic characteristics under normal conditions.
Keywords: Glaucoma, retina, neurodegeneration, hemoglobin, Rac1, proteomics, age
Introduction
There is a tremendous interest and demand in understanding the pathological mechanism of glaucomatous eye at molecular levels, especially at early stage of the disease. Genomic and proteomic studies of retina, trabecular meshwork cells, aqueous humor and tear in donor eyes of human glaucoma patients have revealed dysregulation in genes involved in multiple cellular pathways or processes, such as cytoarchitecture, oxidative stress response, mitochondrial damage, neural degeneration, inflammation, and apoptosis [1-7]. A limitation in human studies, however, is that the analysis is only feasible on post-mortem specimen from patients in whom the stages of glaucoma may vary greatly. Early stages of experimental glaucoma (EG) have been modeled in a number of animal species such as rabbit, swine, rodents, and non-human primate (NHP), by means of induced elevation of intraocular pressure (IOP). High IOP has been considered to be the most impactful factor associated with glaucomatous damages [8] and is the only treatable risk factor to date. These animal models provide the possibility to identify glaucomatous changes at its onset stage. Among the models, induction of chronic high IOP in NHP produces the closest simulation to the development of glaucoma in human.
The impact of high IOP on the retina has been investigated intensively, mostly on rodents with an acute IOP elevation. In a limited number of published studies on NHP, chronic elevation of IOP has been shown to result in retina nerve fiber layer thinning, and progressive loss of retinal ganglion cells (RGC) and axons [9-13]. Changes in expression levels of genes involved in structural remodeling, immune response, as well as regulation of Müller cells and astrocytes activity have been reported for glaucomatous retinas in NHP [14]. To investigate protein actuators of glaucomatous changes in the retina, in a previous study, we published the first characterization of retinal proteomic changes in NHP EG eyes, and compared them with that in retinas from NHP eyes subjected to optic nerve transection (ONT) [15]. We found that retinal proteomic changes are distinct between animals with mild or high IOP elevation or ONT, as was reported in the above-mentioned genomic study. Here, we report a new proteomic analysis of retinas from eyes treated with the same IOP elevation scheme used in the afore-mentioned publication, with the inclusion of animals at additional younger ages, and with a more advanced mass spectrometry (MS) system, by which twice as many proteins were identified and quantified than that in the previous study. Current results suggest an increased presence of a potentially novel signaling network that has not been reported for the retina before, and a disruption of the correlation between age and retinal proteomes in glaucoma.
Materials and methods
Animals
All procedures adhered to the Association for Research in Vision and Ophthalmology’s Statement for the Use of Animals in Ophthalmic and Vision Research, and were approved and monitored by the Institutional Animal Care and Use Committee (IACUC) at Legacy Research Institute. Five adult female rhesus monkeys (Macaca mulatta) were included in the study. They were seven to eleven years old, respectively, at the beginning of the study.
Induction of unilateral experimental glaucoma [16, 17]
Chronic IOP elevation was induced by laser treatment to the trabecular meshwork in one eye of each animal under ketamine and xylazine anesthesia. One hundred eighty degrees of the trabecular meshwork (50 μm spot size, 1.0 second duration, 600 to 750 mW power) were treated in each of 2 separate sessions at least 2 weeks apart. After each treatment, a sub-Tenon’s injection of 0.5 ml of dexamethasone (10 mg/ml) was given in the inferior fornix of the treated eye to prevent possible post-laser inflammatory response. Laser treatments were repeated (but limited to a 45°, 90° or 180° sector) on subsequent occasions as necessary to achieve a sustained IOP elevation. Since lasering of trabecular meshwork was noninvasive and the only procedure to induce chronic IOP elevation, no sham procedure was performed in the contralateral eyes.
For each animal, three to five pre-laser tests were included to establish baseline values of IOP and peripapillary retinal nerve fiber layer thickness (RNFLT). Thereafter, the same measurements were repeated once every two weeks for the duration of the post-laser follow-up. During each test, IOP was measured by a rebound tonometer (Tonopen XL, Reichert Inc) under ketamine and xylazine anesthesia. The peripapillary retinal nerve fiber layer thickness (RNFLT) was measured using a spectral-domain optical coherence tomography (SDOCT) instrument (Spectralis, Heidelberg Engineering GmbH, Germany) to determine the stage and progression of EG. A single circular B-scan with 12-degree diameter was recorded in both eyes of each animal. Nine to sixteen individual sweeps were averaged to comprise the final B-scan recorded at each session.
The follow-up testing was continued either until the RNFLT was reduced up to 20% from the baseline in the optic nerve head or until abnormal hemodynamic regulatory capability in the optic nerve head was detected [18]. The animals were then euthanized under deep anesthesia, and both eyes of each animal were enucleated. Retinas from the enucleated eyes were isolated immediately, frozen on dry ice, and stored at -80°C for further analyses. Optic nerve 2 mm behind globe was obtained and processed for complete (100%) automated axon counts to validate the severity of retinal ganglion cells damage [19,20].
Protein extraction and sample processing prior to MS analysis [15]
All retinal specimens were processed on the same day. Retinas were thawed on ice and individually homogenized in 450 μl RIPA buffer with a hand-held homogenizer. The homogenates were subjected to three cycles of freeze-thaw between a dry ice-ethanol bath and ice followed by centrifugation at 16,000 g for 30 min. The cleared supernatants were subjected to buffer exchange with 50 mM ammonium bicarbonate to removed components of the RIPA buffer, using 3 kDa spin columns (Millipore). Protein concentrations in the exchanged supernatants were determined with the DC Protein Assay (Bio-Rad). For each retinal sample, 200 μg proteins were denatured with 0.05% RapiGest SF (Waters; 10 min at 95°C), reduced with 10 mM dithiothreitol (30 min at 60°C), and alkylated with 40 mM iodoacetamide (1 hr in the dark at 37°C). The denatured proteins were then digested with sequencing-grade trypsin (Promega; 50:1 w/w, overnight at 37°C). The digestion was terminated by the addition of 0.5% trifluoroacetic acid (TFA), which also destroyed RapiGest in digestion mixtures. The digests were centrifuged at 13,000 rpm for 10 min and supernatants were proceeded to quantitative MS analysis.
Protein identification and quantitation by MS, and analysis of MS data
Tryptic peptides from each retina were analyzed with an advanced MS system consisting of a nanoACQUITY ultra performance liquid chromatography (UPLC) coupled to the Synapt G2-S mass spectrometer (Waters). For each MS run, approximately 300 ng tryptic peptides were injected, and the UPLC separation took place over 120 min using mobile phases consisted of 0.1% (v/v) formic acid (FA) in water (A) and 0.1% (v/v) FA in 100% acetonitrile (ACN) (B), respectively. The system was first equilibrated with 97% mobile phase A and 3% mobile phase B, and then a gradient of mobile phase B was applied as follows: from 3% to 28% in the first 90 min, to 40% in the next 30 min, and then a flush at 85% followed by a re-equilibration to the original condition in the next 40 min. A lock mass solution of glu-fibrinopeptide (200 fmol/μl in 25% ACN and 0.1% FA in water) was delivered via the auxiliary solvent manager at 500 nl/min into the reference sprayer of the NanoLockSpray source. Electrospray ionization was performed with an uncoated, pulled fused silica emitter (New Objective) with a potential of 3.0 kV. The mass spectrometer was operated under HDMSE and RESOLUTION mode with a typical mass resolution of 20,000. Alternating scans were used to detect precursor ions and then fragment ions. The masses of the precursors were detected with a 0.8 s scan with a relatively low collision energy (CE) of 4 eV (MS scan), which was followed by a 0.8-s scan during which the CE was ramped from 19 to 45 eV (HDMSE scan). The ion mobility separation (IMS) wave velocity was maintained at 600 m/s. The instrument was calibrated with 13 fragment ions of the MS/MS spectrum of glu-fibrinopeptide. To correct any mass shift that occurred during instrument operation, a single-point calibration was performed against the lock mass compound, doubly-charged glu-fibrinopeptide [mass/charge ratio (m/z) 785.8426], which was sampled every 30 s.
MS data were analyzed with ProteinLynx Global Server (PLGS) software (Waters; version 2.4), using a combined database of macaca and human proteins from the Universal Protein Resource (UniProt, www.UniProt.org). At the time when data analyses were performed, the combined protein database contained a total of 18,433 human and macaca protein sequences and an equal number of reversed sequences for the estimation of false-positive identifications.
Only proteins that met the following criteria were accepted as valid entries for further bioinformatic or biochemical analyses: two fragment ions per peptide and five fragment ions per protein with at least two unique peptides per protein; within 10 and 20 parts per million (ppm) of the theoretical masses for precursor and fragment ions, respectively. The false positive rate of database search was 4% (estimated using reversed sequences). Furthermore, the accepted proteins must be identified in at least two of three technical replicate MS runs of the same sample. Absolute quantitation of identified protein was calculated by comparison of ion chromatographic peak areas relative to that of an internal standard (yeast alcohol dehydrogenase, ADH). The amounts of protein in each sample were then normalized to femtomoles per nanogram (fmol/ng) to take into account different loading amounts of individual samples. The fmol/ng values were compared between the control and EG eyes of the same animal with the Student t-test. Proteins with p values less than 0.05 were considered significantly changed in the EG eyes.
Hierarchical clustering of retinal proteomes of the control and EG eyes was performed using the R v2.11.1 and MultiExperiment Viewer v4.8.1 softwares. Pearson correlation and average linkage clustering were used for calculating distance between clusters [21]. The same analysis was also performed on our previously published retinal proteomic datasets from four pairs of EG eyes [15]. Bioinformatic characterization of retinal proteins that showed a difference in quantities between the control and EG eyes was performed with the assistance of the MetaCore program (Thomson Reuters, Version 6.14).
Western blotting
Western blot analysis was performed following standard protocols [15], with appropriate antibodies specified in the corresponding figure legends under “Results and Discussion” section. Western blotting for α tubulin was performed on the same blots for proteins of interests as a loading control. Signals of appropriate protein bands on Western blotting images were documented with a FujiFilm LAS-4000 system (Fuji). The band intensities were normalized with that of α tubulin and compared between the two eyes of the same animal. The significance of differences was determined with the Student t-test, and a p value less than 0.05 was considered significant.
Results and discussion
Induction of glaucomatous conditions
By the time of sacrificing, the IOP levels in five EG eyes were significantly higher than that in their contralateral control eyes, (32.6 ± 4.1 mmHg and 13.7 ± 2.4 mmHg (mean ± S.E.), respectively; p = 0.015 by paired Student t-test) (Table 1). Accompanying the IOP elevation, an average of 10.1 ± 3.0% loss of RNFLT was observed in the EG eyes. Hence, an early stage of glaucomatous condition was considered successfully modeled in the EG eyes. The study employed a noninvasive procedure of laser treatment, which causes no or only mild temporary inflammatory response within anterior chamber during the first few days after lasering. It is thus not anticipated that such a transient and local response would contribute to retinal proteomic differences between the EG and contralateral control eyes, when comparisons were made at the end of experiments weeks after the laser treatment.
Table 1.
Demography of test subjects
| Animal ID | Sex | Age (yr) | Highest IOP | Duration (days) | Accumulated IOP (IOP x days) | Loss of RNFLT (%) | Ratios of axon count (EG/CTR) |
|---|---|---|---|---|---|---|---|
| 24253 | F | 7 | 18 | 227 | 114 | 17 | 0.79 |
| 29584 | F | 7 | 41 | 279 | 158 | 12 | No data |
| 28675 | F | 8 | 36 | 233 | 93 | 4.5 | 0.91 |
| 23583 | F | 8 | 26 | 107 | 52 | 0 | 1.05 |
| 28318 | F | 11 | 42 | 184 | 79 | 17 | 0.78 |
| Mean | 8.2 | 32.6 | 206 | 99.2 | 10.1 | 0.88 | |
| SE | 0.7 | 4.1 | 29.0 | 17.8 | 3.4 | 0.06 |
The Duration refers to the number of days from 1st laser treatment to sacrifice. Accumulated IOP refers to the accumulation of IOP differences between EG and contralateral control (CTR) eyes of the same animal by days after laser treatment.
Retinal proteomic changes in EG eyes
In this study, the MS analysis of retinal proteins employed an advanced UPLC-MS/MS system with IMS. As a result, a total of 475 proteins were identified in the 5 pairs of retinas. On average, ~260 proteins were identified and quantified for each eye. Table 2 lists the numbers of proteins identified in individual retinas and numbers of proteins that showed a difference in quantity between the control and EG eyes of the same animals. For individual animals, approximately 80% of identified retinal proteins overlapped between the control and EG eyes. It is noticeable that the numbers of identified retinal proteins are limited when compared with that of identified in other neuronal tissues, such as mouse cortices, when analyzed using the same protein extraction and MS analysis protocols (Zhou, unpublished data). Although, to the best of our knowledge, the current numbers are the highest obtained with single UPLC-MS/MS runs of NHP retinas without pre-fractionation or enrichment steps, it remains a challenge to improve the proteome coverage by MS-based proteomic analysis of NHP retinas. Multiple factors may affect the total number of identifiable proteins in a single MS run. Examples of such factors may include the composition of the proteome and the stoichiometry of all extractable proteins. For a proteome highly enr-iched with a few proteins (for example, cytoskeleton proteins in muscle cells), the numbers of identifiable proteins in a single MS run is expected to be less than that of a proteome in which the abundances of cellular proteins are more evenly distributed. It will be the interest of future studies to compare the compositions of different neuronal tissues (NHP retinas vs. brain tissues, for example). Nevertheless, with numbers more than twice as great as those previously published on analysis of NHP retinal proteomes [15], the current datasets allowed the recognition of a potentially novel signaling network that appeared increased in EG eyes (described next). The complete lists of identified retinal proteins in this study are provided in Supplementary Table 1.
Table 2.
Numbers of proteins identified and quantified in individual retinas*
| Animal ID | EG | CTR | Detected in both eyes | Regulated in EG eyes** | |
|---|---|---|---|---|---|
|
| |||||
| Up | Down | ||||
| 24253 | 269 | 256 | 211 | 78 | 73 |
| 29584 | 272 | 281 | 209 | 93 | 109 |
| 28675 | 253 | 277 | 224 | 68 | 66 |
| 23583 | 262 | 272 | 217 | 62 | 80 |
| 28318 | 244 | 262 | 194 | 84 | 79 |
Numbers represent proteins that were confidently identified and quantified. See Methods for criteria for acceptance of MS-detected proteins.
A protein is considered up- or down-regulated in the EG eye if it showed an increase or decreased, respectively, in the EG eye, or was detected only in the EG or control (CTR) eye, respectively, of the same animal.
For proteins that were increased or decreased in quantities in the EG eyes, when compared with control eyes, their association with known biological processes or network was assessed with the assistance of the MetaCore program. As listed in Table 3, cytoskeleton, immune response and cell cycle processes were recognized either in all five or four of the five animals. The processes of apoptosis, cell adhesion and protein folding were recognized in at least three of the five EG eyes. The recognition of the immune response processes in the current study is worth noting. In recent years, there has been an increased interest in significant roles of immune response in neuronal disorders. Little is known about the underlying molecular mechanisms in glaucomatous retinas for such responses. As to be introduced and discussed below, our current proteomic data suggest the involvement of a signaling network that may mediate the retinal response to high IOP conditions, and such response may include the immune response.
Table 3.
Bioinformatics of changed proteins in EG eyes
| Process networks | Animals |
|---|---|
| Increased | |
| Apoptosis | 23583, 28675, 29584 |
| Cell adhesion | 23583, 24253, 28318 |
| Cell cycle | 24253, 28675, 29584 |
| Cytoskeleton | 23583, 24253, 28318, 28675, 29584 |
| DNA damage | 24253, 28675 |
| Immune response | 24253, 28318, 28675, 29584 |
| Inflammation | 24253, 28318, 29584 |
| Muscle contraction | 28318, 29584 |
| Neurophysiological process | 23583, 28675, 29584 |
| Protein folding | 28318 |
| Reproduction | 23583 |
| Response to hypoxia and oxidative stress | 28675 |
| Signal Transduction | 28675 |
| Transcription | 23583, 29584 |
| Transport | 28675, 29584 |
| Decreased | |
| Apoptosis | 24253, 28675, 29584 |
| Cell adhesion | 24253, 28318, 28675 |
| Cell cycle | 23583, 28318, 28675, 29584 |
| Cytoskeleton | 23583, 24253, 28318, 28675, 29584 |
| Development | 24253, 28318, 29584 |
| Immune response | 23583, 28318, 29584 |
| Muscle contraction | 23583 |
| Neurophysiological process | 24253 |
| Protein folding | 23583, 24253, 29584 |
| Response to hypoxia and oxidative stress | 29584 |
| Transcription | 28318, 28675 |
Lists of proteins that showed a change in EG retinas were uploaded onto the MetaCore Program to assess their association with known biological processes or signaling networks. The table lists the top ten most significant processes as determined by the MetaCore Program.
Recognition of a novel signaling network in retinas of NHP EG eyes
By further examining individual proteins that changed in the same direction in EG eyes, as determined by quantitative MS, five proteins were recognized, namely hemoglobin subunit alpha (HBA), Ras related C3 botulinum toxin substrate 1 (Rac1), core histone macro H2A 1 (H2AFY), vesicle fusing ATPase (NSF) and retinol dehydrogenase 12 (RDH12). Of them, HBA and Rac1 showed an increase, whereas H2AFY, NSF and RDH12 showed a decrease in the EG eyes. An increase in HBA protein has been reported in post mortem human glaucomatous eyes, as well as rat eyes subjected to modeled glaucomatous conditions [22]. Such an increase may reflect a protective response towards high IOP-induced hypoxic conditions in the eye. Rac1 is a GTP binding protein of the Ras superfamily. It integrates diverse signaling pathways. Previous in vitro and in vivo studies have suggested that Rac1 activation may improve retina ganglion cell growth and survival [3,23-25]. In the current study, an increase of both Rac1 and HBA proteins in EG eyes led us to consider whether the two proteins may be involved in the same molecular pathways. With the assistance of the MetaCore program and using Rac1 and HBA as root proteins, Protein Interaction Network analysis was performed. The result, as illustrated in Figure 1, revealed a possible regulatory role of Rac1 on HBA through the mitogen-activated protein kinase (MAPK) pathways. Rac1 is known to interact with MAPK signal transduction pathways [26,27]; the activation of the latter has been reported in stress responses including glaucomatous conditions [27]. In mammals, there are three major MAPK pathways: the p38, the extracellular signal-regulated kinase (ERK), and c-Jun NH2-terminal kinase (JNK) pathways [27], respectively. The bioinformatics output from the current analysis directed us to further consider whether there were synergistic increases in levels of MAPK signaling pathway proteins when the levels of Rac1 and HBA proteins were increased in EG eyes, as determined by MS analysis. Accordingly, Western blot analysis was performed for the phosphorylated form of Erk1/2 (p-Erk1/2), Rac1 and HBA using the remaining protein extracts from retinas that were analyzed by MS. The results, as illustrated in Figure 2, showed a trend of increase in EG eyes in the levels of all three proteins. Such results not only validated the MS determinations on HBA and Rac1, but also support the possibility of an enhanced presence of a signaling network involving Rac1, MAPK pathways and HBA in the EG eyes. One may postulate that the increased Rac1 and MAPK pathways play a regulatory role on the increase in HBA levels in EG eyes. This possibility remains to be examined in the future with biochemical and molecular biological approaches. Furthermore, as mentioned earlier, in literature, immune response has been considered as a part of the retinal response to high IOP. In datasets of the current study, Rac1 and Erk1/2 were among proteins that contributed to the recognition of immune response processes by the MetaCore program. We speculate that a signaling cascade involving Rac1/MAPK pathways/HBA proteins exists in the retina and has the potential to mediate retinal glaucomatous changes.
Figure 1.
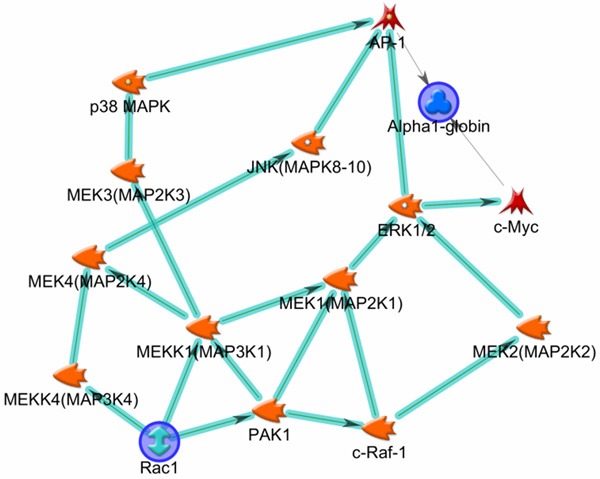
Molecular network involving Rac1, Erk1/2 and HBA. The figure illustrates possible relationships among Rac1, Erk1/2 and HBA proteins, as revealed by the results of Protein Interaction Network analysis performed by the MetaCore program (Version 6.14), using Rac1 and HBA as root proteins (blue circles). The orange objects represent protein kinases; among them those with phosphatase activities are indicated by a hallow dot in the object. The direction of arrow-headed cyan lines indicates outgoing interaction. Accordingly, a possible forward going, regulatory relationship among Rac1, Erk1/2 and HBA is demonstrated.
Figure 2.
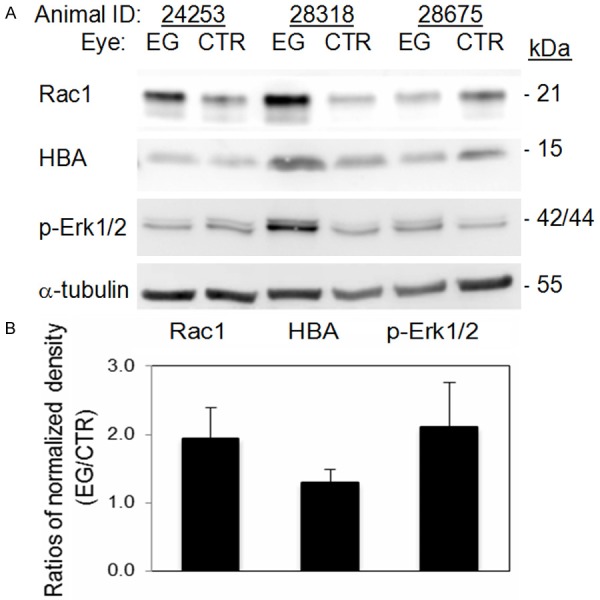
Western blot analysis for Rac 1, HBA and Phosphorylated Erk1/2 (p-Erk1/2). Western blot analysis was performed on protein extracts from the same retinas that were analyzed with quantitative MS. Antibodies against Rac-1 and p-Erk1/2, respectively, were obtained from Cell Signaling, and anti-HBA from Chemicon. HRP-conjugated goat anti-rabbit IgG and goat anti-chicken IgY antibodies were from Santa Cruz and Abcam, respectively. Analysis for α tubulin (antibody from Santa Cruz) was also performed on the same blots as a loading control. A. Images of Western blotting. B. Fold differences in band densities between EG and control (CTR) eyes. Densities of appropriate protein bands in A, after first normalized with that of α tubulin, were compared between EG and control eyes. Due to the limited sample size (n = 3), the differences between EG and control eyes were not statistically significant, but with a clear trend of increase in EG eyes. p = 0.118, 0.165 and 0.202 for Rac1, p-Erk1 and HBA, respectively.
With limited availability of specimen for neuroanatomical analysis in this study, we were unable to determine the retinal cell populations in which the above-mentioned proteomic changes took place in EG eyes. Results of studies on retinal hemoglobin expression in rats and postmortem human glaucomatous eyes suggest that retina ganglion cells and pigment epithelium cells are likely involved [22,28].
Age and retinal proteome under normal and glaucomatous conditions
The association between neurodegenerative conditions and aging has long been recognized. In human, the prevalence of open-angle glaucoma is higher in older than in the relatively younger populations [29]. Little is known about the age-retinal proteome relationship in human and how the relationship may change in retinal disorders. The ages of the five NHPs used in this study ranged from 7 to 11 years. To assess whether the retinal proteomes exhibit age-related differences, hierarchical clustering analysis was used to blindly classify identified proteomic datasets in this study. First, we analyzed retinal proteomes of the five control eyes. As shown in Figure 3A, an age-proteome cluster relationship appears to exist, with the proteome of the oldest animal clearly differentiated from that of the four younger animals (Pearson correlation factor 0.74). We then re-analyzed the previously published retinal proteomic datasets on four NHP animals, aged between 9 and 13 years old [15], noting that the data were generated with different analytical protocols and MS settings. As illustrated in Figure 3B, a similar age-proteome cluster relationship was also seen. Interestingly, when the proteomic datasets of EG eyes were analyzed, no age-proteome cluster relationship was seen in datasets from either the present or the previously published study (Figure 3C, 3D, respectively). In other words, the age-proteome relationship appeared disrupted in retinas of EG eyes. Thus, the retinal proteome not only has changed in response to high IOP conditions, but also lost its correlation to age. Could this implicate the possibility that an attenuated ability to adapting age-related changes, as manifested by proteomic changes, increases the risk of glaucoma? Since the life span of Macaca mulatta is approximately 29 years, the corresponding human ages of NHP animals in this study can be estimated to be between 20 and 35 years old. As mentioned earlier, in human, open-angle glaucoma is more prevalent in older than in relatively younger populations [29]. Very interestingly, in a recent publication on the longitudinal aspect of age-related changes in IOP in human, more substantial changes were reported for populations below 30 years old than those above [30]. A more definite characterization of age-proteome relationship in NHP retinas will rely on future studies to include animals with greater age spans.
Figure 3.
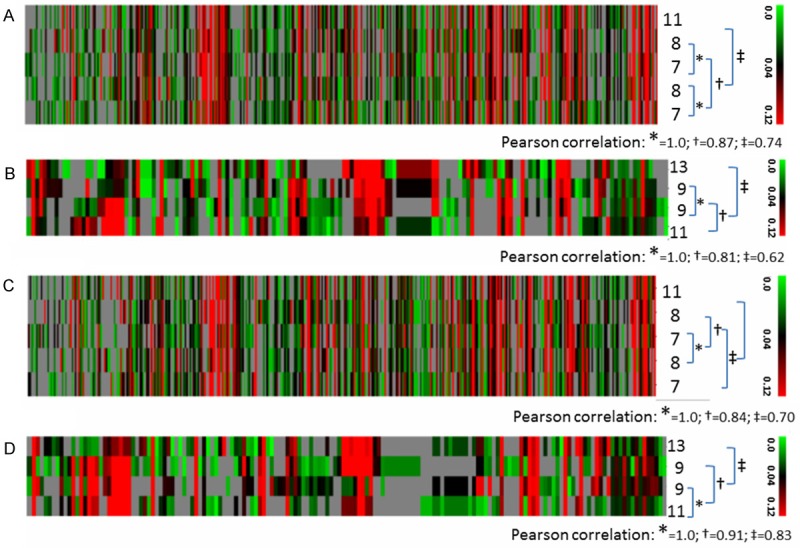
Hierarchical clustering analysis. Colors on the dendrograms indicate protein abundance (green-black-red: low-medium-high; grey: not identified as indicated by the color bar cade on the right of each graph). Each column corresponds to one protein. Each row corresponds to one NHP study subject, with its age marked on the right of the dendrogram. The Pearson correlation factors are listed under the square brackets linking the animals analyzed. A and C: Control and EG eyes, respectively, in the current study. B and D: Control and EG eyes, respectively, in the 2011 Stowell study [15].
Conclusion
Using an advanced, quantitative MS-based proteomic approach, a molecular signaling pathway involving Rac1/MAPK/HBA proteins was recognized in retinas of early glaucoma eyes modeled on NHP. A possible disruption of age-proteome relationship was observed in the retina of glaucomatous eyes as well.
Acknowledgements
The word was supported by NIH R01 grant EY019939 (LW), Legacy Good Samaritan Foundation, Portland, OR (LW), NIMHD G12 Grant 8G12MD007602 and G20 grant RR031196 (to MSM/AZ), and MSM Endowment (MSM/AZ).
Supporting Information
References
- 1.Nikolskaya T, Nikolsky Y, Serebryiskaya T, Zvereva S, Sviridov E, Dezso Z, Rahkmatulin E, Brennan RJ, Yankovsky N, Bhattacharya SK, Agapova O, Hernandez MR, Shestopalov VI. Network analysis of human glaucomatous optic nerve head astrocytes. BMC Med Genomics. 2009;2:24. doi: 10.1186/1755-8794-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izzotti A, Longobardi M, Cartiglia C, Sacca SC. Proteome alterations in primary open angle glaucoma aqueous humor. J Proteome Res. 2010;9:4831–8. doi: 10.1021/pr1005372. [DOI] [PubMed] [Google Scholar]
- 3.Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G. Glaucomatous tissue stress and the regulation of immune response through glial Toll-like receptor signaling. Invest Ophthalmol Vis Sci. 2010;51:5697–707. doi: 10.1167/iovs.10-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tezel G, Yang X, Luo C, Kain AD, Powell DW, Kuehn MH, Kaplan HJ. Oxidative stress and the regulation of complement activation in human glaucoma. Invest Ophthalmol Vis Sci. 2010;51:5071–82. doi: 10.1167/iovs.10-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colak D, Morales J, Bosley TM, Al-Bakheet A, AlYounes B, Kaya N, Abu-Amero KK. Genome-wide expression profiling of patients with primary open angle glaucoma. Invest Ophthalmol Vis Sci. 2012;53:5899–904. doi: 10.1167/iovs.12-9634. [DOI] [PubMed] [Google Scholar]
- 6.Pieragostino D, Agnifili L, Fasanella V, D’Aguanno S, Mastropasqua R, Di Ilio C, Sacchetta P, Urbani A, Del Boccio P. Shotgun proteomics reveals specific modulated protein patterns in tears of patients with primary open angle glaucoma naive to therapy. Mol Biosyst. 2013;9:1108–16. doi: 10.1039/c3mb25463a. [DOI] [PubMed] [Google Scholar]
- 7.Tezel G. A decade of proteomics studies of glaucomatous neurodegeneration. Proteomics Clin Appl. 2014;8:154–67. doi: 10.1002/prca.201300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray K, Mookherjee S. Molecular complexity of primary open angle glaucoma: current concepts. J Genet. 2009;88:451–67. doi: 10.1007/s12041-009-0065-3. [DOI] [PubMed] [Google Scholar]
- 9.Gaasterland D, Tanishima T, Kuwabara T. Axoplasmic flow during chronic experimental glaucoma. 1. Light and electron microscopic studies of the monkey optic nervehead during development of glaucomatous cupping. Invest Ophthalmol Vis Sci. 1978;17:838–46. [PubMed] [Google Scholar]
- 10.Glovinsky Y, Quigley HA, Dunkelberger GR. Retinal ganglion cell loss is size dependent in experimental glaucoma. Invest Ophthalmol Vis Sci. 1991;32:484–91. [PubMed] [Google Scholar]
- 11.Crawford ML, Harwerth RS, Smith EL 3rd, Shen F, Carter-Dawson L. Glaucoma in primates: cytochrome oxidase reactivity in parvo- and magnocellular pathways. Invest Ophthalmol Vis Sci. 2000;41:1791–802. [PubMed] [Google Scholar]
- 12.Hare WA, Ton H, Ruiz G, Feldmann B, Wijono M, WoldeMussie E. Characterization of retinal injury using ERG measures obtained with both conventional and multifocal methods in chronic ocular hypertensive primates. Invest Ophthalmol Vis Sci. 2001;42:127–36. [PubMed] [Google Scholar]
- 13.Fortune B, Reynaud J, Cull G, Burgoyne CF, Wang L. The Effect of Age on Optic Nerve Axon Counts, SDOCT Scan Quality, and Peripapillary Retinal Nerve Fiber Layer Thickness Measurements in Rhesus Monkeys. Transl Vis Sci Technol. 2014;3:2. doi: 10.1167/tvst.3.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyahara T, Kikuchi T, Akimoto M, Kurokawa T, Shibuki H, Yoshimura N. Gene microarray analysis of experimental glaucomatous retina from cynomologous monkey. Invest Ophthalmol Vis Sci. 2003;44:4347–56. doi: 10.1167/iovs.02-1032. [DOI] [PubMed] [Google Scholar]
- 15.Stowell C, Arbogast B, Cioffi G, Burgoyne C, Zhou A. Retinal proteomic changes following unilateral optic nerve transection and early experimental glaucoma in non-human primate eyes. Exp Eye Res. 2011;93:13–28. doi: 10.1016/j.exer.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Bellezza AJ, Rintalan CJ, Thompson HW, Downs JC, Hart RT, Burgoyne CF. Deformation of the lamina cribrosa and anterior scleral canal wall in early experimental glaucoma. Invest Ophthalmol Vis Sci. 2003;44:623–37. doi: 10.1167/iovs.01-1282. [DOI] [PubMed] [Google Scholar]
- 17.Burgoyne CF, Downs JC, Bellezza AJ, Hart RT. Three-dimensional reconstruction of normal and early glaucoma monkey optic nerve head connective tissues. Invest Ophthalmol Vis Sci. 2004;45:4388–99. doi: 10.1167/iovs.04-0022. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Burgoyne CF, Cull G, Thompson S, Fortune B. Static blood flow autoregulation in the optic nerve head in normal and experimental glaucoma. Invest Ophthalmol Vis Sci. 2014;55:873–80. doi: 10.1167/iovs.13-13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cull GA, Reynaud J, Wang L, Cioffi GA, Burgoyne CF, Fortune B. Relationship between orbital optic nerve axon counts and retinal nerve fiber layer thickness measured by spectral domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2012;53:7766–73. doi: 10.1167/iovs.12-10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynaud J, Cull G, Wang L, Fortune B, Gardiner S, Burgoyne CF, Cioffi GA. Automated quantification of optic nerve axons in primate glaucomatous and normal eyes--method and comparison to semi-automated manual quantification. Invest Ophthalmol Vis Sci. 2012;53:2951–9. doi: 10.1167/iovs.11-9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tezel G, Yang X, Luo C, Cai J, Kain AD, Powell DW, Kuehn MH, Pierce WM. Hemoglobin expression and regulation in glaucoma: insights into retinal ganglion cell oxygenation. Invest Ophthalmol Vis Sci. 2010;51:907–19. doi: 10.1167/iovs.09-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linseman DA, Laessig T, Meintzer MK, McClure M, Barth H, Aktories K, Heidenreich KA. An essential role for Rac/Cdc42 GTPases in cerebellar granule neuron survival. J Biol Chem. 2001;276:39123–31. doi: 10.1074/jbc.M103959200. [DOI] [PubMed] [Google Scholar]
- 24.Le SS, Loucks FA, Udo H, Richardson-Burns S, Phelps RA, Bouchard RJ, Barth H, Aktories K, Tyler KL, Kandel ER, Heidenreich KA, Linseman DA. Inhibition of Rac GTPase triggers a c-Jun- and Bim-dependent mitochondrial apoptotic cascade in cerebellar granule neurons. J Neurochem. 2005;94:1025–39. doi: 10.1111/j.1471-4159.2005.03252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzetto E, Ettorre M, Pontelli V, Bolomini-Vittori M, Bolognin S, Zorzan S, Laudanna C, Buffelli M. Rac1 selective activation improves retina ganglion cell survival and regeneration. PLoS One. 2013;8:e64350. doi: 10.1371/journal.pone.0064350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vojtek AB, Cooper JA. Rho family members: activators of MAP kinase cascades. Cell. 1995;82:527–9. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 27.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 28.Tezel TH, Geng L, Lato EB, Schaal S, Liu Y, Dean D, Klein JB, Kaplan HJ. Synthesis and secretion of hemoglobin by retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2009;50:1911–9. doi: 10.1167/iovs.07-1372. [DOI] [PubMed] [Google Scholar]
- 29.Mathews PM, Ramulu PY, Friedman DS, Utine CA, Akpek EK. Evaluation of ocular surface disease in patients with glaucoma. Ophthalmology. 2013;120:2241–8. doi: 10.1016/j.ophtha.2013.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao D, Kim MH, Pastor-Barriuso R, Chang Y, Ryu S, Zhang Y, Rampal S, Shin H, Kim JM, Friedman DS, Guallar E, Cho J. A longitudinal study of age-related changes in intraocular pressure: the Kangbuk Samsung Health Study. Invest Ophthalmol Vis Sci. 2014;55:6244–50. doi: 10.1167/iovs.14-14151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


