Abstract
In the last decade we have witnessed a dramatic increase in the proportion and absolute number of bacterial pathogens resistant to multiple antibacterial agents. Multidrug-resistant bacteria are currently considered as an emergent global disease and a major public health problem. The B-Debate meeting brought together renowned experts representing the main stakeholders (i.e. policy makers, public health authorities, regulatory agencies, pharmaceutical companies and the scientific community at large) to review the global threat of antibiotic resistance and come up with a coordinated set of strategies to fight antimicrobial resistance in a multifaceted approach. We summarize the views of the B-Debate participants regarding the current situation of antimicrobial resistance in animals and the food chain, within the community and the healthcare setting as well as the role of the environment and the development of novel diagnostic and therapeutic strategies, providing expert recommendations to tackle the global threat of antimicrobial resistance.
Keywords: Antibiotic consumption, antibiotic resistance, antibiotic stewardship, antibiotics as growth promoters, drug discovery, infection control measures, multidrug resistant bacteria, self-medication, surveillance, wastewater treatment plants
Facing the problem
In the last decade we have witnessed a dramatic increase both in the proportion and absolute number of bacterial pathogens presenting multidrug resistance to antibacterial agents. Organizations such as the US Centers for Disease Control and Prevention (CDC), the European Centre for Disease Prevention and Control (ECDC) and the World Health Organization (WHO) are considering infections caused by multidrug-resistant (MDR) bacteria as an emergent global disease and a major public health problem.
The emergence of resistant microorganisms, either by mutations or the acquisition of mobile genetic elements carrying resistance genes, may take place irrespective of the presence of antibacterial agents. It is the exposure to these drugs what provides the necessary selective pressure for the rise and spread of resistant pathogens. Therefore, the driving force behind the increasing rates of resistance can ultimately be found in the abuse and misuse of antibacterial agents, whether used in patients and livestock or released into the environment. This is no longer a medical issue. Antimicrobial resistance has become a global health threat that will require the coordinated action of many different stakeholders to tackle antibiotic resistance at its very root.
The aim of the meeting organized jointly by B-Debate (Bio-Cat) and the Barcelona Institute for Global Health (ISGlobal) in partnership with the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and the Spanish Network for Research in Infectious Diseases (REIPI) was to generate debate among the main stakeholders (i.e. policy makers, public health authorities, regulatory agencies, pharmaceutical companies and the scientific community at large) and come up with a coordinated set of strategies to fight antimicrobial resistance in a multifaceted approach. The meeting focused on three major areas: antimicrobial resistance in animals and the food chain; in the environment and the community; and within the healthcare setting. Also discussed was the lack of new therapeutic options.
Antimicrobial resistance in animals and the food chain
The widespread use of antimicrobial agents in animals and the food chain constitutes an important source of antimicrobial resistance, although the impact of such use on human health remains controversial [1]. Massive amounts of antibiotics have been used as growth promoters as well as for prophylaxis and the treatment of infections among farm animals and in aquaculture, increasing the selective pressure on both commensal and pathogenic microorganisms that can spread to humans through direct contact and via the food chain or indirectly from the environmental pollution of farm effluents [2].
Interventions to limit the emergence and spread of resistant bacteria in the animal setting may include the following: (a) banning antibiotic use as growth promoters and limiting its use for other nontherapeutic applications, (b) reducing the dissemination of MDR bacteria through the food chain by improving farm biosecurity and developing alternative treatment strategies and increasing hygienic conditions and practices along the food chain, (c) developing education programs, mainly directed at veterinarians, farmers, and food handlers and (d) linking surveillance systems on antibiotic resistance established for humans and animals.
The European Food Safety Authority (EFSA) is playing an essential role in detecting emerging risks in the area of MDR bacteria within the food industry. Several proposals have been made by EFSA for the harmonization of monitoring and reporting of resistant bacteria, such as: (a) agreeing a comprehensive set of antibacterial agents to be included in the monitoring plans, (b) reinforcing antimicrobial resistance monitoring in sentinel bacteria, (c) conducting active monitoring programs in healthy animals based on randomized sampling plans and (d) harmonizing of epidemiological values [3].
Antibiotics that have become critical for human health should be clearly identified and their use restricted to humans only in order to avoid cross-resistance. In this respect, the WHO has established a list of essential antimicrobial agents for human use to be avoided in nonhuman interventions [4]. Compliance with the WHO recommendations, however, is neither mandatory nor regulated. WHO has also initiated different collaborative programs to tackle foodborne antimicrobial resistance through the promotion of national coordination and integrated surveillance, prevention and control of antibiotic resistance in the food chain and the improvement of awareness on antibiotic use and risk of resistance among veterinarians and the food industry.
Antimicrobial resistance within the community
Antimicrobial resistance in the community has steadily been increasing during the last decades, especially regarding resistance to quinolones, carbapenems and third-generation cephalosporins. The latter relates to the increased prevalence of extended-spectrum β-lactamases among Enterobacteriaceae and with the spread of high-risk clones such as Escherichia coli ST131 [5]. Surveillance studies of antimicrobial resistance and antibiotic consumption have drawn attention to this phenomenon and should be used to drive political campaigns to contain resistance [6].
The task of the CDC/ECDC is crucial in identifying, assessing and communicating current and emerging human health threats on antimicrobial resistance. The latest ECDC report on antimicrobial resistance surveillance in Europe (http://www.ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2012.pdf) showed that methicillin-resistant Staphylococcus aureus prevalence is stabilizing or even decreasing in some countries, while resistance to third-generation cephalosporins in particular and multidrug resistance (three or four antibacterial agents) in general continues to show a sharp and widespread increase in E. coli and Klebsiella pneumoniae[7]. Increasing resistance to carbapenems is also becoming more frequent in a number of countries [8].
Currently, antimicrobial consumption data from the European Union and countries belonging to the European Economic Area/European Free Trade Association are expressed as a number of defined daily doses (DDD) per 1000 inhabitants and per day. Complementary to DDD, the number of packages per 1000 inhabitants and per day are also reported, depending on the availability of data on packages from the national surveillance networks [6]. Information on packages is deemed to improve the understanding and interpretation of differences in the levels and trends of antimicrobial consumption observed between and within countries, as the DDD system cannot take into account changes in package content [9]. In addition, a drug resistance index that aggregates information about antibiotic resistance and antibiotic used into a single composite measure has also been proposed [10]. Such drug resistance index, similar to the way the Dow Jones is used in economics, would allow the continuous quantitation of antibiotic effectiveness overtime in particular geographic areas.
As stated above, antibiotic abuse has greatly contributed to speed up the development of antibiotic resistance, and in this regard human medicine has played a key role [11]. Inappropriate prescribing (whether caused by obsolete guidelines or pharmaceutical pressures), over-the-counter antibiotic availability and self-medication reflect a general lack of awareness on the global threat that antibiotic resistance poses to our society [12]. Educational programs on the rational use of antibiotics addressed to primary care physicians, drug vendors and the community in general must be enforced to ease the pressure on prescribers and reduce antibiotic consumption. Similarly, the prescription of delayed receipts conditioned to the remission or worsening of clinical symptoms might also contribute to such reduction.
Additional measures should include up-to-date local antibiotic prescribing guidelines, active reporting on antibiotic prescribing and consumption and the enforcement of local surveillance programs on antibiotic resistance. The implementation of such measures, however, needs substantial legislation amendments and increased funding, which depend on a strong commitment by policy makers at both national and international scales [13].
Of particular note is the use of antibiotics in low-income countries, where there are additional factors contributing to the emergence of resistance, including (a) less potent activity of some antibacterial agents (including counterfeit drugs), (b) over-the-counter availability, with insufficient dosages, (c) lack of diagnostic laboratories and (d) poor level of sanitation, facilitating the familiar and community spread of resistant organisms [14,15]. Special attention should be paid to global food commercialization and international travel.
The role of the environment
The excessive use of antimicrobial agents to treat both humans and animals has also caused the accumulation of these compounds in the environment, and the impact of such accumulation on the emergence of antibiotic resistance should not be underrated [16].
Antibacterial agents have several routes of entry into the environment, such as sewage from the community or hospitals through manure and water bodies [17,18]. The accumulation of antibacterial agents further selects resistant microorganisms, turning the environment into a gigantic reservoir for antibiotic resistance genes that feeds on the constant and increasing environmental pollution. Wastewater treatment plants have become a hot spot for horizontal gene transfer and the coselection of genetic determinants providing resistance to antibiotics, pollutants, heavy metals, biocides, disinfectants, or detergents. The current legislation on water quality mainly focuses on the presence of indicator microorganisms but does not address the antibiotic concentrations of sewages and treatment plants.
Strategies to mitigate the risks of environmental exposure should be aimed at improving industrial systems for sanitation and decontamination of hospital sewage water [19].
Antimicrobial resistance within the healthcare setting
The concurrence of high antibiotic consumption, critically ill patients and a permanent influx of pathogenic species within the healthcare setting nurtures the development of resistance and provides an ideal scenario for the dissemination of resistant microorganisms and horizontal transfer of resistance genes. The management of MDR microorganisms in healthcare facilities is therefore a key issue.
The degree of antimicrobial resistance in these settings depends on intrinsic factors related to the particular idiosyncrasies of each centre as well as on external factors such as the influx of resistant pathogens that originate in the community. Intrinsic differences in resistance rates between hospitals can be attributed to use of individual rooms vs. two or three bedrooms or open units, staffing, antibiotic stewardship, environmental cleaning, adherence to hand hygiene precautions and infection control programs [20].
Antimicrobial consumption also shows huge interhospital differences that might be partially explained by case mix as well as by differences in the flow of patients carrying resistance bacteria that are transferred from other healthcare facilities [21,22].
In order to minimize the unwanted consequences of antimicrobial use, implementation of antimicrobial stewardship programs should be mandatory [23,24]. Ideally, antimicrobial stewardship should be designed to include the following: (a) passive educational measures (antibiotic guidelines, educational sessions), (b) active interventions (clinical rounds, prospective audits, reassessment of antibiotic perspectives), (c) restrictive measures (limiting antibiotics on the hospital formulary, reporting of susceptibility by the microbiology laboratory, antibiotic order form, preauthorization), and (d) supplemental measures (computer-assisted management programs, multidisciplinary stewardship teams, consultancy services). In addition, infection control measures should be planned according to the microorganisms of interest, case mix of patients and whether endemic or epidemic [20]. Ideally, the universal screening of all patients should be performed at admission.
Risk assessment in target pathogens needs to consider (a) the pathogen itself, as there are different dosing strategies for different species, (b) time of exposure—the duration of treatment should be kept for as short as possible, (c) drug exposure related to risk in an inverse U relationship, (d) patterns of drug exposure, (e) inoculum size, with different dosing for high-load (pneumonia) and low-load infections (surgically treated complicated skin and soft tissue infection) and (f) combination therapy to suppress resistance [25].
Diagnosis and treatment options
We use breakpoints to categorize microorganisms as susceptible (treatable with the agent in question) and resistant (not treatable with the agent) to guide therapy [26]. At the international level, breakpoints are officially determined by two main institutions: the Clinical and Laboratory Standards Institute (CLSI) in the United States, which is a not-for-profit membership organization sponsored by the industry and the medical community; and the European Committee on Antimicrobial Susceptibility Testing (EUCAST), a joint effort among ESCMID, ECDC, and the European Medicines Agency (EMA) [27]. Breakpoints are reviewed at regular intervals because changes in doses, administration modes and indications for therapy occur and new resistance mechanisms are discovered. Differences among breakpoints suggested by CLSI or EUCAST are subtle but significant for certain antimicrobial classes, and such differences reflect the use of divergent criteria as well as different conflicts of interest. In addition, there is also a plethora of likewise national organizations providing breakpoints and guidelines (i.e. BSAC, CA-SFM, CRG, DIN, NWGA, SRGA, among others) that all together contribute to a very poor standardization of procedures. An international harmonization of breakpoints is clearly needed, and in order to achieve it, either all countries must adopt the standards of one of the two main committees (CLSI or EUCAST), or a novel international breakpoint committee must be created that embraces all of the above.
The rapid detection of resistance mechanisms plays an important epidemiological role in surveillance studies to evaluate the potential dissemination of resistance genes. From the therapeutic point of view, however, this information does not exclude the presence of other mechanisms of resistance affecting the same antibacterial agent. Hence, its presence has a high predictive value for resistance but not for susceptibility. The prompt identification of the antimicrobial susceptibility of a microorganism, on the other hand, ensures the administration of the correct treatment and reduces the need for broad-spectrum drugs, limiting the emergence of antimicrobial resistance.
Molecular methods have shortened the time to detect specific resistance mechanisms and the development of next generation sequencing technologies has increased the number of sequenced bacterial genomes at an exponential rate. A better understanding of the molecular basis of antimicrobial resistance has facilitated the development of bioinformatic tools to identify antibiotic resistance genes in bacterial genomes, such as ARG-ANNOT, ResFinder, and the CARD database.
In addition, mass spectrometry techniques have proven extremely useful in the rapid identification of bacterial species and their use in the detection of resistance profiles to certain classes of antibiotics has provided excellent results [28]. Similarly, advanced applications of nanoparticles and bacterial microencapsulation to clinical are very promising and might be fully developed in the years to come [29,30].
The development of novel therapeutic strategies, however, seems to have reached a dead end. Despite the urgent need to find new antibacterial products, many pharmaceutical companies have abandoned antibiotic drug discovery programs. Several factors have contributed to this situation: (a) difficulty predicting the development of resistance adding risk to research and development (R&D) investment, (b) significant scientific bottlenecks, (c) complex and divergent regulatory requirement and (d) the challenge of the commercial model—there is high investment but low returns compared to alternative R&D investment.
Relaunching antimicrobial drug discovery and development should be a global priority, and some initiatives have been suggested to encourage investment by the pharmaceutical industry, such as extending the period of exclusivity for certain antibiotics (i.e. the GAIN (Generating Antibiotics Incentives Now) Act) and amending regulatory requirements to accelerate access to antibiotics for serious infections for which there are few, if any, alternatives.
Promoting research and development of novel antimicrobial drugs needs to address the issue of the challenging commercial model and come up with strategies to reconcile public health needs with an attractive economic model for the pharmaceutical industry [31]. Initiatives such as the 10 × ’20 Initiative, promoted by the Infectious Diseases Society of America (IDSA), that attempts to produce ten new systemic antibiotics by the year 2020 [32], or the European Innovative Medicine Initiative (http://www.imi.europa.eu), supporting collaborative research projects between the pharmaceutical industry and the academia, combine public and private funding to invigorate antimicrobial drug research. One of these research projects to tackle antimicrobial resistance is COMBACTE (Combatting Bacterial Resistance in Europe), which aims to give antibiotic drug development a much-needed boost by pioneering new ways of designing and implementing efficient clinical trials for novel antibiotics.
Fortunately, there are also a handful of small- and medium-size companies as well as multiple research groups that are investing in antimicrobial drug discovery, although such initiatives will eventually have to rely on larger corporations to proceed through the expensive phase 2/3 of clinical development.
In this regard, efforts are being made by EMA to relax the regulations for clinical trials, a measure thought to cheapen and accelerate the release of new drugs to the market.
Table 1 summarizes the different strategies that are being used to discover and develop drugs to fight bacteria.
Table 1.
Strategies for discovery and development of novel antibacterial drugs
| Strategy | Description | Reference |
|---|---|---|
| Drug derivatives | Modification of the basic structure of known antimicrobial agents or development of inhibitors of a specific mechanism of resistance (i.e. new β-lactamases or efflux pump inhibitors). | [34,35] |
| Discovery of new antimicrobial agents | Classical or whole-cell antibacterial assay to find antibiotics produced by microorganism of different sources. | [36] |
| Genomic or target-base antibiotic discovery with the use of new tools such as combinatorial chemistry and genomics. | [34] | |
| Antivirulence drugs | Antibodies or compounds blocking or inhibiting virulence factors. | [37] |
| Nanoparticles | Development of antibacterial peptides or peptidomimetics. | [30] |
| Bacteriophages or enzybiotics | Delivery of bacteriophages or phage-lytic enzymes. | [38,39] |
| Ecology/evolutionary biology approaches | Aimed at targeting the ecology and evolution of antibiotic resistance, including inhibitors of plasmid transfer of resistance, and gene-silencing antisense oligomers. | [40] |
Conclusions
The current global threat of antimicrobial resistance and the urgent need to control it and to find new antibacterial products has prompted the different stakeholders to take action in integrating research and public health, and in maintaining and promoting the national and international antimicrobial resistance research community. The Joint Programming Initiative on Antimicrobial Resistance is a collaborative research initiative supported by 18 European countries plus Canada as a response to this threat. It has defined a strategic research agenda under the assumption that only a collaborative effort will provide the necessary critical mass and scientific expertise to answer the most important and urgent research questions related to antimicrobial resistance. Other actions promoted by nonprofit entities such as the World Alliance Against Antibiotic Resistance (WAAAR, France), Antibiotic Action (UK), ReAct (Sweden) and the Antibiotic Resistance Initiative (ISGlobal, Spain), among others, are also playing an important role in this process.
To summarize, the following measures can be taken to prevent the emergence and spread of antibiotic resistance worldwide: (a) rational use of antibiotics in all settings, (b) implementation of infection control measures in healthcare settings, (c) development of strategies to mitigate the risks of environmental exposure, (d) development of rapid diagnostic tests, (e) promotion of research on antibacterial resistance prevention and surveillance, (f) promotion of research and development of novel antimicrobial strategies and antibacterial agents and (g) improved general awareness of antibiotic use and risk of increasing resistance. The flow of resistance and the main interventions that are needed to prevent the threat of antibiotic resistance at specific key points are summarized in Fig. 1.
Fig. 1.
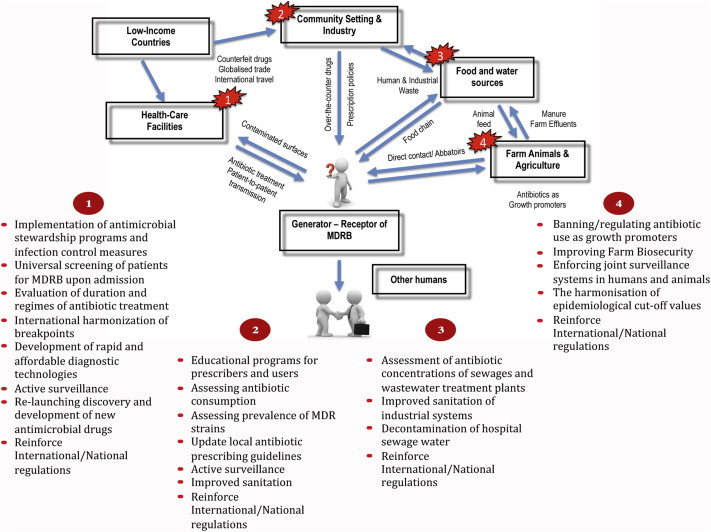
Intervention measures to tackle antibiotic resistance. Flow of antibiotic resistance genes, antimicrobial agents and pathogens ultimately generating multidrug-resistant bacteria and reaching a human host. Measures to prevent the emergence and dissemination of antibiotic resistance are shown at different key points.
The factors that have led us to the current situation and the measures proposed to circumvent it are no novelty to the scientific community, and much has been written about it. Implementing those measures will demand a concerted action of all stakeholders involved (policy makers, public health authorities, regulatory agencies, pharmaceutical companies and the scientific community at large), but above all, it will require strong regulatory modifications and a great deal of investment. This is the real bottleneck.
In the long run, the treatment of patients infected with drug-resistant pathogens is much more expensive as a result of longer hospitalization times and the use of more expensive last-resort drugs. The annual economic burden associated with the treatment of antibiotic-resistant infections has been estimated to be between $21,000 and $34,000 million in the United States alone, and around €1500 million in Europe, which includes the economic impact associated with the number of days of lost productivity, estimated to be approximately €450 million each year in Europe [33].
A global and coordinated initiative to tackle antibiotic resistance will be needed to persuade the general population and policy makers of the advantages, both medical and economic, of combating the threat of antimicrobial resistance.
Conflict of interest
None declared.
Acknowledgements
This article stems from the critical, ongoing efforts of experts who attended the B-Debate meeting, “The Global Threat of Antimicrobial Resistance: Science for Intervention,” 10–13 November 2013, Barcelona, Spain, cosponsored by B-Debate (and initiative of Biocat and “la Caixa” Foundation), ISGlobal, ESCMID and REIPI. The research leading to these results has received funding from the IMI (Innovation Medicine Initiatives) Project Combatting Bacterial Resistance in Europe—COMBACTE under grant 115523. The presentation slides used during the B-Debate meeting are available online (http://www.bdebate.org/en/moreinfo/3065).
References
- 1.Marshall B.M., Levy S.B. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebana E., Carattoli A., Coque T.M., Hasman H., Magiorakos A.P., Mevius D. Public health risks of enterobacterial isolates producing extended-spectrum beta-lactamases or AmpC beta-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis. 2013;56:1030–1037. doi: 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 3.European Centre for Disease Prevention and Control (ECDC), European Food Safety Authority (EFSA), European Medicines Agency (EMEA) 2009. Joint opinion on antimicrobial resistance (AMR) focused on zoonotic infections.http://www.efsa.europa.eu/en/scdocs/doc/1372.pdf Available at: [accessed 8.5.15] [Google Scholar]
- 4.World Health Organization . 2011. WHO list of critically important antimicrobials (CIA), 3rd revision.http://www.who.int/foodsafety/publications/antimicrobials-third/en/ Available at: [accessed 8.5.15] [Google Scholar]
- 5.Colpan A., Johnston B., Porter S., Clabots C., Anway R., Thao L. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis. 2013;57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruyndonckx R., Hens N., Aerts M., Goossens H., Molenberghs G., Coenen S. Measuring trends of outpatient antibiotic use in Europe: jointly modelling longitudinal data in defined daily doses and packages. J Antimicrob Chemother. 2014;69:1981–1986. doi: 10.1093/jac/dku063. [DOI] [PubMed] [Google Scholar]
- 7.European Centre for Disease Prevention and Control (ECDC), European Antimicrobial Surveillance Network (EARS-NET) 2013. Technical report.http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2012.pdf Antimicrobial resistance surveillance in Europe, 2012. Available at: [accessed 8.5.15] [Google Scholar]
- 8.Voulgari E., Poulou A., Koumaki V., Tsakris A. Carbapenemase-producing Enterobacteriaceae: now that the storm is finally here, how will timely detection help us fight back? Future Microbiol. 2013;8:27–39. doi: 10.2217/fmb.12.130. [DOI] [PubMed] [Google Scholar]
- 9.Coenen S., Gielen B., Blommaert A., Beutels P., Hens N., Goossens H. Appropriate international measures for outpatient antibiotic prescribing and consumption: recommendations from a national data comparison of different measures. J Antimicrob Chemother. 2014;69:529–534. doi: 10.1093/jac/dkt385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laxminarayan R., Klugman K.P. Communicating trends in resistance using a drug resistance index. BMJ Open. 2011;1:e000135. doi: 10.1136/bmjopen-2011-000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spellberg B., Guidos R., Gilbert D., Bradley J., Boucher H.W., Scheld W.M. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 12.Coenen S., Francis N., Kelly M., Hood K., Nuttall J., Little P. Are patient views about antibiotics related to clinician perceptions, management and outcome? A multi-country study in outpatients with acute cough. PLoS One. 2013;8:e76691. doi: 10.1371/journal.pone.0076691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spellberg B., Blaser M., Guidos R.J., Boucher H.W., Bradley J.S., Eisenstein B.I. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis. 2011;52(Suppl. 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laxminarayan R., Heymann D.L. Challenges of drug resistance in the developing world. BMJ. 2012;344:e1567. doi: 10.1136/bmj.e1567. [DOI] [PubMed] [Google Scholar]
- 15.Morgan D.J., Okeke I.N., Laxminarayan R., Perencevich E.N., Weisenberg S. Non-prescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis. 2011;11:692–701. doi: 10.1016/S1473-3099(11)70054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wellington E.M., Boxall A.B., Cross P., Feil E.J., Gaze W.H., Hawkey P.M. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 17.Finley R.L., Collignon P., Larsson D.G., McEwen S.A., Li X.Z., Gaze W.H. The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis. 2013;57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 18.Heuer O.E., Kruse H., Grave K., Collignon P., Karunasagar I., Angulo F.J. Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis. 2009;49:1248–1253. doi: 10.1086/605667. [DOI] [PubMed] [Google Scholar]
- 19.Pruden A., Larsson D.G., Amezquita A., Collignon P., Brandt K.K., Graham D.W. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ Health Perspect. 2013;121:878–885. doi: 10.1289/ehp.1206446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacconelli E., Cataldo M.A., Dancer S.J., De Angelis G., Falcone M., Frank U. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect. 2014;20(Suppl. 1):1–55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 21.Berger A., Edelsberg J., Oster G., Huang X., Weber D.J. Patterns of initial antibiotic therapy for community-acquired pneumonia in US hospitals, 2000 to 2009. Am J Med Sci. 2014;347:347–356. doi: 10.1097/MAJ.0b013e318294833f. [DOI] [PubMed] [Google Scholar]
- 22.Blommaert A., Marais C., Hens N., Coenen S., Muller A., Goossens H. Determinants of between-country differences in ambulatory antibiotic use and antibiotic resistance in Europe: a longitudinal observational study. J Antimicrob Chemother. 2014;69:535–547. doi: 10.1093/jac/dkt377. [DOI] [PubMed] [Google Scholar]
- 23.Cisneros J.M., Neth O., Gil-Navarro M.V., Lepe J.A., Jimenez-Parrilla F., Cordero E. Global impact of an educational antimicrobial stewardship programme on prescribing practice in a tertiary hospital centre. Clin Microbiol Infect. 2014;20:82–88. doi: 10.1111/1469-0691.12191. [DOI] [PubMed] [Google Scholar]
- 24.Gyssens I.C., Kern W.V., Livermore D.M. The role of antibiotic stewardship in limiting antibacterial resistance among hematology patients. Haematologica. 2013;98:1821–1825. doi: 10.3324/haematol.2013.091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischbach M.A. Combination therapies for combating antimicrobial resistance. Curr Opin Microbiol. 2011;14:519–523. doi: 10.1016/j.mib.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leclercq R., Canton R., Brown D.F., Giske C.G., Heisig P., MacGowan A.P. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013;19:141–160. doi: 10.1111/j.1469-0691.2011.03703.x. [DOI] [PubMed] [Google Scholar]
- 27.Kahlmeter G., Brown D.F., Goldstein F.W., MacGowan A.P., Mouton J.W., Osterlund A. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J Antimicrob Chemother. 2003;52:145–148. doi: 10.1093/jac/dkg312. [DOI] [PubMed] [Google Scholar]
- 28.Croxatto A., Prod'hom G., Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- 29.Frima H.J., Gabellieri C., Nilsson M.I. Drug delivery research in the European Union's Seventh Framework Programme for Research. J Control Release. 2012;161:409–415. doi: 10.1016/j.jconrel.2012.01.044. [DOI] [PubMed] [Google Scholar]
- 30.Pelgrift R.Y., Friedman A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliv Rev. 2013;65:1803–1815. doi: 10.1016/j.addr.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 31.So A.D., Gupta N., Brahmachari S.K., Chopra I., Munos B., Nathan C. Towards new business models for R&D for novel antibiotics. Drug Resist Updat. 2011;14:88–94. doi: 10.1016/j.drup.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Boucher H.W., Talbot G.H., Benjamin D.K., Jr, Bradley J., Guidos R.J., Jones R.N. 10 × '20 Progress—development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis. 2013;56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.European Centre for Disease Prevention and Control (ECDC), European Medicines Agency (EMEA) 2009. Technical report.http://www.ema.europa.eu/docs/en_GB/document_library/Report/2009/11/WC500008770.pdf The bacterial challenge: time to react. Available at: [accessed 8.5.15] [Google Scholar]
- 34.Walsh C.T., Wencewicz T.A. Prospects for new antibiotics: a molecule-centered perspective. J Antibiot (Tokyo) 2014;67:7–22. doi: 10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 35.Goemaere E., Melet A., Larue V., Lieutaud A., Alves de Sousa R., Chevalier J. New peptide deformylase inhibitors and cooperative interaction: a combination to improve antibacterial activity. J Antimicrob Chemother. 2012;67:1392–1400. doi: 10.1093/jac/dks058. [DOI] [PubMed] [Google Scholar]
- 36.Singh S.B., Young K., Miesel L. Screening strategies for discovery of antibacterial natural products. Expert Rev Anti Infect Ther. 2011;9:589–613. doi: 10.1586/eri.11.81. [DOI] [PubMed] [Google Scholar]
- 37.Waldor M.K. Disarming pathogens—a new approach for antibiotic development. N Engl J Med. 2006;354:296–297. doi: 10.1056/NEJMcibr054591. [DOI] [PubMed] [Google Scholar]
- 38.Pastagia M., Schuch R., Fischetti V.A., Huang D.B. Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol. 2013;62:1506–1516. doi: 10.1099/jmm.0.061028-0. [DOI] [PubMed] [Google Scholar]
- 39.Bragg R., van der Westhuizen W., Lee J.Y., Coetsee E., Boucher C. Bacteriophages as potential treatment option for antibiotic resistant bacteria. Adv Exp Med Biol. 2014;807:97–110. doi: 10.1007/978-81-322-1777-0_7. [DOI] [PubMed] [Google Scholar]
- 40.Mosqueda N., Gato E., Roca I., López M., de Alegría C.R., Fernández-Cuenca F. Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J Antimicrob Chemother. 2014;69:2629–2633. doi: 10.1093/jac/dku179. [DOI] [PubMed] [Google Scholar]


