Abstract
The conventional hemagglutinin (HA)- and neuraminidase (NA)-based influenza vaccines need to be updated most years and are ineffective if the glycoprotein HA of the vaccine strains is a mismatch with that of the epidemic strain. Universal vaccines targeting conserved viral components might provide cross-protection and thus complement and improve conventional vaccines. In this study, we generated DNA plasmids and recombinant vaccinia viruses expressing the conserved proteins nucleoprotein (NP), polymerase basic 1 (PB1), and matrix 1 (M1) from influenza virus strain A/Beijing/30/95 (H3N2). BALB/c mice were immunized intramuscularly with a single vaccine based on NP, PB1, or M1 alone or a combination vaccine based on all three antigens and were then challenged with lethal doses of the heterologous influenza virus strain A/PR/8/34 (H1N1). Vaccines based on NP, PB1, and M1 provided complete or partial protection against challenge with 1.7 50% lethal dose (LD50) of PR8 in mice. Of the three antigens, NP-based vaccines induced protection against 5 LD50 and 10 LD50 and thus exhibited the greatest protective effect. Universal influenza vaccines based on the combination of NP, PB1, and M1 induced a strong immune response and thus might be an alternative approach to addressing future influenza virus pandemics.
INTRODUCTION
The conventional influenza vaccines that are available currently to prevent seasonal flu outbreaks depend mainly on the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) (1, 2). However, HA- and NA-based conventional influenza vaccines sometimes fail to prevent flu epidemics because the HA and/or NA in the vaccine strains is a mismatch with that in circulating virus strains (3–7). Universal influenza vaccines (UIVs) that induce effective and long-term cross-protection and address the risk of mismatch may overcome the shortcomings of conventional influenza vaccines. Therefore, the development of a UIV capable of inducing long-term immunity and cross-protection remains a priority in influenza vaccine research (8).
Influenza viruses are classified as type A, B, or C based on their nucleoprotein (NP) and matrix protein (M). Among the three subtypes, influenza A virus has been the target of UIVs, because the diverse influenza A strains frequently trigger influenza epidemics and pandemics. A previous study indicated that humans mount a good response to the highly conserved internal proteins NP, M1, and polymerase basic 1 (PB1) of influenza A virus (9); therefore, these highly conserved influenza A virus antigens are the basis of UIVs. Multiple studies have investigated the potential of NP (10–13), matrix protein 1 (M1) (14–17), and ion channel (M2, mainly M2e) (18–27) as alternative vaccine antigens for the prevention of seasonal and pandemic flu outbreaks. PB1 has also shown protective potential but requires further investigation for inclusion in UIVs. Košík et al. (28) constructed a DNA vaccine based on PB1, which provided some protective immunity in a mouse model. We previously constructed DNA vaccines based on PB1 and PB2 from influenza virus strains A/PR/8/34 (H1N1) (PR8) and A/Beijing/30/95 (H3N2) (BJ95). Mice immunized with DNA vaccines based on PB1 from PR8 or BJ95 were protected against sublethal PR8 challenge, whereas mice immunized with PB2-based DNA vaccines were not. These data suggest that the influenza viral structural protein PB1 shows promise for inclusion in a DNA vaccine against the influenza A virus (29). Recent studies suggested that the injection of vaccines based on NP, M1, M2, or PB1 conferred protection in mice challenged with a lethal virus dose. In many cases, UIVs were developed by combining several antigens or epitopes to induce a comprehensive immune response. For example, NP was often used in combination with M1 or M2e to provide protection superior to that conferred by either antigen alone (30–43). In addition, UIVs containing influenza virus HA, M1, and/or NP provided effective cross-protection against a lethal challenge of influenza virus (38, 44–47). Jeon, Ben-Yedidia, and Arnon (48) fused the oligonucleotides coding for three epitopes, HA91–108 (B-cell epitope), NP55–69 (Th-cell epitope), and NP147–158 (CD8+ T-cell epitope), of influenza virus in tandem with the flagellin protein of Salmonella. Mucosal immunization with the fusion vaccine protected BALB/c mice against influenza virus of a different subtype. Therefore, UIVs based on multiple components can induce a strong immune response and might represent an alternative approach to preventing future influenza virus pandemics.
UIVs based on multiple proteins or epitopes were designed using various types of vaccines, including DNA, recombinant subunit, and virus-vectored vaccines, to elicit comprehensive cross-protection immune responses against influenza viruses. In some cases, alternate prime-boost strategies were used to develop combination vaccines for stable and long-term protection. For example, Lo et al. (30) constructed NP-based DNA and adenovirus vaccines and showed that a DNA prime-adenovirus boost strategy greatly improved the cross-protection induced by NP-based vaccines. Luo et al. (49) found that the immune response induced in mice by the NP DNA vaccine was enhanced by an intranasal boost with recombinant NP (rNP) protein. The prime-boost strategy provided protection not only against the homologous virus but also cross-protection against a heterosubtypic H9N2 strain. A UIV based on a fusion of NP and M1 was constructed in recombinant modified vaccinia virus Ankara (MVA) and adenovirus (Ad), and various immunization regimens were assessed. The alternate Ad prime-MVA boost vaccination regimen induced the strongest immune responses in BALB/c mice and chickens and therefore was the most immunogenic regime (38, 41).
In the present study, we generated UIVs based on NP, M1, and PB1 in DNA plasmids and recombinant vaccinia virus (rVV). We then investigated the ability of these vaccines to induce immunity and provide cross-protection against PR8 challenge in mice immunized with DNA vaccines, rVV vaccines, or DNA prime-rVV boost regimens based on a sole antigen or a combination of the three antigens.
MATERIALS AND METHODS
Influenza A viruses.
The influenza A viruses A/PR/8/34 (H1N1) (PR8) and A/Beijing/30/95 (H3N2) (BJ95) were used in this study. PR8 is a mouse-adapted influenza strain that was propagated on 9-day-old chick embryos at 34°C for 48 h; the allantoic fluid was then collected and stored at −70°C until use. The 50% lethal dose (LD50) titer of PR8 was assessed in BALB/c mice before the challenge experiments. One LD50 is equal to 103.6 50% tissue culture infective doses (TCID50). Influenza virus BJ95 was used as the template to obtain the target genes in this study. Moreover, allantoic fluid containing BJ95 was concentrated by centrifugation, diluted with diethyl ether, and then used to coat enzyme-linked immunosorbent assay (ELISA) plates to detect immunization-induced specific IgGs in mice.
Vaccinia virus Tiantan strain and relevant vector.
Vaccinia virus Tiantan strain is replication competent. It was developed by Chinese scientists as a vaccination agent against smallpox in China and was documented by the WHO to successfully prevent smallpox (50). It played a decisive role in the eradication of smallpox in China and was used to inoculate hundreds of millions of individuals for ∼50 years until the end of the smallpox vaccination program in 1980. Vaccinia virus Tiantan is a characteristic strain with a wide host range, powerful multiplication capacity, abundant regions containing unnecessary genes, a high capacity for foreign gene insertion, noncarcinogenicity, lasting immunity, and cytoplasmic replication, and a long history of applications in humans. It has been used widely as a recombinant vaccine vector for many years, and multiple recombinant vaccinia viruses based on the Tiantan strain have been reported (51–55). Previous studies suggested that the vaccinia virus Tiantan strain-based vector system is suitable for expressing foreign genes and constructing recombinant vaccines, and the resulting recombinant vaccines are safe and effective in animals.
Construction of DNA plasmids.
Viral RNA from influenza virus BJ95 was isolated using the TRIzol reagent. The universal forward primer for influenza A virus, 5′-AGCAAAAGCAGG-3′, and total RNA were used to generate full-length cDNA transcripts for influenza A virus BJ95 using a ThermoScript reverse transcriptase PCR (RT-PCR) system (Gibco, Grand Island, NY, USA), according to the manufacturer's instructions. PCR amplification was then performed to acquire the target genes using full-length cDNA transcripts as the template. The cDNA corresponding to the NP gene was generated using the forward primer 5′-ACGGATCCATCATGGCGTCCCAAGGCAC-3′, which contains a BamHI restriction enzyme site (underlined), and the reverse primer 5′-TTGGATCCTTAATTGTCGTACTC-3′, which contains a stop codon and a BamHI restriction enzyme site (underlined). The cDNA corresponding to the PB1 gene was generated using the forward primer 5′-AAGGATCCCGAATGGATGTCAATCC-3′, which contains a BamHI restriction site (underlined), and the reverse primer 5′-AGGGATCCTCATTATTTTTGCCGTCT-3′, which contains a stop codon and a BamHI restriction site (underlined). The cDNA corresponding to the M1 gene was generated using the forward primer 5′-GATCCCGGGACCATGAGCCTTCTAACCGA-3′, which contains an SmaI restriction site (underlined), and the reverse primer 5′-ACACCCGGGTCACTTGAATCGTTGC-3′, which contains a stop codon and an SmaI restriction site (underlined). The entire open reading frames of the NP, PB1, and M1 genes were then digested using BamHI, BamHI, and SmaI, respectively, and inserted into the shuttle vector pMD18T. Subsequently, the cDNAs of the NP, PB1, and M1 genes in pMD18T were inserted into the plasmid expression vector pSCA. The plasmids were propagated in Escherichia coli strain DH5α cells, purified using Qiagen-tip 500 kits (Qiagen, Düsseldorf, Germany), and stored at −20°C.
The expression of NP, PB1, and M1 proteins by pSCA-NP, pSCA-PB1, and pSCA-M1, respectively, was confirmed using indirect immunofluorescence. First, MDCK cells were transiently transfected with the plasmids, and the expression of NP, PB1, and M1 was confirmed 18 to 24 h later using mouse monoclonal influenza A virus NP-specific (ViroStat, Portland, ME, USA), goat polyclonal influenza A virus PB1-specific (Santa Cruz, Dallas, TX, USA), and mouse polyclonal influenza A virus M1-specific (Santa Cruz) antibodies. The signals were then visualized using fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse, rabbit anti-goat, and goat anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA) secondary antibodies.
Construction of recombinant vaccinia virus.
The pJSA1175 vector, the homologous recombinant plasmid of the vaccinia virus Tiantan strain, contains two back-to-back promoters (p11 and p7.5). The plasmid contains the lacZ gene downstream of the late p11 promoter, whereas the early and late promoter p7.5 was empty loaded for the insertion of foreign genes. Because there is a vaccinia virus early transcription termination signal TTTTTNT sequence (56, 57), the 1493-TTTTTTT-1499 sequence in the PB1 gene from pMD18-PB1 was PCR mutated to 1493-TTCTTCT-1499 to avoid early transcription termination of the PB1 gene in recombinant vaccinia virus (rVV). The mutated PB1 gene was named PB1(m) and inserted into the shuttle vector pMD18-T, similar to PB1 in pMD18-PB1. Next, the entire open reading frames of the NP gene in pMD18-NP, the PB1 gene in pMD18-PB1(m), and the M1 gene in pMD18-M1 were digested with BamHI, BamHI, and SmaI, respectively, as appropriate, smoothed, and then inserted into pJSA1175 vector.
To generate recombinant vaccinia virus via homologous recombination, primary chicken embryo fibroblast (CEF) cells were infected with a multiplicity of infection (MOI) of 0.01 to 0.1 of vaccinia virus Tiantan strain and then transfected with pJSA1175-NP, pJSA1175-PB1, and pJSA1175-M1 plasmids, as described previously (58). Subsequently, pure recombinant vaccinia viruses containing the NP, PB1, and M1 genes were generated using plaque purification in CEFs. Mock recombinant vaccinia virus (rVV-c) was generated using the same method with empty pJSA1175 vector. The recombinant vaccinia viruses were amplified in CEF cells.
The expression of NP, PB1, and M1 proteins by rVV-NP, rVV-PB1, and rVV-M1, respectively, was confirmed using indirect immunofluorescence. First, HeLa cells were infected with rVV at an MOI of 0.001 to 0.002, and the expression of NP, PB1, and M1 was confirmed 24 to 36 h later using the method described above for DNA plasmids. Meanwhile, the expression of the target proteins was confirmed using immunoblotting. CEF cells were infected with rVV at an MOI of 0.1 to 0.2, and the expression of NP, PB1, and M1 was confirmed 24 h to 48 h later using rabbit polyclonal NP-specific antiserum (obtained by immunizing rabbits with purified NP1–167 aa [aa, amino acids] in our laboratory), goat polyclonal PB1-specific antibodies (Santa Cruz), and goat polyclonal M1-specific antibodies (Santa Cruz), respectively. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Sigma-Aldrich), rabbit anti-goat IgG (Sigma-Aldrich), and rabbit anti-goat IgG (Sigma-Aldrich) secondary antibodies, respectively, were then used. Cellular proteins were extracted from the infected CEF cells by resuspending cell pellets with buffer containing 250 mM Tris-HCl, 5% β-mercaptoethanol, 2% SDS, 0.0025% bromophenol blue, and 10% glycerol, followed by clearance by centrifugation at 12,000 rpm for 10 min.
Vaccination and challenge.
Specific-pathogen-free (SPF) female BALB/c mice (5 to 6 weeks old) were purchased from the Institute of Experimental Animals, Chinese Academy of Medical Sciences (Beijing, China). All mice were bred under specific-pathogen-free conditions at the Institute for Occupational Health and Poison Control (IOHPC), Chinese Center for Disease Control and Prevention (China CDC). The mouse study was conducted in strict accordance with the recommendations in the guide for the care and use of laboratory animals of the China CDC. Groups of mice were intramuscularly (i.m.) inoculated with vaccines in the bilateral gastrocnemius without the use of any reagent or equipment (Table 1), as described below.
TABLE 1.
Mouse groups according to immunogen
| Vector | Group | Antigena | Immunogen (dose)b at wk: |
|||
|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | |||
| DNA | 1 | None | pSCA-c (10) | pSCA-c (10) | pSCA-c (10) | None |
| 2 | NP | pSCA-NP (10) | pSCA-NP (10) | pSCA-NP (10) | None | |
| 3 | PB1 | pSCA-PB1 (10) | pSCA-PB1 (10) | pSCA-PB1 (10) | None | |
| 4 | M1 | pSCA-M1 (10) | pSCA-M1 (10) | pSCA-M1 (10) | None | |
| 5 | NP + PB1 + M1 | pSCA-NP/PB1/M1 (10:10:10) | pSCA-NP/PB1/M1 (10:10:10) | pSCA-NP/PB1/M1 (10:10:10) | None | |
| rVV | 6 | None | None | rVV-c (107) | None | rVV-c (107) |
| 7 | NP | None | rVV-NP (107) | None | rVV-NP (107) | |
| 8 | PB1 | None | rVV-PB1 (107) | None | rVV-PB1 (107) | |
| 9 | M1 | None | rVV-M1 (107) | None | rVV-M1 (107) | |
| 10 | NP + PB1 + M1 | None | rVV-NP/PB1/M1 (107:107:107) | None | rVV-NP/PB1/M1 (107:107:107) | |
| DNA + rVV | 11 | None | pSCA-c (10) | pSCA-c (10) | pSCA-c (10) | rVV-c (107) |
| 12 | NP | pSCA-NP (10) | pSCA-NP (10) | pSCA-NP (10) | rVV-NP (107) | |
| 13 | PB1 | pSCA-PB1 (10) | pSCA-PB1 (10) | pSCA-PB1 (10) | rVV-PB1 (107) | |
| 14 | M1 | pSCA-M1 (10) | pSCA-M1 (10) | pSCA-M1 (10) | rVV-M1 (107) | |
| 15 | NP + PB1 + M1 | pSCA-NP/PB1/M1 (10:10:10) | pSCA-NP/PB1/M1 (10:10:10) | pSCA-NP/PB1/M1 (10:10:10) | rVV-NP/PB1/M1 (107:107:107) | |
None, without exogenous antigen (vector control, such as pSCA-c, rVV-c, or pSCA-c + rVV-c).
Dosage values reflect either 10 μg or 107 PFU. None, mice were immunized without immunogen at the indicated time.
The first group was immunized with DNA alone (DNA). Three doses of 10 μg of DNA plasmid in a 100-μl volume of phosphate-buffered saline (PBS) were given on weeks 0, 2, and 4. The combined immunization group was injected with 10 μg of pSCA-NP, 10 μg of pSCA-PB1, and 10 μg of pSCA-M1 mixed in a total volume of 100 μl of PBS. Mice immunized with the empty pSCA vector were used as the control group (pSCA-c or DNA-c).
The second group was immunized with rVV alone (rVV). Two doses of 107 PFU of rVV in a 100-μl volume of PBS were given on weeks 2 and 6. The combined immunization group was immunized with 107 PFU of rVV-NP, 107 PFU of rVV-PB1, and 107 PFU of rVV-M1 in a total volume of 100 μl of PBS. Mice immunized with rVV-c were used as the control group (rVV-c).
The third group was immunized with a DNA prime-rVV boost (DNA + rVV). Mice were primed with three doses of 10 μg of DNA plasmids at weeks 0, 2, and 4 and then boosted with one dose of 107 PFU of rVV at week 6. The combined immunization group received three doses of prime immunization with 10 μg of pSCA-NP, 10 μg of pSCA-PB1, and 10 μg of pSCA-M1 in a volume of 100 μl of PBS, followed by boost immunization with 107 PFU of rVV-NP, 107 PFU of rVV-PB1, and 107 PFU of rVV-M1 in a volume of 100 μl of PBS. Mice immunized with DNA-c (pSCA-c) and rVV-c were used as the control group (DNA-c + rVV-c).
Blood samples were collected from all mice in each group on week 8, the sera were separated by centrifugation, and the serum IgG titers were determined. In addition, three mice from each group were sacrificed, and the spleens were removed aseptically and ground through a 200-mesh sieve. Spleen mononuclear cells (SMNCs) were obtained after depletion of erythrocytes from spleen cells using ammonium-chloride-potassium (ACK) lysis buffer (0.15 mol/liter NH4Cl, 0.01 mol/liter KHCO3, and 0.1 mol/liter Na2-EDTA·2H2O [pH 7.2 to 7.4]). In addition, mice were anesthetized using sodium pentobarbital (10 mg/ml) at a dose of 60 mg/kg of body weight and then challenged with 50 μl of 1.7, 5, or 10 LD50 of PR8 by intranasal administration. Body weight and mortality were monitored daily for 2 to 3 weeks after challenge. Mice that lost 30% of their initial weight were euthanized and recorded as having died (18, 59, 60).
ELISA.
The concentration of IgG against BJ95 was measured using ELISA. First, the wells of ELISA plates were coated with 200 ng/well of lysed concentrated influenza A virus BJ95. Next, serum from each mouse in the immunization group was diluted serially and added to the plate. Next, a 1,000-fold dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG was added. Finally, bound antibodies were detected using 3,3′,5,5′-tetramethylbenzidine (TMB) substrate. The antibody levels in serum were expressed as endpoint titers and had an optical density at 450 nm (OD450) that was >2.1-fold higher than the mean OD450 of serum samples from the unvaccinated mice.
ELISPOT assays.
Spleen cells were isolated from mice at week 8 and processed for gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assays, as described previously (61). SMNC suspensions were treated with 4 μg/ml NP147–155 (TYQRTRALV), PB1317–325 (MFLAMITYI), or M195–106 (KAVKLYRKLKRE) for 30 h at 37°C. The choice of peptides used was based on previous reports and our previous experiments (data not shown). Spots were counted using an ELISPOT image analyzer (Bioreader 4000 Pro-X; Bio-Sys GmbH, Karben, Germany). The number of peptide-reactive cells was presented as the number of spot-forming cells (SFC) per 106 SMNCs and was calculated by subtracting the number of spots in the unstimulated control wells from that in specific peptide-containing wells. The results for each group are presented as means ± standard deviations (SD).
Statistical analysis.
The SPSS (version 17.0) and Prism (version 5.0a) software packages were used for all statistical analyses. Antibody titers were converted to log10 values before analysis. Differences in antibody titers and ELISPOT results among groups were analyzed using one-way analysis of variance (ANOVA). Weight change curves were analyzed using paired t tests. Survival curves were analyzed using log rank (Mantel-Cox) tests. Differences were considered to be significant at a P value of <0.05.
RESULTS
Generation and analysis of recombinant DNA plasmids.
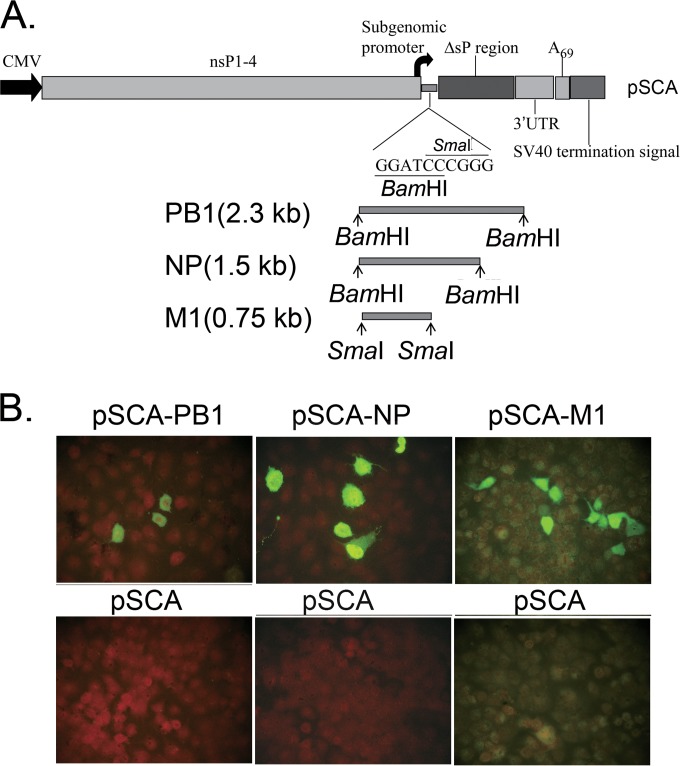
To assess whether recombinant DNA plasmids expressing influenza A virus NP, PB1, and M1 from H3N2 BJ95 virus could protect against influenza A virus PR8 challenge in mice, individual expression plasmids were constructed based on the pSCA vector. The plasmids pSCA-NP, pSCA-PB1, and pSCA-M1 were generated using the NP, PB1, and M1 cDNAs from pMD18T-NP, pMD18T-PB1, and pMD18T-M1, respectively (Fig. 1A). The plasmids were verified using sequencing and restriction enzyme digestion (data not shown). The expression of NP, PB1, and M1 in MDCK cells transiently transfected with pSCA-NP, pSCA-PB1, and pSCA-M1, respectively, was confirmed using indirect immunofluorescence assays (Fig. 1B). BJ95 NP, PB1, and M1 proteins were detected in the corresponding plasmid-transfected cells but not in mock-transfected cells.
FIG 1.
Genetic organization of DNA vaccines based on PB1, NP, and M1 in pSCA vector and the resulting protein expression in MDCK cells transfected with recombinant plasmids. (A) Schematic diagram of the genetic organization of the pSCA DNA vector (top). The bottom shows the cDNA fragments of influenza viruses BJ95 PB1, NP, and M1 flanked by BamHI, BamHI, and SmaI restriction sites, respectively. CMV, cytomegalovirus. (B) Indirect immunofluorescence showing the expression of influenza PB1, NP, and M1 in MDCK cells transfected with pSCA-PB1 (left), pSCA-NP (middle), and pSCA-M1 (right) recombinant plasmids (top) stained with the polyclonal or monoclonal antibodies (Abs) indicated in Materials and Methods. The bottom images show the results of MDCK cells mock transfected with pSCA plasmid and detected using the same Abs as in the top images.
Generation and analysis of recombinant vaccinia virus.
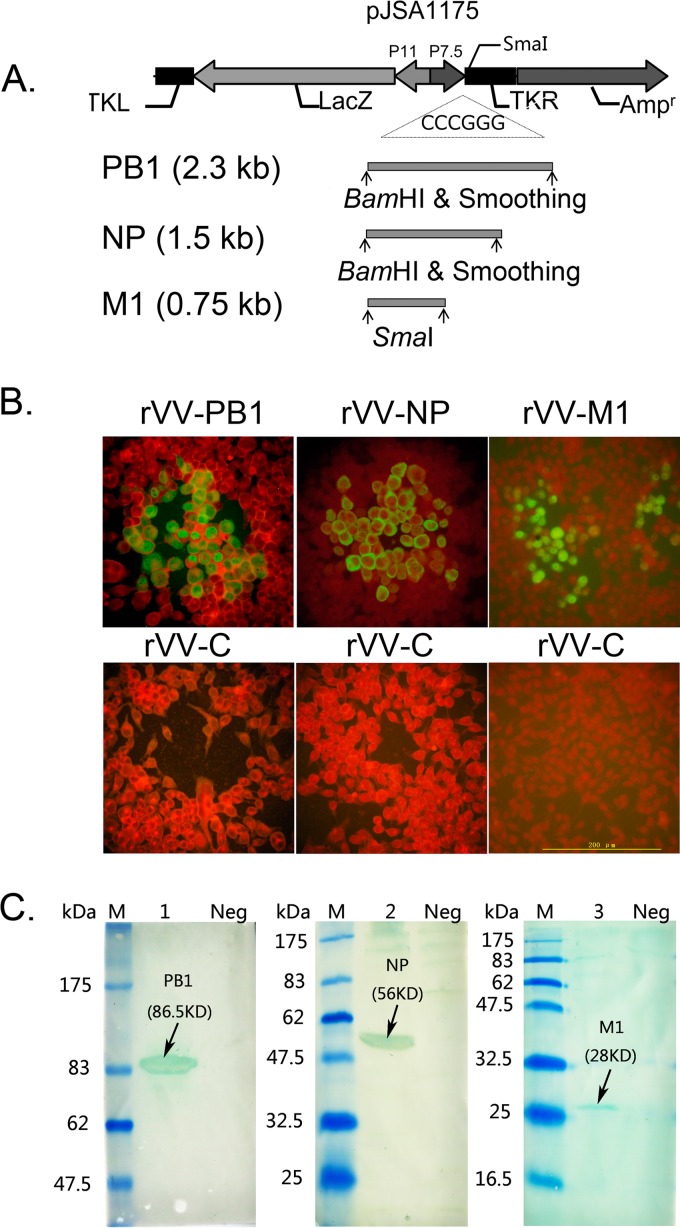
To assess whether recombinant vaccinia virus expressing influenza A virus NP, PB1, and M1 from H3N2 BJ95 virus could protect against influenza A virus PR8 challenge in mice, the H3N2 BJ95 viral NP, PB1, and M1 genes were inserted into the pJSA1175 shuttle vector (Fig. 2A). The recombinant vaccinia viruses were then generated by homologous recombination with vaccinia virus Tiantan strain with the corresponding plasmids in CEFs. The plasmids containing influenza A virus NP, PB1, and M1 were verified by sequencing and restriction enzyme digestion (data not shown). Infectious rVV-NP, rVV-PB1, and rVV-M1 were plaque purified, and the insertion of the target genes in the recombinant vaccinia viruses was confirmed by PCR sequencing (data not shown). One plaque-purified clone matching the exact cDNA of the target gene sequence was used in all subsequent experiments. The expression of NP, PB1, and M1 in rVV-NP-, rVV-PB1-, and rVV-M1-infected cells, respectively, was confirmed by indirect immunofluorescence (Fig. 2B) and immunoblotting (Fig. 2C). BJ95 NP, PB1, and M1 proteins were detected in recombinant vaccinia viruses-infected cells but not in mock-infected cells (Fig. 2B). Immunoblotting revealed the presence of proteins with molecular masses of 56, 86.5, and 28 kDa that were recognized by rabbit polyclonal NP-specific antiserum, goat polyclonal PB1-specific antibodies, and goat polyclonal M1-specific antibodies, respectively, which corresponded to influenza virus BJ95 NP, PB1, and M1, respectively (Fig. 2C).
FIG 2.
Genetic organization of PB1-, NP-, and M1-expressing pJSA1175 recombinant plasmids containing homologous sequences from vaccinia virus and the resulting protein expression in HeLa cells and CEFs infected with recombinant vaccinia virus. (A) Schematic diagram of the genetic organization of the pJSA1175 DNA vector (top). The bottom shows the cDNA fragments of influenza virus BJ95 (H3N2) PB1, NP, and M1 after restriction digestion with BamHI or SmaI, as appropriate, and smoothing, respectively. (B) Indirect immunofluorescence showing the expression of influenza PB1, NP, and M1 in plaques formed in rVV-PB1 (left)-, rVV-NP (middle)-, and rVV-M1 (right)-infected HeLa cells (upper) and stained with the polyclonal or monoclonal Abs indicated in Materials and Methods. The bottom images show the results of HeLa cells mock infected with rVV and analyzed using the same Abs as in the top images. (C) Western blots showing the expression of influenza PB1, NP, and M1 from rVV-PB1 (lane 1)-, rVV-NP (lane 2)-, and rVV-M1 (lane 3)-infected CEF cell lysates stained with the polyclonal or monoclonal antiserum indicated in Materials and Methods. M, molecular weight marker; Neg, cell lysates from CEFs mock infected with rVV-c.
Humoral and cellular immune responses were induced by immunization with DNA, rVV, or DNA prime-rVV boost.
To investigate whether the NP-, PB1-, or M1-based vaccine generated influenza A virus-specific antibodies and cellular immune responses in vivo, mice were vaccinated intramuscularly with the DNA plasmids, rVV, or DNA prime-rVV boosts; empty vector pSCA-c, mock rVV-c, and pSCA-c + rVV-c served as the respective negative controls (Table 1).
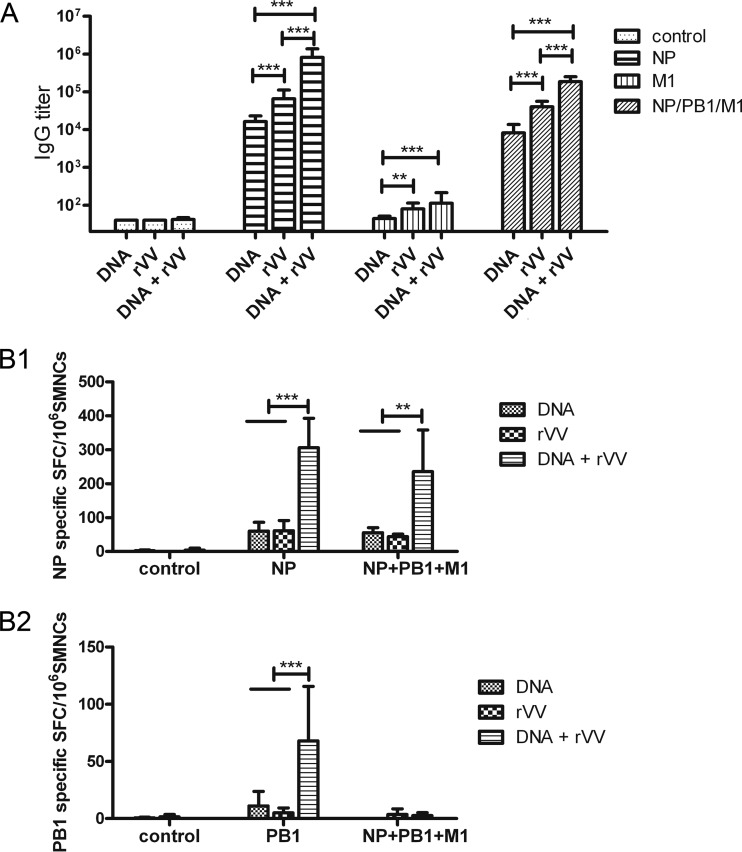
Blood samples were collected on week 8, and the sera were prepared. Concentrated BJ95 influenza A virus was lysed using diethyl ether and used to coat 96-well plates, and the serially diluted serum samples were added to the plates. The results (Fig. 3A) showed that pSCA-NP and rVV-NP vaccination induced robust serum IgG titers compared with those of the control groups (P < 0.001). In addition, rVV-NP vaccination induced higher ELISA IgG titers than did pSCA-NP (P < 0.001). Importantly, pSCA-NP prime-rVV-NP boost immunization induced a higher IgG titer than that of the two above-mentioned groups (P < 0.001). However, no obvious IgG response was induced by the PB1-based vaccine, regardless of whether it was applied alone or in the prime-boost immunization strategy (data not shown). In addition, rVV-M1 immunization induced a marked ELISA IgG response compared with that of the control group (P < 0.01), whereas pSCA-M1 immunization did not. Meanwhile, pSCA-M1 prime-rVV-M1 boost immunization induced a slightly, but not significantly, higher ELISA IgG titer than that of rVV-M1 alone (P > 0.05; Fig. 3A). When NP-, PB1-, and M1-based vaccines were applied in a combination immunization, DNA and rVV vaccines induced obvious humoral immune responses, and the combined rVV vaccines induced a significantly higher IgG titer than did the combined DNA vaccines. Furthermore, DNA prime-rVV boost immunization induced the highest IgG titer among the three groups.
FIG 3.
Humoral and cellular immune responses induced by influenza virus PB1-, NP-, and M1-based vaccines. Mice were immunized intramuscularly with influenza virus PB1-, NP-, and M1-based DNA or rVV vaccines, according to the immunization schedule described in Table 1. (A) Humoral immune response in influenza virus PB1, NP, and M1 vaccine-immunized mice. A serum sample was obtained from each mouse at week 8, and the presence of IgG antibodies specific for influenza A virus was analyzed using ELISA. The bars show the geometric mean antibody titers, and the error bars indicate 95% confidence intervals (n = 14 mice/group). (B) Cellular immune responses in influenza virus PB1, NP, and M1 vaccine-immunized mice. Mice were sacrificed at week 8, and the spleens were separated under aseptic conditions and ground to isolate SMNCs (n = 3 mice/group). Next, 4 μg/ml NP147–155 (TYQRTRALV) (B1) and PB1317–325 (MFLAMITYI) (B2) were used as stimulants in ELISPOT assays. The numbers of SMNCs that produced IFN-γ after stimulation with peptides for 30 h are presented as spot-forming cells (SFCs)/106 SMNCs. The bars show mean SFCs/106 SMNCs, and the error bars indicate standard deviations. Lines above two or more groups indicate comparable results. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 using one-way ANOVA.
To examine the effect of immunization with NP-, PB1-, and M1-based vaccines on the cell-mediated immune responses, three mice in each group were euthanized at week 8, and IFN-γ ELISPOT assays were performed. The data revealed that both pSCA-NP (60 ± 25.9 [mean ± SD] SFC/106 SMNCs) and rVV-NP (61.3 ± 30 SFC/106 SMNCs) induced a cellular immune response. However, pSCA-NP prime-rVV-NP boost immunization induced a stronger cellular immune response (306 ± 86.8 SFC/106 SMNCs, P < 0.001) (Fig. 3B1). Among the PB1-based vaccines, only pSCA-PB1 prime-rVV-PB1 boost immunization, but not the pSCA-PB1 and rVV-PB1 vaccines, induced a PB1-specific cellular immune response (68 ± 47.7 SFC/106 SMNCs) (Fig. 3B2). M1-based vaccines did not increase any of the monitored parameters compared with the negative controls at week 8 (data not shown). NP-PB1-M1 combination vaccines induced only NP-specific cellular immune responses, and DNA prime-rVV boost immunization induced the strongest immune response (P < 0.01). In contrast, NP-PB1-M1 combination-based DNA and rVV vaccines and DNA prime-rVV boost immunization induced responses comparable to those of the NP-based vaccine alone, as determined by ELISPOT assays (Fig. 3B).
NP-, PB1-, and M1-based vaccines protect mice against influenza A virus PR8 infection.
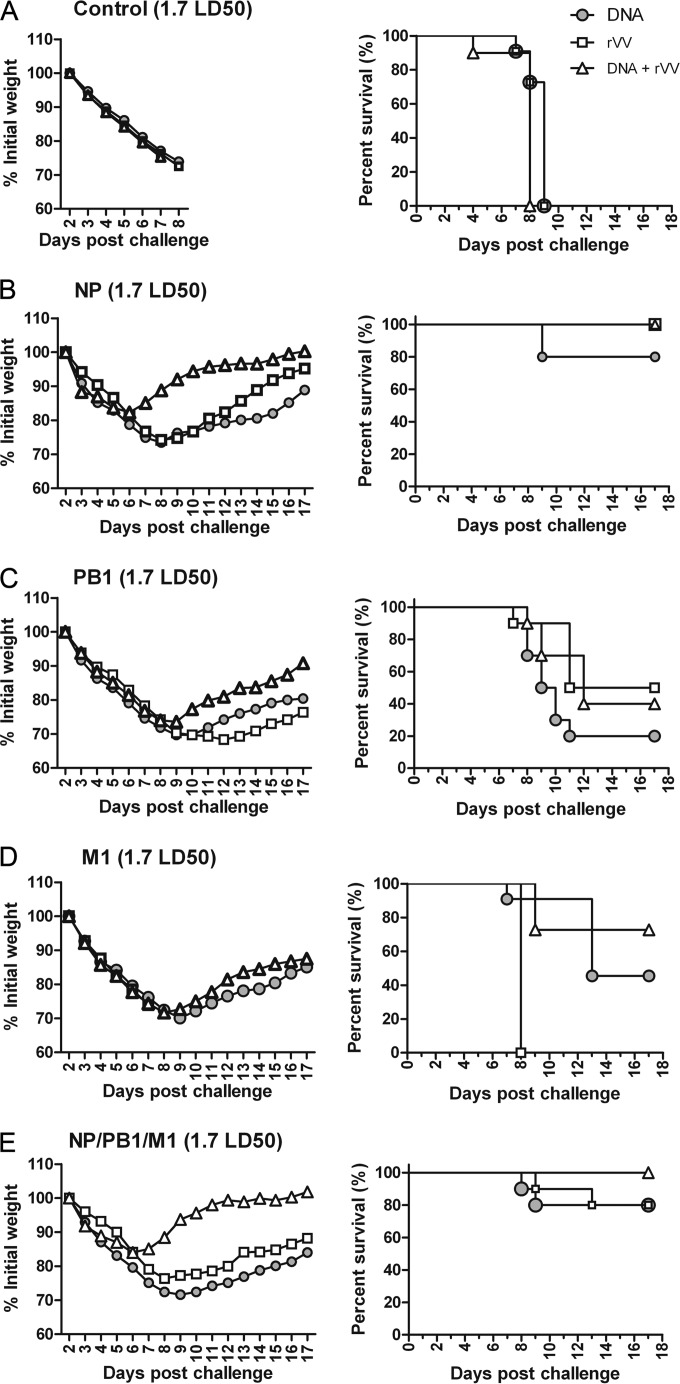
To investigate the potential of the NP-, PB1-, and M1-based vaccines to protect against influenza virus infection, mice were immunized with the vaccines and challenged with 1.7 and 5 LD50 of PR8 virus at week 8. The survival rate and changes in body weight, which served as indicators of the progression of viral infection, were monitored. Standard controls were included.
First, 1.7 LD50 of PR8 was applied in a challenge experiment. All mice in the control groups treated with pSCA-c, rVV-c, or pSCA-c + rVV-c exhibited significant weight loss beginning on day 4 and peaking on day 9 postchallenge, when all mice had died (Fig. 4A). In the NP-based vaccine groups, mice immunized with pSCA-NP, rVV-NP, and pSCA-NP + rVV-NP had 80%, 100%, and 100% survival rates, respectively, which were not significantly different (P > 0.05) (Fig. 4B, right). Mild weight loss occurred in the pSCA-NP + rVV-NP (18%), pSCA-NP (25%), and rVV-NP (25%) groups. Specifically, mice immunized with pSCA-NP + rVV-NP began to gain weight on day 6 postchallenge, which was 2 to 3 days earlier than those immunized with pSCA-NP or rVV-NP (Fig. 4B, left).
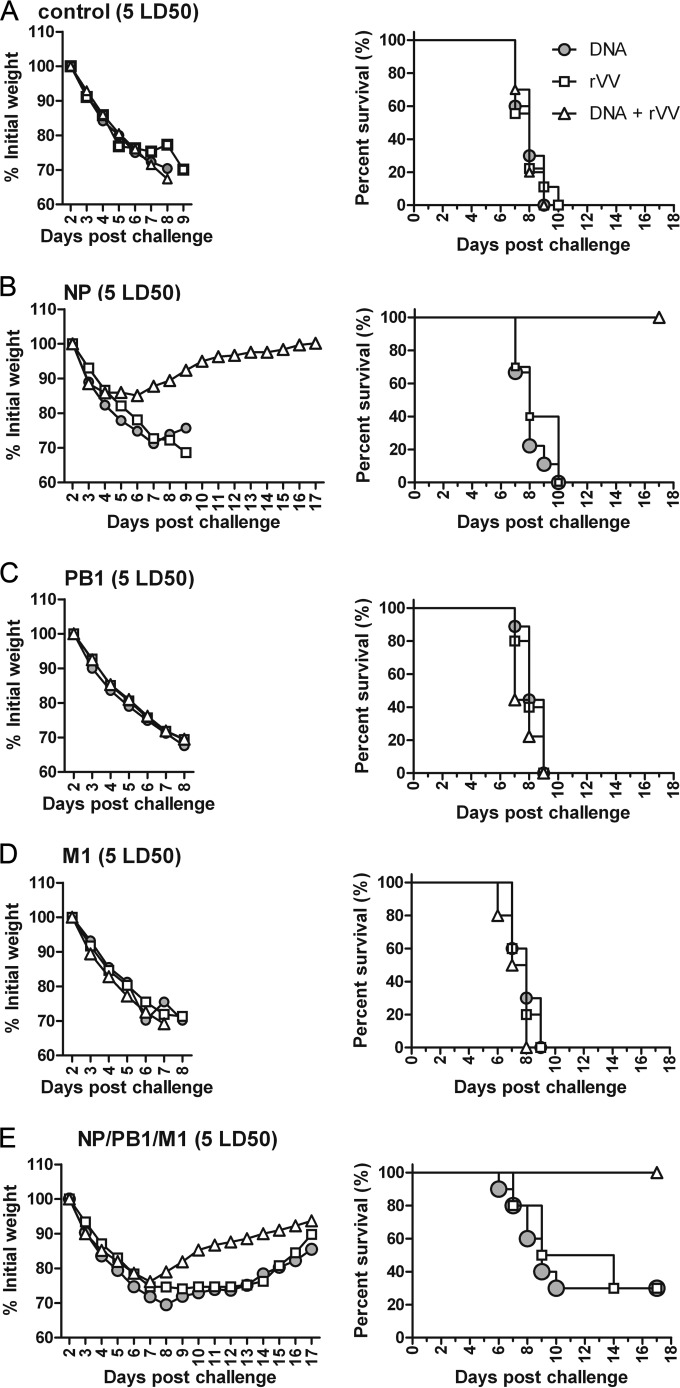
FIG 4.
Protective efficacy of influenza A virus PB1, NP, and M1 vaccines against 1.7 LD50 of PR8. Fifteen groups of mice were immunized with the vaccine control (A), NP (B), PB1 (C), M1 (D), or a combination of the NP, PB1, and M1 (E) DNA, rVV, or prime-boost DNA-rVV vaccines (Table 1) at the times indicated in Fig. 3. Mice were challenged with 1.7 LD50 of influenza virus PR8 at week 8 and were monitored daily for 17 days after challenge (n = 10 mice/group). The mice were weighed daily to detect morbidity (left graphs). The mean weights in each treatment group were followed for the duration of the study, and the percentage of the original body weight was calculated based on the mean weight of each group at day 0. The survival rates (right graphs) were calculated following challenge.
In the PB1-based vaccine groups, mice immunized with pSCA-PB1, rVV-PB1, and pSCA-PB1 + rVV-PB1 had survival rates of 20%, 50%, and 40%, respectively, which were not significantly different (P > 0.05) (Fig. 4C, right). All mice immunized with PB1-based vaccines exhibited serious weight loss, among which those immunized with pSCA-PB1 + rVV-PB1 began to gain weight at day 9 postchallenge; this was ∼2 to 3 days earlier than those immunized with pSCA-PB1 or rVV-PB1 (Fig. 4C, left).
In the M1-based vaccine groups, all mice experienced serious weight loss (Fig. 4D, left), and all those immunized with rVV-M1 succumbed to infection. The mice immunized with pSCA-M1 had a 40% survival rate, which was significantly higher than that of those immunized with rVV-M1. Mice immunized with pSCA-M1 + rVV-M1 had the highest survival rate, at 70%; however, this was not significantly different from that of those immunized with rVV-M1 (P > 0.05) (Fig. 4D, right).
When mice were immunized with the NP-PB1-M1 combination vaccine, those immunized with DNA and rVV vaccines experienced weight loss of ∼25 to 30%, whereas mice immunized DNA + rVV experienced less weight loss (∼16%) and began to gain weight at day 6 postchallenge, which was ∼2 to 3 days earlier than the other two groups (Fig. 4E, left). A total of 80%, 80%, and 100% of the mice immunized with combined DNA, rVV, and DNA+rVV survived after challenge with 1.7 LD50 of PR8, respectively; these differences were not significant (P > 0.05) (Fig. 4E, right).
Taken together, these data suggest that among the DNA vaccines, pSCA-PB1 had the least protective efficacy, followed by pSCA-M1; optimal protection was afforded by pSCA-NP immunization. Meanwhile, combination immunization with pSCA-NP, pSCA-PB1, and pSCA-M1 induced strong protection. Regarding rVV vaccines, rVV-M1 exhibited poorer protective efficacy than did rVV-PB1, whereas the highest protective efficacy was induced by rVV-NP, which resulted in a 100% survival rate. Meanwhile, combination immunization with rVV-NP, rVV-PB1, and rVV-M1 induced some protection, as evidenced by an 80% survival rate.
For the DNA prime-rVV boost, PB1-based vaccines resulted in the lowest survival rate, of 40%, whereas mice immunized with M1-based vaccines had a 70% survival rate. However, there was no significant difference between the two groups (P > 0.05). NP-based vaccines protected 100% of mice against challenge with 1.7 LD50 of PR8, which was significantly higher than that induced by PB1-based vaccines (P < 0.01) but comparable to that with M1-based vaccines (P > 0.05). Moreover, immunization with NP-PB1-M1 combination vaccines induced survival rates similar to those of NP-based vaccines. In summary, the PB1- and M1-based vaccines conferred partial protective immunity against infection with 1.7 LD50 of influenza A virus PR8 in mice; however, the protective efficacy was poorer than that of the NP-based vaccines. Moreover, vaccination with the three-antigen combination induced strong protection.
Next, to further investigate the protective efficacy of NP-, PB1-, and M1-based vaccines, 5 LD50 of PR8 was applied in the challenge experiment. The mice in the control groups (Fig. 5A), those immunized with PB1-based vaccines (Fig. 5C), and those immunized with M1-based vaccines (Fig. 5D) showed significant weight loss and succumbed to infection by day 9 after challenge. In the NP-based vaccine immunization groups, all mice immunized with pSCA-NP or rVV-NP exhibited serious weight loss and died, whereas those immunized with pSCA-NP + rVV-NP showed only 15% weight loss and began to gain weight by day 6 postchallenge; 100% of the mice in this group survived, which was significantly higher than the survival rates in the other two groups (P < 0.001) (Fig. 5B). When mice were immunized with NP-PB1-M1 combination vaccines, those receiving DNA and rVV vaccines had serious weight loss and a 30% survival rate. In contrast, those immunized with DNA + rVV experienced less weight loss, and all mice in this group survived a lethal challenge with 5 LD50, which was significantly more than that in the DNA and rVV groups (P < 0.01) (Fig. 5E). These data suggest that DNA or rVV vaccines based on a single NP, PB1, or M1 antigen did not protect mice against lethal challenge with 5 LD50 of PR8. With the DNA prime-rVV boost strategy, only pSCA-NP + rVV-NP immunization resulted in optimal protection and achieved a 100% survival rate. When the combination NP-PB1-M1 vaccine was applied, both DNA and rVV vaccines conferred partial protective immunity against lethal challenge with PR8. However, only the DNA + rVV vaccination strategy completely protected the mice.
FIG 5.
Protective efficacy of immunization with influenza virus PB1-, NP-, and M1-based vaccines against 5 LD50 of PR8. Fifteen groups of mice were immunized with the vaccine control (A), NP (B), PB1 (C), M1 (D), or a combination of the NP, PB1, and M1 (E) DNA, rVV, or prime-boost DNA-rVV vaccines (Table 1) at the times indicated in Fig. 3. Mice were challenged with 5 LD50 of influenza virus PR8 at week 8 and monitored daily for 17 days after challenge (n = 10 mice per experimental group). The mice were weighed daily to detect morbidity (left graphs). The mean weights in each treatment group were followed for the duration of the study, and the percentage of the original body weight was calculated based on the mean weight of each group at day 0. The survival rates (right graphs) were calculated following challenge.
Overall, the PB1- and M1-based DNA and rVV vaccines, regardless of whether they were applied individually or in a prime-boost strategy, conferred weak immunity against infection with influenza A virus PR8 in mouse models. Both pSCA-NP and rVV-NP protected mice against challenge with 1.7 LD50 of PR8. In addition, pSCA-NP prime-rVV-NP boost immunization conferred complete protection against challenge with 1.7 and 5 LD50 of PR8. Combination immunization with the three-antigen-based DNA or rVV vaccines conferred fair protection against challenge with 1.7 LD50 of PR8, as well as a degree of protection against challenge with 5 LD50 of PR8. Finally, DNA prime-rVV boost immunization protected mice completely against challenge with 1.7 LD50 and 5 LD50 of PR8.
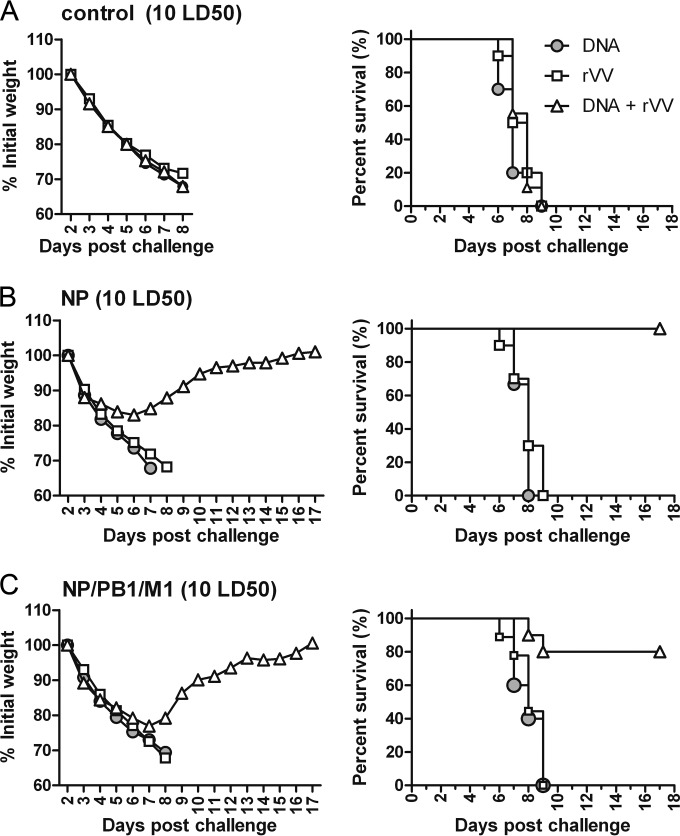
Next, to investigate the protective potency of NP-based and combination vaccines, 10 LD50 of PR8 was used in a challenge experiment. After challenge, all mice in the control groups succumbed to infection (Fig. 6A), as did mice immunized with pSCA-NP or rVV-NP (Fig. 6B). This suggests that pSCA-NP or rVV-NP immunization did not protect against challenge with 10 LD50 in mice. In contrast, mice immunized with pSCA-NP + rVV-NP exhibited 17% weight loss but then began to gain weight on day 6 postchallenge; all mice in this group survived (Fig. 6B). All mice immunized with a DNA or rVV combination NP-PB1-M1 vaccine succumbed to infection with 10 LD50 of PR8, suggesting that these did not protect against challenge with 10 LD50 of PR8. However, mice immunized with the combination vaccines using a prime-boost strategy suffered only 22% weight loss and then began to gain weight on day 7 postchallenge. Around 80% of the mice in this group survived (Fig. 6C), similar to the group immunized with pSCA-NP prime-rVV-NP boost (P > 0.05). In summary, only the DNA prime-rVV boost strategy based on NP or a combination of antigens protected against challenge with 10 LD50 of PR8.
FIG 6.
Protective efficacy of immunization with influenza virus NP-based vaccines against 10 LD50 of PR8. Nine groups of mice were immunized with the vaccine control (A), NP (B), or a combination of the NP, PB1, and M1 (C) DNA, rVV, or prime-boosted DNA-rVV vaccines (Table 1) at the times indicated in Fig. 3. Mice were challenged with 10 LD50 of influenza virus PR8 at week 8 and monitored daily for 17 days after challenge (n = 10 mice/group). The mice were weighed daily to detect morbidity (left graphs). The mean weights in each treatment group were followed for the duration of the study, and the percentage of the original body weight was calculated based on the mean weight of each group at day 0. The survival rates (right graphs) were calculated following challenge.
Taken together, these data suggest that among the three antigens NP, PB1, and M1, only NP showed optimal protection against challenge with a high dose of PR8. In addition, a combination of the three antigens induced good protection against challenge with 10 LD50 of PR8.
DISCUSSION
The development of a universal influenza vaccine capable of inducing broad-spectrum and long-term immunity is important. Identification of the conserved influenza virus genes that could be used to develop a combination vaccine that induces stable and long-term protection is critical. In the present study, the protective potency of a universal influenza vaccine based on the conserved NP, PB1, and M1 proteins was assessed. In theory, NP-, PB1-, and/or M1-based universal influenza vaccine strategies have several advantages. These universal vaccines are raised against highly conserved proteins that are clearly defined components. The current study assessed the immunogenicity and protective potency of NP-, PB1-, and M1-based vaccines against heterologous influenza virus PR8 challenge. The data demonstrate the feasibility of strategies based on NP, PB1, and M1 candidate universal vaccines. Among the three conserved antigens, NP resulted in the strongest protection against PR8 challenge, whereas PB1- and M1-based vaccines induced only limited protection against challenge with 1.7 LD50 of PR8 but none against challenge with 5 LD50 of PR8. This suggests that NP is an important antigen. Consistent with this, NP is the preferred candidate antigen in the research and development of UIVs. For example, the universal influenza vaccine N8295 produced by Dynavax (62) and MVA-NP+M1 by Oxford (14, 42, 44) include NP.
Previous studies found that influenza vaccines based on a single conserved protein, such as M1, M2, or NP, did not provide adequate protection, and vaccines based on two or more antigens induced higher protection than those based on a single antigen (8). To improve the protective efficacy of UIVs, combination vaccines using multiple target antigens have been developed. In previous studies, the whole genes or epitopes of HA, M1, and/or NP were used for development of combination vaccines; these induced effective cross-protection (32, 38, 44–48). In our previous study, the whole NP gene and the extracellular domain of M2 (M2e) from BJ95 were fused and expressed in E. coli. The fusion protein NM2e elicited a more robust immune response and induced greater protection against challenge with 20 LD50 of PR8 in mice than NP alone (61). The above-mentioned studies suggested the development of universal influenza vaccines by combining multiple conserved antigens to be a feasible strategy for preventing influenza pandemics.
In the present study, we investigated the feasibility of developing a universal influenza vaccine by combining the NP, PB1 and M1 antigens. The data reveal that the combination NP-PB1-M1 vaccines induced certain protective effects in BALB/c mice. Specifically, 80% of the mice immunized with NP-PB1-M1 combination vaccines using the DNA prime-rVV boost strategy survived challenge with 10 LD50 of PR8, which was comparable to the survival rate of mice immunized with NP-based vaccines. Therefore, the protective efficacy of the combination vaccines must be improved further, as they did not induce a comprehensive immune response. Moreover, mice immunized with the combination vaccines experienced more serious weight loss than did those immunized with NP-based vaccines following challenge with 5 or 10 LD50 of PR8, which suggests that the addition of PB1 and M1 reduced the immune response and protection induced by NP. There are several possible reasons for this.
The first possibility is interference among the candidate antigens. The expression of PB1 and M1 might influence the expression or presentation of NP and thereby weaken the immune response induced by NP immunization. Moreover, competition might occur among the antigens. The NP antigen is easily recognized by the immune system and so was likely the primary inducer of an immune response upon administration in combination with PB1 and M1; therefore, PB1 and M1 are not easily recognized by the immune system. This was confirmed by the fact that immunization with the NP-PB1-M1 combination vaccine using a DNA prime-rVV boost did not induce a marked cellular response against PB1, whereas immunization with PB1 alone using pSCA-PB1 prime-rVV-PB1 boost did.
Another possible reason for the greater efficacy of the NP antigen is interference by the vector. The combination vaccines were applied using a dose of vector 3-fold higher than that for the single vaccine; therefore, adverse effects resulting from the increased concentration of vector cannot be ruled out. Such effects might have diminished the immune response to the antigens. This could be overcome by maintaining the target antigen dose while decreasing that of the vector. In addition, interference among vectors could be decreased by developing vector-based vaccines using fusion antigens. Several combination vaccines that yielded less interference have been generated. For example, Donnelly et al. (63) constructed a DNA vaccine by fusing influenza virus HA, M1, and NP genes. In contrast, Jeon, Ben-Yedidia, and Arnon (48) fused the HA91–108 (B-cell epitope), NP55–69 (Th-cell epitope), and NP147–158 (CD8+ T-cell epitope) influenza virus epitopes with Salmonella flagellin. Zhou et al. (32) constructed adenovirus-based vaccines by fusing M2e and NP. In addition, the MVA-based universal influenza vaccine MVA-NP+M1, which has been tested in human phase 1 clinical trials, contains a fusion of NP to M1 (39). Finally, an rVV-based vaccine that contains multiple T epitopes of the M1, nonstructural 1 (NS1), NP, PB1, and polymerase acidic (PA) influenza virus proteins has been developed (64). All of the above-mentioned vaccines induced fair immune responses in animal models or humans. This suggests that the interference among antigens and the background effect of the DNA or virus vector could be minimized by constructing a universal vaccine with an appropriate vector by fusing the target antigens NP, PB1, and M1 or expressing two or more proteins using a dual promoter in a single vector. Various UIVs have been developed using several vaccine forms based on the combination of multiple antigens or epitopes, including recombinant subunit vaccines, DNA vaccines, and virus-vectored vaccines. In many cases, the immunization strategy of DNA prime-virus-vectored vaccine boosts was preferred because it induced a strong immune response (65, 66). More importantly, the NP-based DNA prime-adenovirus boosting strategy significantly improves the NP-induced cross-protective effect (30). In addition, DNA plasmids are good priming regimens in heterologous prime-boost vaccination against influenza A viruses in both animals and humans (67, 68). In the current study, the DNA prime-rVV boost strategy induced effective protection against PR8 challenge. The PB1 antigen induced a stronger cellular immune response in mice via the pSCA-PB1 prime-rVV-PB1 boost strategy than that with either pSCA-PB1 or rVV-PB1 alone. Moreover, immunization using the NP-based vaccines with the pSCA-NP prime-rVV-NP boost strategy induced stronger cellular and humoral immune responses than did pSCA-NP or rVV-NP immunization alone. Furthermore, the pSCA-NP prime-rVV-NP boost strategy afforded protection against 10 LD50 of PR8 challenge in mice, whereas neither pSCA-NP nor rVV-NP alone did. The M1 antigen was only weakly immunogenic, and although the pSCA-M1 prime-rVV-M1 boost immunization did not induce a marked cellular immune response, it enhanced the M1-specific humoral immune response and conferred a degree of protection in mice against challenge with 1.7 LD50 of PR8. Finally, the combination vaccines based on NP-PB1-M1 administered using the DNA prime-rVV boost strategy also showed protection superior to that of the DNA or rVV vaccine alone. Accordingly, the DNA prime-rVV boost immunization strategy is superior to single DNA or rVV vaccine immunization. Previous data suggested that the prime-boost strategy not only increased the number of memory CD8+ T cells but also enhanced the function of CD8+ T cells (69).
Immunization with NP, PB1, or M1 does not elicit the production of neutralizing antibodies. Therefore, the protective immunity is mediated mainly by NP-, PB1-, or M1-specific cytotoxic T lymphocyte (CTL) immune responses and nonneutralizing functions of specific antibodies (8). In the current study, NP immunization induced specific humoral and cellular immune responses in mice, whereas PB1 immunization induced only a specific cellular response, and M1 immunization induced only a marked humoral immune response. This suggests that the NP-specific humoral and cellular immune responses, PB1-related cellular response, and M1-related humoral response might play roles in protection against heterologous challenge. This might explain the superior protection and immune response induced by NP immunization, as well as the poor protection and incomplete immune response induced by immunization with PB1 or M1. Thus, the immunogenicity of PB1 and M1 vaccines needs to be improved further to induce comprehensive immune responses and elicit cross-protection.
In addition, although some exciting progress has been made in the current study, the immunogenicity of NP-, PB1-, and M1-based DNA vaccines and vaccinia virus vector-based vaccines remains limited, and further optimization is needed regarding vaccine design, immunization strategies, delivery systems, and detection methods.
The NP, M1 and PB1 antigens exhibit different immunogenicities; thus, an inappropriate mouse strain might have been used. Previously, we investigated the immune response and protective efficacy of NP-based universal vaccines in BALB/c mice (61, 70, 71). To maintain consistency with the previous experiments, BALB/c mice were also used in the present study to confirm the immunogenicity and protective efficacy of the NP-, M1-, and PB1-based universal vaccines. It is worth considering that although commonly used mouse strains generate strong T-cell responses to NP, BALB/c mice do not generate CTL responses against PB1 or M1 (72). Other mouse strains, such as CBA and C57/BL6, mount stronger responses against PB1, making it likely that PB1- or M1-based universal vaccines might induce better immune responses and protective effects in these mouse strains. Thus, CBA and C57/BL6 strains should be used in future experiments.
A multiple-vaccination regimen was used in the current study, such as three doses of DNA plus a vaccinia virus boost vaccine at 2-week intervals or two doses of vaccinia virus vaccine at 4-week intervals. However, such a protracted immunization schedule is not feasible for routine use in humans. Our primary aim was to confirm the immunogenicity of the NP, M1, and PB1 antigens. Once the appropriate antigen or antigen combination has been confirmed, the immunization schedule should be optimized for routine use in a future experiment using a single immunogen with a single vaccination.
In the current experiment, DNA vaccines induced poor immune responses, which was likely caused by the immunization route. Previous studies have used an electroporation machine (49, 73) or a gene gun (74) to enhance the immunogenicity and/or decrease the dose of the antigen required. However, in the current study, DNA was injected directly into the muscles without the use of any transfection reagent or equipment. Therefore, a future study should utilize a reagent or equipment to enhance the immunogenicity of the NP, PB1, and M1 antigens. Previous studies of recombinant vaccinia virus vaccines used single intraperitoneal inoculations in mice (64, 75). In our previous study, vaccinia virus-vectored vaccines induced a stronger immune response with intraperitoneal than with intramuscular administration (data not shown); however, intramuscular inoculation is preferred for routine use in humans. In previous clinical studies, the vaccinia virus-vectored vaccine was generally safe and well tolerated, with significantly fewer local side effects after intramuscular administration than after intradermal administration; thus, the intramuscular injection was preferred and administered to volunteers (39, 44, 76). As such, future experiments should preferentially use intramuscular inoculation of vaccinia virus-vectored vaccines.
Ether-split influenza virus was used as a target antigen for assessing the antibody responses; therefore, it is possible that the magnitude of the PB1-specific antibody response (particularly compared with that of NP and M1) was underestimated significantly, because there is comparatively little PB1 present in influenza virions compared with that in NP or M1. In future experiments, it would be more appropriate to use recombinant proteins to assess this possibility and optimize the detection methods.
In conclusion, DNA and rVV vaccines based on influenza NP, PB1, and M1 showed protective effects. NP was the most effective of the three antigens and conferred protection against heterologous PR8 challenge. UIVs based on a combination of NP, PB1, and M1 would likely induce strong immune responses; therefore, they might be an alternative approach for preventing future influenza virus pandemics.
ACKNOWLEDGMENT
This work was supported by a grant from the National High Technology Research and Development Program of China (863 Program) (grant 2006AA02A203).
REFERENCES
- 1.Slepushkin VA, Katz JM, Black RA, Gamble WC, Rota PA, Cox NJ. 1995. Protection of mice against influenza A virus challenge by vaccination with baculovirus-expressed M2 protein. Vaccine 13:1399–1402. doi: 10.1016/0264-410X(95)92777-Y. [DOI] [PubMed] [Google Scholar]
- 2.Fiers W, De Filette M, El Bakkouri K, Schepens B, Roose K, Schotsaert M, Birkett A, Saelens X. 2009. M2e-based universal influenza A vaccine. Vaccine 27:6280–6283. doi: 10.1016/j.vaccine.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Gerdil C. 2003. The annual production cycle for influenza vaccine. Vaccine 21:1776–1779. doi: 10.1016/S0264-410X(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 4.Palese P. 2006. Making better influenza virus vaccines? Emerg Infect Dis 12:61–65. doi: 10.3201/eid1201.051043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manzoli L, Schioppa F, Boccia A, Villari P. 2007. The efficacy of influenza vaccine for healthy children: a meta-analysis evaluating potential sources of variation in efficacy estimates including study quality. Pediatr Infect Dis J 26:97–106. doi: 10.1097/01.inf.0000253053.01151.bd. [DOI] [PubMed] [Google Scholar]
- 6.Jefferson T, Di Pietrantonj C, Rivetti A, Bawazeer GA, Al-Ansary LA, Ferroni E. 2007. Vaccines for preventing influenza in healthy adults. Cochrane Database Syst Rev (7):CD001269. doi: 10.1002/14651858.CD001269.pub4. [DOI] [PubMed] [Google Scholar]
- 7.Fedson DS. 2005. Preparing for pandemic vaccination: an international policy agenda for vaccine development. J Public Health Policy 26:4–29. doi: 10.1057/palgrave.jphp.3200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, Luo J, Chen Z. 2014. Development of UIVs based on influenza virus M and NP genes. Infection 42:251–262. doi: 10.1007/s15010-013-0546-4. [DOI] [PubMed] [Google Scholar]
- 9.Lee LY, Ha do LA, Simmons C, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest 118:3478–3490. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha S, Yoshida S, Ohba K, Matsui K, Matsuda T, Takeshita F, Umeda K, Tamura Y, Okuda K, Klinman D, Xin KQ, Okuda K. 2006. A fused gene of nucleoprotein (NP) and herpes simplex virus genes (VP22) induces highly protective immunity against different subtypes of influenza virus. Virology 354:48–57. doi: 10.1016/j.virol.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. 2007. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol 18:529–536. doi: 10.1016/j.copbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, Nabel GJ. 2005. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 23:5404–5410. doi: 10.1016/j.vaccine.2005.04.047. [DOI] [PubMed] [Google Scholar]
- 13.Zhirnov OP, Isaeva EI, Konakova TE, Thoidis G, Piskareva LM, Akopova II, Kartashov A, Altstein AD, Ilyinskii PO, Shneider AM. 2007. Protection against mouse and avian influenza A strains via vaccination with a combination of conserved proteins NP, M1 and NS1. Influenza Other Respir Viruses 1:71–79. doi: 10.1111/j.1750-2659.2007.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antrobus RD, Lillie PJ, Berthoud TK, Spencer AJ, McLaren JE, Ladell K, Lambe T, Milicic A, Price DA, Hill AV, Gilbert SC. 2012. A T cell-inducing influenza vaccine for the elderly: safety and immunogenicity of MVA-NP+M1 in adults aged over 50 years. PLoS One 7:e48322. doi: 10.1371/journal.pone.0048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atsmon J, Kate-Ilovitz E, Shaikevich D, Singer Y, Volokhov I, Haim KY, Ben-Yedidia T. 2012. Safety and immunogenicity of multimeric-001–a novel universal influenza vaccine. J Clin Immunol 32:595–603. doi: 10.1007/s10875-011-9632-5. [DOI] [PubMed] [Google Scholar]
- 16.Adar Y, Singer Y, Levi R, Tzehoval E, Perk S, Banet-Noach C, Nagar S, Arnon R, Ben-Yedidia T. 2009. A universal epitope-based influenza vaccine and its efficacy against H5N1. Vaccine 27:2099–2107. doi: 10.1016/j.vaccine.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Ilyinskii PO, Meriin AB, Gabai VL, Zhirnov OP, Thoidis G, Shneider AM. 2008. Prime-boost vaccination with a combination of proteosome-degradable and wild-type forms of two influenza proteins leads to augmented CTL response. Vaccine 26:2177–2185. doi: 10.1016/j.vaccine.2008.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tompkins SM, Zhao ZS, Lo CY, Misplon JA, Liu T, Ye Z, Hogan RJ, Wu Z, Benton KA, Tumpey TM, Epstein SL. 2007. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis 13:426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eliasson DG, El Bakkouri K, Schön K, Ramne A, Festjens E, Lowenadler B, Fiers W, Saelens X, Lycke N. 2008. CTA1-M2e-DD: a novel mucosal adjuvant targeted influenza vaccine. Vaccine 26:1243–1252. doi: 10.1016/j.vaccine.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Huleatt JW, Nakaar V, Desai P, Huang Y, Hewitt D, Jacobs A, Tang J, McDonald W, Song L, Evans RK, Umlauf S, Tussey L, Powell TJ. 2008. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 26:201–214. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 21.Wu F, Huang JH, Yuan XY, Huang WS, Chen YH. 2007. Characterization of immunity induced by M2e of influenza virus. Vaccine 25:8868–8873. doi: 10.1016/j.vaccine.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 22.Ernst WA, Kim HJ, Tumpey TM, Jansen AD, Tai W, Cramer DV, Adler-Moore JP, Fujii G. 2006. Protection against H1, H5, H6 and H9 influenza A infection with liposomal matrix 2 epitope vaccines. Vaccine 24:5158–5168. doi: 10.1016/j.vaccine.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Fan J, Liang X, Horton MS, Perry HC, Citron MP, Heidecker GJ, Fu TM, Joyce J, Przysiecki CT, Keller PM, Garsky VM, Ionescu R, Rippeon Y, Shi L, Chastain MA, Condra JH, Davies ME, Liao J, Emini EA, Shiver JW. 2004. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 22:2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Zhou C, Zhou L, Chen YH. 2012. Immunization with high epitope density of M2e derived from 2009 pandemic H1N1 elicits protective immunity in mice. Vaccine 30:3463–3469. doi: 10.1016/j.vaccine.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 25.De Filette M, Martens W, Roose K, Deroo T, Vervalle F, Bentahir M, Vandekerckhove J, Fiers W, Saelens X. 2008. An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J Biol Chem 283:11382–11387. doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turley CB, Rupp RE, Johnson C, Taylor DN, Wolfson J, Tussey L, Kavita U, Stanberry L, Shaw A. 2011. Safety and immunogenicity of a recombinant M2e-flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 29:5145–5152. doi: 10.1016/j.vaccine.2011.05.041. [DOI] [PubMed] [Google Scholar]
- 27.Talbot HK, Rock MT, Johnson C, Tussey L, Kavita U, Shanker A, Shaw AR, Taylor DN. 2010. Immunopotentiation of trivalent influenza vaccine when given with VAX102, a recombinant influenza M2e vaccine fused to the TLR5 ligand flagellin. PLoS One 5:e14442. doi: 10.1371/journal.pone.0014442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Košík I, Krejnusová I, Práznovská M, Poláková K, Russ G. 2012. A DNA vaccine expressing PB1 protein of influenza A virus protects mice against virus infection. Arch Virol 157:811–817. doi: 10.1007/s00705-012-1238-6. [DOI] [PubMed] [Google Scholar]
- 29.Li RQ, Lu N, Deng Y, Wang WL, Xin W, Zhang XM, Ruan L. 2006. Cross protective immunity against influenza A virus between subtypes induced by influenza polymerase protein PB1 in mice. Chin J Microbiol Immunol 26:322–327. doi: 10.3760/j:issn:0254-5101.2006.04.008. [DOI] [Google Scholar]
- 30.Lo CY, Wu Z, Misplon JA, Price GE, Pappas C, Kong WP, Tumpey TM, Epstein SL. 2008. Comparison of vaccines for induction of heterosubtypic immunity to influenza A virus: cold-adapted vaccine versus DNA prime-adenovirus boost strategies. Vaccine 26:2062–2072. doi: 10.1016/j.vaccine.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez GS, Planchon R, Wei Q, Rusalov D, Geall A, Enas J, Lalor P, Leamy V, Vahle R, Luke CJ, Rolland A, Kaslow DC, Smith LR. 2007. Vaxfectin-formulated influenza DNA vaccines encoding NP and M2 viral proteins protect mice against lethal viral challenge. Hum Vaccin 3:157–164. doi: 10.4161/hv.3.5.4175. [DOI] [PubMed] [Google Scholar]
- 32.Zhou D, Wu TL, Lasaro MO, Latimer BP, Parzych EM, Bian A, Li Y, Li H, Erikson J, Xiang Z, Ertl HC. 2010. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol Ther 18:2182–2189. doi: 10.1038/mt.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soboleski MR, Gabbard JD, Price GE, Misplon JA, Lo CY, Perez DR, Ye J, Tompkins SM, Epstein SL. 2011. Cold-adapted influenza and recombinant adenovirus vaccines induce cross-protective immunity against pH1N1 challenge in mice. PLoS One 6:e21937. doi: 10.1371/journal.pone.0021937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price GE, Soboleski MR, Lo CY, Misplon JA, Pappas C, Houser KV, Tumpey TM, Epstein SL. 2009. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine 27:6512–6521. doi: 10.1016/j.vaccine.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 35.Price GE, Soboleski MR, Lo CY, Misplon JA, Quirion MR, Houser KV, Pearce MB, Pappas C, Tumpey TM, Epstein SL. 2010. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS One 5:e13162. doi: 10.1371/journal.pone.0013162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lalor PA, Webby RJ, Morrow J, Rusalov D, Kaslow DC, Rolland A, Smith LR. 2008. Plasmid DNA-based vaccines protect mice and ferrets against lethal challenge with A/Vietnam/1203/04 (H5N1) influenza virus. J Infect Dis 197:1643–1652. doi: 10.1086/588431. [DOI] [PubMed] [Google Scholar]
- 37.Vitelli A, Quirion MR, Lo CY, Misplon JA, Grabowska AK, Pierantoni A, Ammendola V, Price GE, Soboleski MR, Cortese R, Colloca S, Nicosia A, Epstein SL. 2013. Vaccination to conserved influenza antigens in mice using a novel simian adenovirus vector, PanAd3, derived from the bonobo Pan paniscus. PLoS One 8:e55435. doi: 10.1371/journal.pone.0055435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd AC, Ruiz-Hernandez R, Peroval MY, Carson C, Balkissoon D, Staines K, Turner AV, Hill AV, Gilbert SC, Butter C. 2013. Towards a universal vaccine for avian influenza: protective efficacy of modified vaccinia virus Ankara and adenovirus vaccines expressing conserved influenza antigens in chickens challenged with low pathogenic avian influenza virus. Vaccine 31:670–675. doi: 10.1016/j.vaccine.2012.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthoud TK, Hamill M, Lillie PJ, Hwenda L, Collins KA, Ewer KJ, Milicic A, Poyntz HC, Lambe T, Fletcher HA, Hill AV, Gilbert SC. 2011. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis 52:1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullarkey CE, Boyd A, van Laarhoven A, Lefevre EA, Veronica Carr B, Baratelli M, Molesti E, Temperton NJ, Butter C, Charleston B, Lambe T, Gilbert SC. 2013. Improved adjuvanting of seasonal influenza vaccines: preclinical studies of MVA-NP+M1 coadministration with inactivated influenza vaccine. Eur J Immunol 43:1940–1952. doi: 10.1002/eji.201242922. [DOI] [PubMed] [Google Scholar]
- 41.Lambe T, Carey JB, Li Y, Spencer AJ, van Laarhoven A, Mullarkey CE, Vrdoljak A, Moore AC, Gilbert SC. 2013. Immunity against heterosubtypic influenza virus induced by adenovirus and MVA expressing nucleoprotein and matrix protein-1. Sci Rep 3:1443. doi: 10.1038/srep01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antrobus RD, Berthoud TK, Mullarkey CE, Hoschler K, Coughlan L, Zambon M, Hill AV, Gilbert SC. 2014. Coadministration of seasonal influenza vaccine and MVA-NP+M1 simultaneously achieves potent humoral and cell-mediated responses. Mol Ther 22:233–238. doi: 10.1038/mt.2013.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antrobus RD, Coughlan L, Berthoud TK, Dicks MD, Hill AV, Lambe T, Gilbert SC. 2014. Clinical assessment of a novel recombinant simian adenovirus ChAdOx1 as a vectored vaccine expressing conserved influenza A antigens. Mol Ther 22:668–674. doi: 10.1038/mt.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lillie PJ, Berthoud TK, Powell TJ, Lambe T, Mullarkey C, Spencer AJ, Hamill M, Peng Y, Blais ME, Duncan CJ, Sheehy SH, Havelock T, Faust SN, Williams RL, Gilbert A, Oxford J, Dong T, Hill AV, Gilbert SC. 2012. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin Infect Dis 55:19–25. doi: 10.1093/cid/cis327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu K, Ling ZY, Sun L, Xu Y, Bian C, He Y, Lu W, Chen Z, Sun B. 2011. Broad humoral and cellular immunity elicited by a bivalent DNA vaccine encoding HA and NP genes from an H5N1 virus. Viral Immunol 24:45–56. doi: 10.1089/vim.2010.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandey A, Singh N, Vemula SV, Couëtil L, Katz JM, Donis R, Sambhara S, Mittal SK. 2012. Impact of preexisting adenovirus vector immunity on immunogenicity and protection conferred with an adenovirus-based H5N1 influenza vaccine. PLoS One 7:e33428. doi: 10.1371/journal.pone.0033428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Gabbard JD, Mooney A, Chen Z, Tompkins SM, He B. 2013. Efficacy of parainfluenza virus 5 mutants expressing hemagglutinin from H5N1 influenza A virus in mice. J Virol 87:9604–9609. doi: 10.1128/JVI.01289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeon SH, Ben-Yedidia T, Arnon R. 2002. Intranasal immunization with synthetic recombinant vaccine containing multiple epitopes of influenza virus. Vaccine 20:2772–2780. doi: 10.1016/S0264-410X(02)00187-1. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, Zheng D, Zhang W, Fang F, Wang H, Sun Y, Ding Y, Xu C, Chen Q, Zhang H, Huang D, Sun B, Chen Z. 2012. Induction of cross-protection against influenza A virus by DNA prime-intranasal protein boost strategy based on nucleoprotein. Virol J 9:286. doi: 10.1186/1743-422X-9-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID, World Health Organization . 1988. Smallpox and its eradication. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/39485/1/9241561106.pdf?ua=1. [Google Scholar]
- 51.Bian T, Wan Y, Lu Z, Ye Z, Zhao L, Ren J, Zhang H, Ruan L, Tian H. 2008. Human papillomavirus type 16 L1E7 chimeric capsomeres have prophylactic and therapeutic efficacy against papillomavirus in mice. Mol Cancer Ther 7:1329–1335. doi: 10.1158/1535-7163.MCT-07-2015. [DOI] [PubMed] [Google Scholar]
- 52.Yan K, Tan W, Wang H, Wang Y, Zhang X, Li Y, Ruan L. 2009. SARS-CoV spike proteins expressed by the vaccinia virus Tiantan strain: secreted sq protein induces robust neutralization antibody in mice. Viral Immunol 22:57–66. doi: 10.1089/vim.2008.0064. [DOI] [PubMed] [Google Scholar]
- 53.Deng Y, Zhang K, Tan W, Wang Y, Chen H, Wu X, Ruan L. 2009. A recombinant DNA and vaccinia virus prime-boost regimen induces potent long-term T-cell responses to HCV in BALB/c mice. Vaccine 27:2085–2088. doi: 10.1016/j.vaccine.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao L, Liu B, Ren J, Feng J, Pang Z, Gao J, Zhang H, Tan W, Tian H, Ruan L. 2011. Immunogenicity in mice and rhesus monkeys vaccinated with recombinant vaccinia virus expressing bivalent E7E6 fusion proteins from human papillomavirus types 16 and 18. Virol J 8:302. doi: 10.1186/1743-422X-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H, Chuai X, Deng Y, Wen B, Wang W, Xiong S, Ruan L, Tan W. 2012. Optimisation of prime-boost immunization in mice using novel protein-based and recombinant vaccinia (Tiantan)-based HBV vaccine. PLoS One 7:e43730. doi: 10.1371/journal.pone.0043730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohamed MR, Niles EG. 2003. UUUUUNU stimulation of vaccinia virus early gene transcription termination. Oligonucleotide sequence and structural requirements for stimulation of premature termination in vitro. J Biol Chem 278:39534–39541. doi: 10.1074/jbc.M306048200. [DOI] [PubMed] [Google Scholar]
- 57.Mohamed MR, Niles EG. 2003. UUUUUNU oligonucleotide stimulation of vaccinia virus early gene transcription termination, in trans. J Biol Chem 278:11794–11801. doi: 10.1074/jbc.M213263200. [DOI] [PubMed] [Google Scholar]
- 58.Wang WL, Huang BY, Deng Y, Wang XP, Tan WJ, Ruan L. 2007. Expression of influenza A3 virus (H3N2) M2 gene in vaccinia virus Tiantan strain. Bing Du Xue Bao 24:377–383. (In Chinese.) doi: 10.3321/j:issn:1000-8721.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 59.Wang G, Zhou F, Buchy P, Zuo T, Hu H, Liu J, Song Y, Ding H, Tsai C, Chen Z, Zhang L, Deubel V, Zhou P. 2014. DNA prime and virus-like particle boost from a single H5N1 strain elicits broadly neutralizing antibody responses against head region of H5 hemagglutinin. J Infect Dis 209:676–685. doi: 10.1093/infdis/jit414. [DOI] [PubMed] [Google Scholar]
- 60.Patterson DP, Rynda-Apple A, Harmsen AL, Harmsen AG, Douglas T. 2013. Biomimetic antigenic nanoparticles elicit controlled protective immune response to influenza. ACS Nano 7:3036–3044. doi: 10.1021/nn4006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, Huang B, Jiang T, Wang X, Qi X, Gao Y, Tan W, Ruan L. 2012. Robust immunity and heterologous protection against influenza in mice elicited by a novel recombinant NP-M2e fusion protein expressed in E. coli. PLoS One 7:e52488. doi: 10.1371/journal.pone.0052488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janssen R, Ervin J, Martin T. Clinical evaluation of N8295, a universal influenza A vaccine containing M2e and NP antigens conjugated to an oligonucleotide immunostimulatory sequence. In 7th WHO Meet Eval Pandemic Influenza Vaccines Clin Trials, 17–18 February 2011, Geneva World Health Organization, Geneva, Switzerland: http://www.who.int/immunization/research/meetings_workshops/flumtg_17_18Feb11/en/index1.html. [Google Scholar]
- 63.Donnelly JJ, Friedman A, Ulmer JB, Liu MA. 1997. Further protection against antigenic drift of influenza virus in a ferret model by DNA vaccination. Vaccine 15:865–868. doi: 10.1016/S0264-410X(96)00268-X. [DOI] [PubMed] [Google Scholar]
- 64.Goodman AG, Heinen PP, Guerra S, Vijayan A, Sorzano COS, Gomez CE, Esteban M. 2011. A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus. PLoS One 6:e25938. doi: 10.1371/journal.pone.0025938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butterfield LH, Economou JS, Gamblin TC, Geller DA. 2014. Alpha fetoprotein DNA prime and adenovirus boost immunization of two hepatocellular cancer patients. J Transl Med 12:86. doi: 10.1186/1479-5876-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarimo EA, Munseri P, Aboud S, Bakari M, Mhalu F, Sandstrom E. 2014. Experiences of social harm and changes in sexual practices among volunteers who had completed a phase I/II HIV vaccine trial employing HIV-1 DNA priming and HIV-1 MVA boosting in Dar es Salaam, Tanzania. PLoS One 9:e90938. doi: 10.1371/journal.pone.0090938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, Nabel GJ. 2010. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 68.Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, Bailer R, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS, VRC 306 Study Team . 2011. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomized clinical trials. Lancet Infect Dis 11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masopust D, Ha SJ, Vezys V, Ahmed R. 2006. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J Immunol 177:831–839. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- 70.Huang B, Wang W, Li R, Wang X, Jiang T, Qi X, Gao Y, Tan W, Ruan L. 2012. Influenza A virus nucleoprotein derived from Escherichia coli or recombinant vaccinia (Tiantan) virus elicits robust cross-protection in mice. Virol J 9:322. doi: 10.1186/1743-422X-9-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang W, Huang B, Jiang T, Wang X, Qi X, Tan W, Ruan L. 2014. Maximal immune response and cross protection by influenza virus nucleoprotein derived from E. coli using an optimized formulation. Virology 468–470:265–273. doi: 10.1016/j.virol.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 72.Reay PA, Jones IM, Gotch FM, McMichael AJ, Brownlee GG. 1989. Recognition of the PB1, neuraminidase, and matrix proteins of influenza virus A/NT/60/68 by cytotoxic T lymphocytes. Virology 170:477–485. doi: 10.1016/0042-6822(89)90439-X. [DOI] [PubMed] [Google Scholar]
- 73.Alexander J, Bilsel P, del Guercio MF, Stewart S, Marinkovic-Petrovic A, Southwood S, Crimi C, Vang L, Walker L, Ishioka G, Chitnis V, Sette A, Assarsson E, Hannaman D, Botten J, Newman MJ. 2010. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine 28:664–672. doi: 10.1016/j.vaccine.2009.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dégano P, Schneider J, Hannan CM, Gilbert SC, Hill AV. 1999. Gene gun intradermal DNA immunization followed by boosting with modified vaccinia virus Ankara: enhanced CD8+ T cell immunogenicity and protective efficacy in the influenza and malaria models. Vaccine 18:623–632. [DOI] [PubMed] [Google Scholar]
- 75.Konishi E, Kurane I, Mason PW, Shope RE, Ennis FA. 1997. Poxvirus-based Japanese encephalitis vaccine candidates induce JE virus-specific CD8+ cytotoxic T lymphocytes in mice. Virology 227:353–360. doi: 10.1006/viro.1996.8331. [DOI] [PubMed] [Google Scholar]
- 76.Powell TJ, Peng Y, Berthoud TK, Blais ME, Lillie PJ, Hill AV, Rowland-Jones SL, McMichael AJ, Gilbert SC, Dong T. 2013. Examination of influenza specific T cell responses after influenza virus challenge in individuals vaccinated with MVA-NP+M1 vaccine. PLoS One 8:e62778. doi: 10.1371/journal.pone.0062778. [DOI] [PMC free article] [PubMed] [Google Scholar]