Abstract
Mediator of DNA damage checkpoint protein 1 (MDC1) is essential for DNA damage response. However, the role of MDC1 in modulating gene transcription independently of DNA damage and the underlying mechanisms have not been fully defined. Androgen receptor (AR) is the central signaling pathway in prostate cancer (PCa) and its target genes are involved in both promotion and suppression of PCa. Here, we functionally identified MDC1 as a co-activator of AR. We demonstrate that MDC1 facilitates the association between AR and histone acetyltransferase GCN5, thereby increasing histone H3 acetylation level on cis-regulatory elements of AR target genes. MDC1 knockdown promotes PCa cells growth and migration. Moreover, depletion of MDC1 results in decreased expression of a subset of the endogenous androgen-induced target genes, including cell cycle negative regulator p21 and PCa metastasis inhibitor Vinculin, in AR positive PCa cell lines. Finally, the expression of MDC1 and p21 correlates negatively with aggressive phenotype of clinical PCa. These studies suggest that MDC1 as an epigenetic modifier regulates AR transcriptional activity and MDC1 may function as a tumor suppressor of PCa, and provide new insight into co-factor-AR-signaling pathway mechanism and a better understanding of the function of MDC1 on PCa.
INTRODUCTION
The androgen receptor (AR), a member of the nuclear receptor (NR) superfamily of ligand-dependent transcription factors, is required for the normal prostate growth and maintenance. It is well accepted that AR plays a crucial role in development of prostate cancer (PCa) as well as progression to castrate-resistant prostate cancer (CRPC) (1–3). The primary role of AR in PCa is believed to regulate expression of AR responsive genes that are essential for prostate tumorigenesis and progression. In addition to promoting PCa proliferation, androgen signaling through AR can also lead to apoptosis in PCa cells via inducing the expression of p21(WAF1/CIP1), a cyclin-dependent kinase inhibitor (4). Moreover, it is recently reported that AR-induced expression of cytoskeletal genes including Vinculin promote epithelial differentiation and inhibit metastasis (5). Therefore, identification of the detailed molecular mechanisms underlying the modulation of AR activity is essential for the development of novel pharmaceutical targets for PCa.
As a transcription factor, the protein structures of AR mainly contains activation function 1 (AF-1) and activation function 2 (AF-2). AF-1 functions in a ligand-independent manner, whereas activity of AF-2 needs cognate ligand binding. AR activity and specificity are controlled by specific co-regulator complexes (6) at multiple levels, including chromatin modifications involved in regulation of target gene transcription via the alteration of chromatin structure (7,8).
An increasing number of AR co-factors have been identified that they aberrantly expressed in PCa leading to a deregulated AR transcriptional network. Among them, AR co-activators including LSD1, p68, RNF6, JARID1B, ARD1 and FLH2 (9–14) become over-expressed in PCa suggesting their function on cancer cell proliferation. However, mounting evidence suggests that some of AR co-activators with reduced expression in PCa were involved in tumor suppression, including ART-27, ARA70, BRCA1, p44 and TBLR1 (4,15–18). On the other hand, HOXB13 or DACH1 acting as a co-repressor of AR induces growth suppression of PCa (19,20), while, it was recently proved that NR co-repressors including βArrestin2, HDAC, EZH2 or MTA1 play crucial roles in progression of PCa or breast cancer through inhibition of NR action (5,21,22). Thus alterations in epigenetic mechanism of AR co-factors in transcriptional regulation may influence the selective expression of AR target genes and thereby govern the tumor proliferation or suppression. The discovery of new co-regulators of steroid receptor will expand our knowledge of their actions.
MDC1/NFBD1 contains tandem BRCA1 C-terminal (BRCT) domains as well as a forkhead-associated domain and a repeat region, which mediate protein interaction. MDC1 is essential for DNA damage response (DDR) (23–25) and has an anti-apoptosis activity through the regulation of p53 (26). MDC1-null mice displayed some phenotypes including ionizing radiation (IR) sensitivity, male infertility, increase of tumor incidence, gross genomic instability and so on (27). However, the function of MDC1 in modulation of NR-induced transcription or PCa is still unknown and the mechanisms underlying the function have not been fully defined.
In previous study, we generated a Drosophila experimental system to isolate AR co-regulators involving in the modulation of AR-induced transcriptional activity via alteration of chromatin structure in vivo (8,28,29). USP22 was identified as a co-activator of AR through counteracting heterochromatin silencing (8). In the current studies, we functionally identified mutator protein (mu2) as a co-activator of AR with the Drosophila system and further investigated the role of MDC1, a human homolog of mu2 (30), in modulation of AR-mediated transactivation and PCa progression. Our studies reveal that MDC1 facilitates the association between AR and histone acetyltransferase (HAT) GCN5, thereby increasing histone H3 acetylation level on cis-regulatory elements of AR target genes. Downregulation of MDC1 expression promotes PCa cells growth and migration. In addition, knockdown MDC1 decreased a subset of androgen-induced target gene expressions. Accordingly, the expression of MDC1 and p21 identified as an AR target gene correlates negatively with aggressive phenotype of clinical PCa. These studies demonstrate a crucial role of MDC1 in regulating AR action and suppression of PCa progression. Our findings may provide new insight into co-factor-AR-signaling pathway mechanism and a better understanding of the function of MDC1 on PCa beyond maintenance of genomic integrity.
MATERIALS AND METHODS
Drosophila stocks, generation of transgenic flies and genetics
Drosophila stocks were maintained at 25°C on standard cornmeal sucrose-based media. The yw stain was used as wild-type in all experiments. ARAF-1-associated PEV experimental models were generated as previous reported (8). For production of UAS-mu2 expression construct, cDNA sequences were amplified by polymerase chain reaction (PCR) and subcloned into pCaSpeR3. A Flag tag was inserted at the N terminus of mu2 cDNA in pCaSpeR3 constructs. The UAS-mu2 expression construct was sent to EMBL Drosophila Injection Service for generation of transgenic flies. For expression of UAS-mu2 in salivary glands, UAS-mu2 transgenic fly lines were crossed with pnr-GAL4 driver lines. Other fly stocks were obtained from Bloomington Stock Center. Similar age flies were used for all comparisons.
To test the effect of mu-2 on ARAF-1-associated PEV experimental models, the male hemizygous for mutants (loss of function and gain of function) were crossed to ARAF-1-associated PEV female. The non-TM3 progeny possessing the mutant allele and mosaic red eye were picked up for determination of the effects of mutants on ARAF-1-associated PEV.
Fluorescent immunostaining, histology and eye pigmentation measurement
Immunostaining analysis of polytene chromosomes and eye disc histology analysis in Drosophila was performed as previously described (8,28). Immunostaining analysis are presented in the Supplementary ‘Materials and Methods’ section. To measure eye pigmentation, the heads of 40 female flies (2–3 days old; raised at 25°C) of each phenotype were homogenized in 1 ml of methanol (acidified with 0.1% HCl). Eye pigmentation was represented as the absorbance of the supernatant at 480 nm.
Plasmids
A Drosophila mu2 cDNA clone was produced by OPEN biosystems (Clone ID LD44171). Human MDC1 cDNA coding sequence was amplified by PCR using KIAA0170 plasmid from the HUGE database (generous gift from Dr T. Nagase) as a template. PCR products for MDC1 full length or several truncated mutants were cloned in a pcDNA3.1 (Invitrogen) derivate containing a sequence encoding a Flag epitope upstream of the cloning site, to generate pcFlag-MDC1.
Antibodies
The antibodies used in this study were: anti-Flag (M2 or rabbit, Sigma), anti-trimethyl H3-K9, anti-AcH3, anti-AcH4, anti-trimethyl H3-K27, anti-AcH4-K16, anti-AcH3-K9 (Upstate Biotechnology), anti-HP1 C1A9 (Developmental Studies Hybridoma Bank at the University of Iowa), anti-GCN5, anti-AR (N-20) (Santa Cruz Biotechnology), anti-MDC1 (Abcam), anti-MYST1 (Bethyl laboratories), anti-AR 441, anti-p21WAF1 Ab-11 (Thermo scientific), anti-GAPDH (Shanghai Kangchen).
Cell culture and treatment
The CWR22Rv1, LNCaP and DU145 cells were maintained in RPMI1640 (GIBCO-BRL). The PC3 cell was maintained in F12. HEK293T cells were maintained in Dulbercco's modified Eagle's medium. All the culture media were supplemented with 10% fetal calf serum (FBS), 2 mM glutamine, 100 units/ml streptomycin and penicillin. Luciferase assay was conducted as previously described (28,31) and presented in Supplementary Materials and Methods.
siRNA transfection, lentiviral production and infection
siRNA against MDC1 and a control siRNA (Ambion) were transfected in CWR22Rv1 cells using Lipofectamine™ 2000 (Invitrogen) following the manufacturer's instructions. Sequence of siMDC1: 5′-GUCUCCCAGAAGACAGUGAdTdT-3′ (32). Sequence of siAR: 5′- GACCUACCGAGGAGCUUUCdTdT-3′ (33). Further experiments were performed with the cells followed by incubation for 24–48 h to allow degradation of targeted mRNA.
For lentiviral production and infection, control shRNA (shCtrl) lentivirus and shRNA against MDC1 (shMDC1) lentivirus targeting the same sequence as siMDC1 as above were purchased from Shanghai GeneChem Company. MDC1-stably-silencing and control cell lines were selected with puromycin (2 μg/ml) after lentivirus infection.
RNA isolation, reverse transcription and quantitative real-time PCR
Total RNA was isolated using the Trizol reagent (Invitrogen). Reverse Transcription was performed using SuperScriptII (Invitrogen) and random hexamers according to the manuscription's instructions. cDNAs were quantified by quantitative real-time PCR using SYBR Premix Ex Taq (TaKaRa) on a Mx3000P instrument (Agilent StrataGene). Primers used to detect mRNA expression were shown in Supplemental Table S1. Gene expression levels were calculated relative to the housekeeping gene b-Actin using Stratagene Mx3000P software.
GST pull-down, immunoprecipitation and western blot analysis
In vitro GST pull-down have been described previously (34). For immunoprecipitation experiments, the Flag-MDC1 and its truncated mutations expression plasmids were transiently transfected into HEK293T and LNCaP cells using lipofectamine (Invitrogen) and whole cell extracts were prepared 48 h after transfection as described and equal protein amounts were immunoprecipitated with anti-Flag M2 resin. The immunoprecipitated protein complexes were washed three times with IP buffer (25 mM Tris–HCl, pH8.0; 10% glycerol; 0.1% NP40; 0.5 mM DL-Dithiothreitol (DTT); 5mM MgCl2 and protease inhibitor) containing 0.5M KCl and twice with IP buffer containing 100 mM KCl. The crude extracts and immune complexes were analyzed by western blotting using the indicated primary antibodies and chemiluminescence detection was performed according to manufacturer's instructions (GE-Healthcare). The protein concentration of the whole cell extracts was determined using standard Bradford assays.
ChIP and ChIP re-ChIP
ChIP procedures were described in Supplementary Materials and Methods. The primers used in PCR or real-time PCR were shown in Supplemental Table S2. ChIP re-ChIP experiments were performed essentially as described previously (28). Complexes were eluted from the primary IP by incubation with 10 mM DTT at 37°C for 30 min and diluted 1:50 in buffer (1% Triton X-100, 2 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 20 mM Tris–HCl [pH 8.1]) followed by re-ChIP with the antibodies as indicated.
Cell proliferation and migration assay
CWR22Rv1 or LNCaP cells with stably knockdown of MDC1 or control cells were plated in 96-well plates (2500 cells per well) and incubated in the absence or presence of dihydrotestosterone (DHT). DU145 cells with stably knockdown of MDC1 or control cells were plated in 96-well plates (1000 cells per well). And cell number was assayed at various times using CCK8 assay by absorbance at 450 nm.
Cell-migration assays were carried out using Boyden chambers as described (21). Cells were plated on the upper well of a Boyden chamber at a concentration of 5 × 104 cells per well in 100 μl serum-free culture medium, the lower compartments were filled with 600 μl culture medium containing 10% serum. After incubating at 37°C for indicated time, non-invaded cells were removed from the upper surface of the filter with a cotton swab and the invaded cells on the lower surface of the filter were fixed, stained and photographed.
Immunohistochemical analysis of prostate cancer and testis tissues
Formalin-fixed paraffin-embedded sections of prostate tissue specimens were prepared from clinical prostatectomy specimens in the first hospital of China Medical University. Multicentre ethical approval for data collection and tissue use was granted by the Human Research Ethics Committee of the above hospital. A total of 131 tissues, including 33 BPH tissues and 98 PCa tissues, were collected to perform immunohistochemical staining. For detecting MDC1 expression levels in prostate tissues, anti-rabbit polyclonal MDC1 antibody (abcam, ab11169, 1:100) and avidin-biotin-conjugated second antibodies were used. The signals were visualized with diaminobenzidine and the nuclei were counterstained with hematoxylin as previously described (34).
Statistics
All statistical analysis was performed using the SPSS (17.0) statistical software program. For real-time PCR and luciferase assay, two-sided Student's t-test was used to determinate the significant difference. For immunohistochemistry, Mann–Whitney U test was used to determinate the significant difference between benign prostate hyperplasia (BPH) group and Gleason score (GS) <7 or between BPH group and GS ≥ 7 group.
RESULTS
Functional isolation of mutator2 (mu2) as an androgen receptor co-activator in Drosophila
The transactivation function of NRs is mediated by a number of co-regulators and co-regulator complexes (35,36). Some fly homologs of human NR co-regulators are functionally conserved across species and share properties such as histone modifying enzyme activity (37). To isolate new co-regulators involved in modulation of AR-induced transactivation, we previously developed a novel AR-associated position effect variegation (AR-PEV) model in Drosophila (8,28). In this system, the AR or a truncated AR harboring the ligand-independent AF-1 domain (ARAF-1, 1–720 aa of AR) is expressed in the Drosophila eye using the glass multimer reporter (GMR) gene promoter (Figure 1A). A reporter construct, which contains the white gene and a gene encoding the green fluorescent protein (GFP) controlled by eight androgen responsive elements (AREs), was inserted into a heterochromatic region leading to a mosaic red eye phenotype (Figure 1A and C). The white gene mRNA expression was proved by RT-PCR experiments (Figure 1B). Three AR-PEV experimental models were generated and inverse PCR was performed to verify the localization of chromosomal locus of the reporter gene in each AR-PEV model (Figure 1C). Among them, the model containing the reporter construct translocated in pericentric region (80C2, C3L) was used for further screening. In this system, the AR counteracted silencing by heterochromatin spreading in a ligand-dependent manner (Figure 1C). The ligand-independent ARAF-1 led to higher transactivation of the reporter genes and was hereafter used to analyze the AR-dependent transcriptional activation. Consequently, we genetically screened the co-regulators to identify those modulating AR function in Drosophila. Two different mutations of mutator2 (mu21 and Df(3L)ED4284) gave significant decrease in the pigment area in this AR-dependent PEV model (Figure 1D).
Figure 1.
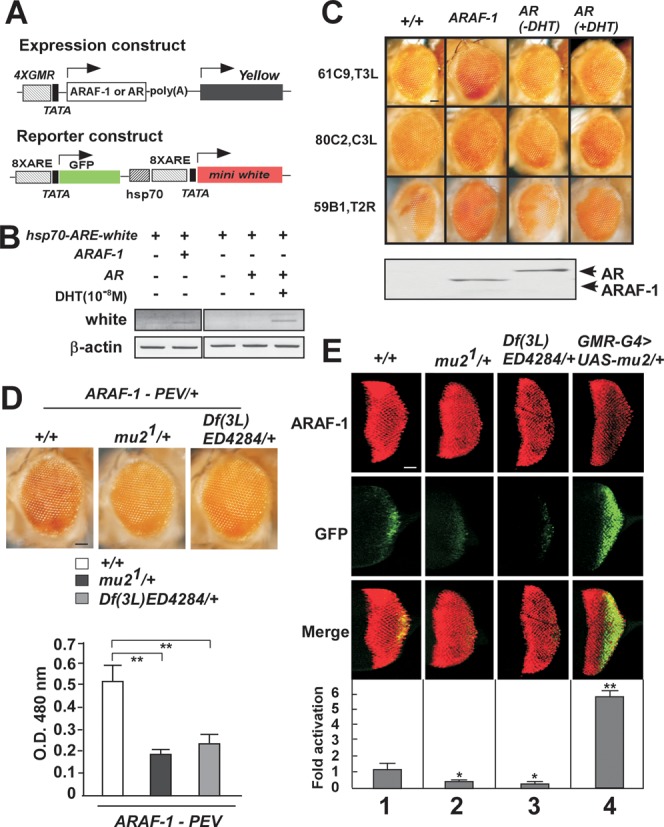
Drosophila mu2 enhances ARAF-1-induced transactivation. (A) Schematic representation of the expression and reporter constructs in ARAF-1-PEV fly lines as previous reported. The expression constructs include the human androgen receptor (AR) or ARAF-1 driven by four copies of GMR binding sites. The reporter construct harbors GFP and white reporter genes individually controlled by the hsp70 promoter in which eight AREs were introduced. (B) The expression and reporter constructs were co-transfected into S2 cells, mRNA transcription levels of white gene were detected by RT-PCR. (C) Eye phenotypes of three kinds of fly lines for AR-PEV experimental models. Cytogenetic localization of the reporter gene carrying hsp70-ARE-white in Drosophila chromosome was analyzed by inverse PCR. Scale bar, 100 μm. (D) Two different fly lines carrying mu2 loss of function (Mu21/+) or difficiency line including mu2 gene location (Df[3L]ED4284) or wild-type flies (+/+) were crossed with ARAF-1-PEV flies. Modification of PEV was analyzed by areas of eye pigmentation in the progeny (upper panels) and by optical density (OD) measurement at 480nm (lower panels). Average values of more than three independent measurements are shown with SD. Scale bar, 100 μm. **, P < 0.01. (E) Flies expression ARAF-1 in the eye with a GMR-GAL4 driver and carrying an ARE-GFP reporter in pericentric heterochromatin were crossed with lines harboring mu2 loss of function (mu21, Df[3L]ED4284) or gain of function (UAS-mu2) mutants as indicated. Expression of ARAF-1 was assessed by immunostaining with an anti-AR antibody (upper panels). The effect of mu2 mutations and overexpression on ARAF-1-induced transactivation was assessed by examination GFP expression (middle panels). Merge images were shown in lower panels. Scale bar, 100 μm. *, P < 0.05; **, P < 0.01.
We further analyzed the effects of mutator2 (mu2) on AR-dependent gene activation in vivo by testing ARAF-1-mediated transactivation on the GFP reporter gene expression system. In order to be able to detect GFP expression in the eye disc of flies, we used additional transgenic lines expressing ARAF-1 in the Drosophila eye with a GMR-GAL4 driver in a GAL4-UAS system. The ARE-GFP reporter was inserted in euchromatic region and further mobilized into pericentric heterochromatin leading to variegated expression of GFP reporter (28). In agreement with our results obtained in the above ARAF-1-PEV model, this reporter system confirmed again that mu2 loss-of-function mutations, mu21 and Df(3L)ED4284, significantly reduced ARAF-1 transactivation of the reporter transgene inserted in pericentric heterochromatin (Figure 1E, lane 1, 2 and 3). In contrast, but consistent with the effects of mu2 loss-of-function mutations, ectopic expression of mu2 (UAS-mu2) dominantly increased GFP transactivation by ARAF-1 (Figure 1E, lane 1 and 4).
We then examined the Flag-tagged mu2 localization on polytene chromosome. In good agreement with previous report (30), mu2 was strongly stained at centromere (Supplementary Figure S1A). Besides centromere, we observed that mu2 also accumulated in euchromatic regions. These results suggested mu2 may have roles in both heterochromatin and euchromatin. Moreover, compared with the ectopic expression of mu2, we observed that heterochromatin protein 1 (HP1) and H3K27met3, which were used as the biomarkers of the centromere, were not able to accumulate in centromere in the fly containing loss-of-function mu2 mutation (mu21) (Supplementary Figure S1B). The results suggested that mu2 might play an important role in maintenance of heterochromatin and chromosome integrity in centromere; on the other hand, mu2 might be involved in the active transcription in euchromatin.
Taken together, these results indicated that mu2 is an AR co-activator in Drosophila.
MDC1 physically associates with AR in mammalian cells
To study the function of mu2 on AR-induced transactivation in mammalian cells, we first generated expression plasmids encoding mediator of DNA damage checkpoint 1 (MDC1), which is the human homolog of mu2 and its truncated mutants with different domains (Figure 2A). We then test the association between MDC1 and AR by Co-immunoprecipitation (Co-IP). HEK293T cells were co-transfected with AR and Flag-tagged MDC1 expression plasmids, results shown in Figure 2B indicated that exogenous AR associates with Flag-tagged MDC1 and the association between two proteins was stronger in the presence of DHT (Figure 2B). We then ask whether endogenous MDC1 interactions with AR, to this end, we turned to CWR22Rv1 PCa cells in which full-length (FL) AR and COOH-terminal truncated AR splice variants are constitutively co-expressed and performed co-IP experiments. Results shown in Figure 2C demonstrated that MDC1 associated with AR FL and AR variants, and the interaction between MDC1 and AR FL was stronger in the presence of DHT, whereas the interaction between MDC1 and AR variants was mildly influenced by DHT treatment (Figure 2C). Furthermore, the results of immunofluoresence experiments showed that MDC1 compartments were distributed in the nucleus and AR was mainly compartmentalized in the nucleus in the presence of DHT (Figure 2D). RNA polymerase II, which is required for the selective initiation of transcription, was used as a positive control (Figure 2E). Previous reports showed that MDC1 knockout mice displayed many phenotypes including male infertility (27,38), we thus examine the distribution of MDC1 or AR in human testis tissue, the representative image shown in Figure 2F and Supplementary Figure S2 demonstrated that MDC1 and AR were mainly stained in Leydig cells.
Figure 2.
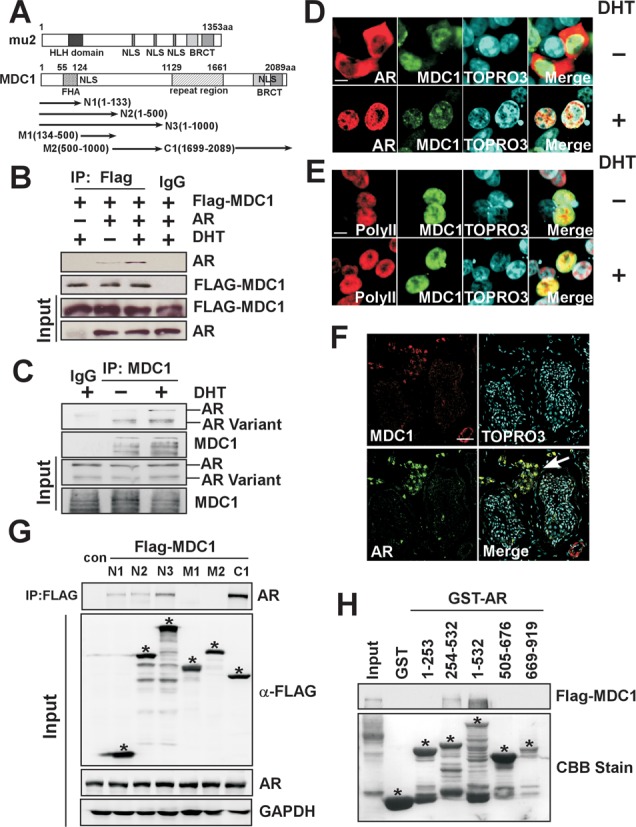
Interaction between MDC1 and AR. (A) Schemetic representation of Drosophila mu2, its ortholog in human (MDC1) and the truncated mutations of MDC1 (N1, N2, N3, M1, M2 and C1). HLH domain, NLS and BRCT domain; FHA domain, NLS, repeat sequence and BRCT domain were individually positioned in the mu2 and MDC1 proteins. (B) Exogenous MDC1 associates with AR invivo. HEK293T cells co-transfected with Flag-MDC1 and AR expression plasmids were immunoprecipitated using anti-Flag antibody or IgG as a control with or without DHT treatment for 48 h. Precipitated proteins were examined by immunoblotting using antibody against AR N-20. A fraction (5%) of the input cell lysate before immunoprecipitation was loaded as a control. (C) Endogenous MDC1 interacts with AR and N-terminal AR variants in CWR22Rv1 cells. With or without DHT treatment for 24 h, CWR22Rv1 cells were collected and cell lysates were immunoprecipitated with MDC1 antibodies or IgG. Precipitates were analyzed by western blot using the indicated antibodies. (D and E) Confocal fluorescence analysis of subcellular distribution of MDC1 and AR. HEK293T cells were co-transfected with plasmids expressing Flag-MDC1 and AR in the absence or presence of DHT (10−8 M) and then were stained with TOPRO3 to visualize the nucleus (blue), Flag antibody (green), AR (N-20) antibody (red in D) or RNA polymerse II antibody (red in E). Merged images were shown as indicated. Scale bar, 5 μm. (F) Distribution of the endogenous MDC1 and AR in normal human testis. Immunofluorescence analyze of normal testis section was shown. The normal testis tissue was stained with TOPRO3 to visualize DNA (Blue), anti-AR (N-20) (green) and anti-MDC1 (red) antibodies. The arrow in the merged image showed the co-compartmentalization of AR and MDC1. Scale bar, 100 μm. (G) The truncated mutants of MDC1 associate with the endogenous AR. Immunoprecipitation was performed with the indicated LNCaP cell lysates using an anti-Flag antibody. Precipitated proteins and the input of cell lysates were analyzed by immunoblotting with the indicated antibodies. * was placed to indicate the position of the specific FLAG-MDC1 truncated mutant proteins. (H) Identification of binding domains in AR for MDC1 interaction. In GST pull-down experiments, Flag-MDC1 protein synthesized by transcription and translation Kit invitro was incubated with GST and GST-AR deletion mutants as indicated. Bound proteins were analyzed by immunoblotting using anti-Flag antibody and equal loading of GST-AR deletion mutants was assessed by coomassie brilliant blue staining. * was placed to indicate the position of the specific GST-AR deletion mutant proteins.
Having demonstrated association between MDC1 and AR, we sought to identify the interaction domains in MDC1 and AR, to this end, LNCaP cells were transfected with expression plasmids encoding MDC1 truncated mutants for co-IP (Figure 2G). Our results showed that AR was strongly precipitated with Flag-tagged MDC1 N3 (1–1000 aa) and C1 (1699–2089 aa), weakly precipitated with N1 (1–133 aa) and N2 (1–500 aa), but not with M1 (134–500 aa) or M2 (500–1000 aa), suggesting that the N1 comprising 1–133 aa or C-terminal fragment comprising 1699–2089 aa mediates AR interaction with MDC1. In addition, the interaction between N3 (1–1000 aa) and AR is stronger than that between N1 (1–133 aa) and AR, suggesting that 500–1000 aa fragment of MDC1 may facilitate MDC1–AR interaction. Meanwhile, different interacting pattern between MDC1 and NBS1 was detected when NBS1, which has been reported to interact with 210–460 aa in N-terminus of MDC1 (39), was subjected to parallel analysis (Supplementary Figure S3). Moreover, GST pull-down experiments were performed with several GST-AR fragments and Flag-MDC1 as shown in Figure 2H, our results demonstrated that the 254–532 aa of AR was responsible for interaction between MDC1 and AR (Figure 2H). Based on the above results, we conclude that MDC1 physically associates with AR in vitro and in vivo.
MDC1 co-activates AR and AR variants-mediated transactivation in human cells
To investigate whether MDC1 is able to enhance AR-induced transactivation in human cells, a series of luciferase assays were performed in human cells. Our results demonstrated that in LNCaP cells, MDC1 full length (FL), MDC1 N3 and C1, but not MDC1 N1 and N2, significantly enhanced AR-mediated transactivation in the presence of DHT (Figure 3A), suggesting that MDC1 co-activates AR-mediated transactivation in a ligand-dependent manner and its functional co-activation domains are located in N-terminal 500–1000 aa fragment or 1699–2089 aa at the C-terminus. In CWR22Rv1 cells which constitutively carry AR FL and ligand-independent AR variants, knockdown of MDC1 repressed AR-mediated transactivation with or without DHT treatment, suggesting MDC1 may participate in upregulation of AR FL or AR variants-induced transactivation (Figure 3B). To confirm these results, we further separately detected effects of MDC1 on the truncated AR harboring the ligand-independent AF-1 domain (ARAF-1) or ligand-dependent AF-2 domain (ARAF-2). The results demonstrated that ARAF-1-mediated transactivation was constitutively enhanced by MDC1 in an androgen-independent manner (Figure 3C). And the transactivation function of ARAF-2 was enhanced by MDC1 in the presence of DHT (Figure 3D). Moreover, the transactivational activity of AR-V7, which is well-studied N-terminal variant of AR, was enhanced by MDC1 without DHT (Supplementary Figure S4). These data indicated that MDC1 is involved in regulation of AR FL and AR variant functions. In addition, as shown in Figure 3E, MDC1 enhances not only AR, but also ERα and GR-induced transactivation in a ligand-dependent manner, suggesting that MDC1 may be involved in up-regulation of a series of NRs-mediated functions.
Figure 3.
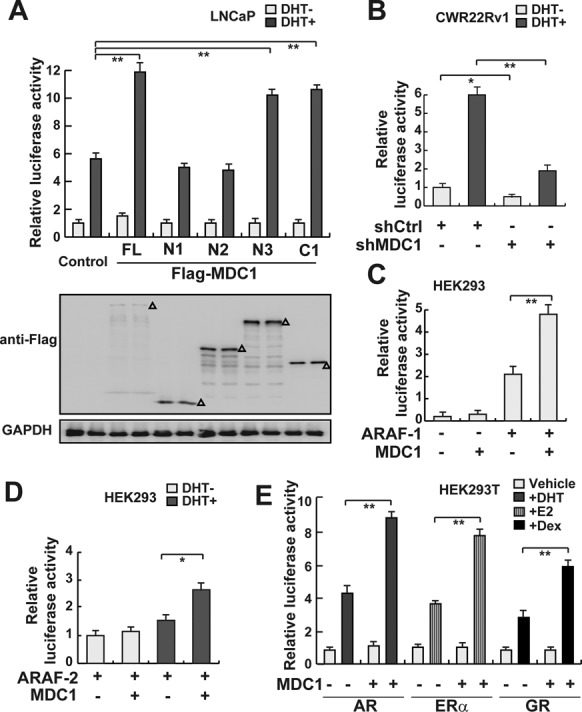
MDC1 co-activates AR-mediated transactivation. (A) MDC1 enhances AR-mediated transactivation and both 500–1000 aa fragment and 1699–2089 aa fragment are required for its function. LNCaP cells were co-transfected with ARE-luc and pRL-TK, together with the indicated expression plasmids in the absence or presence of DHT. The expression of MDC1 truncated mutations were evaluated by western blot as indicated by Δ. (B) MDC1 knockdown represses AR-mediated transactivation in CWR22Rv1 cells. CWR22Rv1 cells with knockdown of MDC1 by shRNA were co-transfected with ARE-luc and pRL-TK in the absence or presence of DHT. (C) MDC1 enhances ARAF-1-mediated transactivation. HEK293 cells were co-transfected with ARE-luc and pRL-TK, together with the indicated expression plasmids in the absence or presence of DHT. (D) MDC1 enhances ARAF-2-mediated transactivation in the presence of DHT. HEK293 cells were co-transfected with ARE-luc and pRL-TK, together with the indicated expression plasmids in the absence or presence of DHT. (E) MDC1 enhances AR, ERα or GR-mediated transactivation. HEK293T Cells were co-transfected with plasmids expressing MDC1 and pRL-TK, together with AR, ERα or GR expression plasmids and their reporter gene plasmids in the absence (white bars) or presence (shadow bars) of relevant ligands. After 24 h of ligand treatment, cells were collected and assayed for luciferase activity. Relative luciferase units shown are the mean value at least three times. In A–E, error bars represent mean ± SD. *P < 0.05; **P < 0.01.
MDC1 depletion impairs GCN5 recruitment to androgen responsive elements of AR target genes
In order to determine MDC1 is recruited to the putative cis-regulatory elements (androgen responsive elements, AREs), which are in PSA promoter (AREI/II), PSA enhancer (AREIII) and KLK2 promoter, and further to gain some insights into the epigenetic mechanism through which MDC1 regulates AR-induced transcriptional activity, chromatin immunoprecipitation (ChIP) assay experiments were performed with siRNA against MDC1 to examine whether MDC1 influences the level of histone modifications at the cis-regulatory elements of AR target genes in CWR22Rv1 cells. It has been shown that MYST1 (MOF), which is a specific HAT for the acetylation of histone H4 at lysine 16 (H4K16Ac), modulates the recruitment of MDC1 to play an important role in DNA damage repair (40). We therefore tested H4K16Ac, H4Ac, H3Ac, H3K9Ac and H3K9Met3 levels at AREs, and found that MDC1 influenced H3K9Ac and H3Ac at AREI/II in PSA promoter and KLK2 promoter, whereas no obvious changes were observed in H4Ac and H4K16Ac. MDC1 knockdown did not alter the recruitment of MYST1, either. We then asked whether MDC1 influences the recruitment of GCN5, which was reported as a specific HAT for the acetylation of H3K9, to cis-regulatory elements of AR target genes. Our data from ChIP assay experiments demonstrated that MDC1 depletion reduced the recruitment of GCN5 to ARE I/II in PSA promoter and KLK2 promoter (Supplementary Figure S5A and S5B). On ARE III in PSA enhancer, MDC1 had mild effects on the acetylation level of H3Ac, H3K9Ac and the recruitment of GCN5 (Supplementary Figure S5A).
In addition, to further confirm the above results, we repeated the ChIP experiments using real-time qPCR in MDC1 knockdown cells induced by lentivirus infection. The results showed that MDC1 knockdown led to a significant decrease of H3K9Ac and H3Ac at AREI/II in PSA promoter, whereas a slight decrease at AREIII in PSA enhancer (Figure 4A and B). Moreover, the recruitment of GCN5 to AREI/II was obviously impaired by MDC1 knockdown, and the recruitment of GCN5 to AREIII was slightly impaired. No significant association of AR and MDC1 with chromatin at GAPDH promoter could be detected, further demonstrating the specificity (Figure 4A and B). However, depletion of MDC1 did not influence the recruitment of AR. Taken together, these results indicated that MDC1 facilitates the recruitment of HAT GCN5 to cis-regulatory elements of AR target genes for co-activating AR-mediated transactivation.
Figure 4.
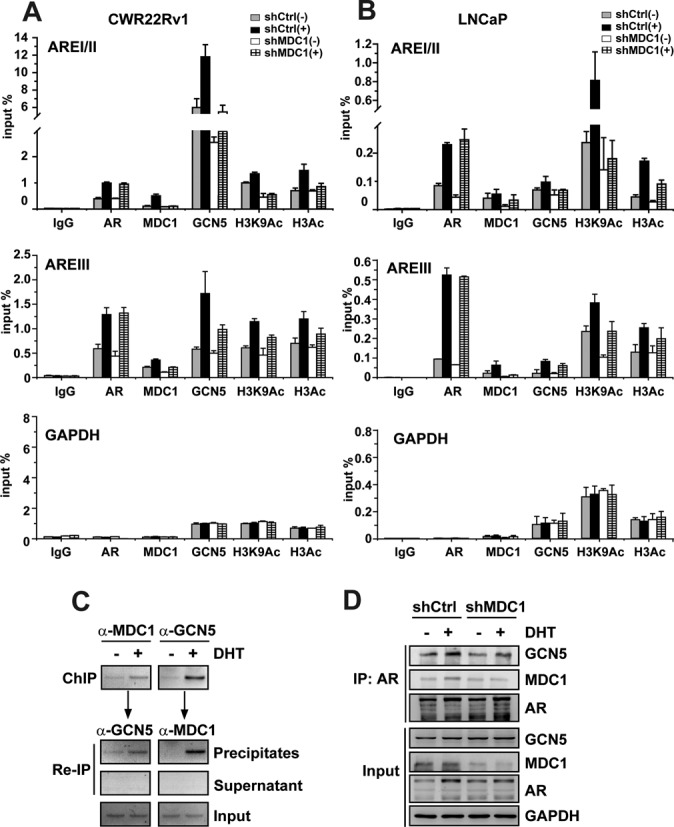
Down-expression of MDC1 attenuates GCN5 recruitment to cis-regulatory elements of AR target genes. (A and B) MDC1 knockdown reduces the epigenetic modification and GCN5 recruitment at androgen responsive elements (AREI/II and AREIII) of AR target genes. CWR22Rv1 or LNCaP cells with knockdown of MDC1 by shRNA were incubated with or without DHT for 4 h and were subject to ChIP assay with antibodies indicated. The precipitated androgen responsive elements of PSA were normalized to input DNA signal as a percentage. Data represents the mean values (±SD) of triplicate real-time PCR. IgG was used as a nonspecific control ChIP. GAPDH promoter regions were used as negative control. (C) MDC1 and GCN5 are predominantly recruited to cis-regulatory elements of KLK2 in the presence of DHT. ChIP/re-ChIP experiments were performed using specific antibodies against MDC1 and GCN5 as indicated. (D) MDC1 knockdown reduces the GCN5 recruitment to AR. CWR22Rv1 cells were immunoprecipitated using anti-AR antibody. Precipitated protein complex were immunoblotting using anti-MDC1, anti-GCN5 or anti-AR antibodies to detect the endogenous proteins. A fraction of the input cell lysate before immunoprecipitation was loaded as a control.
Association of MDC1 with AR and GCN5, and they are predominantly recruited to cis-regulatory elements of AR target genes upon ligand induction
Having established that MDC1 depletion impairs the recruitment of GCN5 to cis-regulatory elements of AR target genes, we further investigated whether MDC1, GCN5 and AR form a complex recruited to AR target genes. Double consecutive ChIP assays were performed in CWR22Rv1 cells. ChIP assays were first performed with antibodies against MDC1 or GCN5 in the soluble chromatin derived from CWR22Rv1 cells in the presence or absence of DHT (Figure 4C, upper panel). Then, the precipitates or supernatants from ChIP were individually subjected to re-IP with antibodies against a second protein. As shown in Figure 4C, the presence of MDC1 and GCN5 at cis-regulatory elements of KLK2 upon ligand induction was detected, suggesting that MDC1 and GCN5 acted in a combinatorial fashion on cis-regulatory elements of KLK2.
In order to further examine the physical association of MDC1 with GCN5 and AR, Co-IP experiments were performed. Without knockdown of MDC1, GCN5 protein was detected in AR precipitate, suggesting that MDC1, GCN5 and AR form a complex in vivo (Figure 4D, lanes 1 and 2). On the contrary, when MDC1 protein was knockdown by shRNA against MDC1, the association of GCN5 with AR was decreased (Figure 4D, lanes 3 and 4). In addition, we detected that GCN5 directly interacted with the 254–532 aa of AR by GST pull-down (Supplementary Figure S7A). And GCN5 associated with the N-terminal 1–500 aa fragment of MDC1 (Supplementary Figure S7B). These results suggest that MDC1 may act as a scaffolding protein for facilitating the association between GCN5 and AR.
We then set out to investigate whether the effects of MDC1 and GCN5 on AR action are additive or synergistic. As shown in Supplementary Figure S6, MDC1 or GCN5 separately enhanced AR-induced transactivation by 1.8–2-folds, co-transfection of MDC1 and GCN5 increased AR function by about four-folds, indicating that MDC1 and GCN5 additively enhanced AR-mediated transactivation. Taken together, it appears that MDC1 forms an AR co-activator complex with GCN5 to the promoter of AR target genes for enhancing AR action.
The function of MDC1 on suppression of cell growth and migration of PCa is related to AR
To determine a physiological role of MDC1 in PCa cells, we analyzed the impact of MDC1 on the growth and motility characteristics of PCa cells with shRNA against MDC1 in CWR22Rv1, LNCaP and DU145 PCa cells. Growth curve analyses showed that MDC1 knockdown promoted the cell proliferation in AR-positive PCa cells, CWR22Rv1 and LNCaP, especially in the presence of DHT (Figure 5A and B). In contrast, mild effect was seen in AR-negative PCa cells DU145 (Figure 5C). In addition, to examine the potential effect of MDC1 on the migration of PCa cells, transwell experiments were performed using Boyden chamber and similar effects were observed. Control shRNA showed low migration, however shMDC1 exhibited a significantly increased migration especially with the treatment of DHT in CWR22Rv1 and LNCaP cells (Figure 5D and E). But the migration of DU145 cells was not affected by shMDC1 (Figure 5F).
Figure 5.
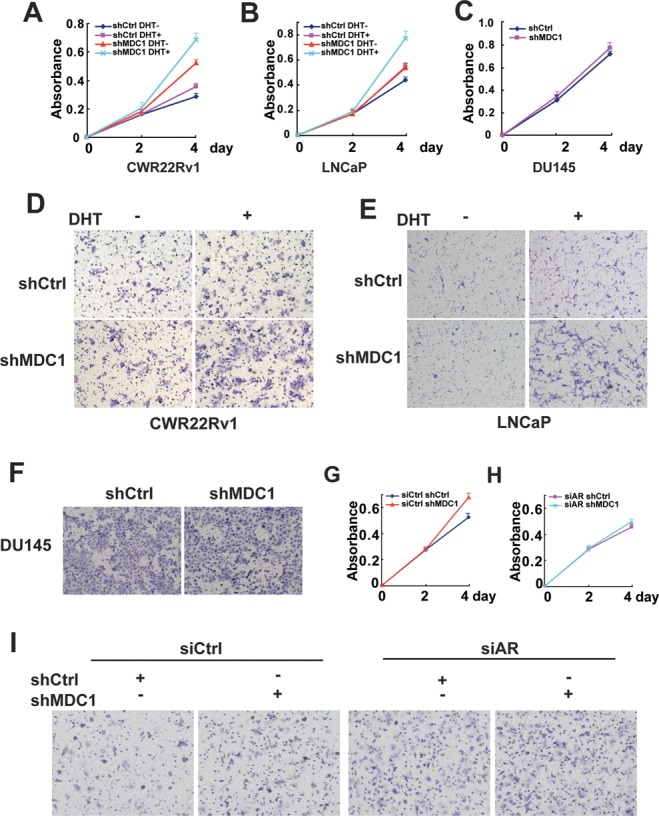
Knockdown MDC1 promotes prostate cancer cell growth and migration. (A and B) Knockdown of MDC1 promotes cell proliferation of CWR22Rv1 and LNCaP. (C) Knockdown of MDC1 mildly promotes cell proliferation DU145. In A–C, absorbance at 450 nm was plotted. Points, mean of three replicates; bars, SD. (D, E and F) Knockdown of MDC1 promotes migration of CWR22Rv1, but not DU145. In D and E, CWR22Rv1 or LNCaP cells with stably knockdown of MDC1 or control cells were plated into transwell chamber by DHT treatment or not, and detected after 20 or 40 h. DU145 cells with stably knockdown of MDC1 or control cells were plated into transwell chamber and detected after 16 h. Scale bar, 100 μm. (G and H) Enhanced proliferation of CWR22Rv1 cells by MDC1 knockdown was impaired by AR knockdown. CWR22Rv1 with stably knockdown of MDC1 or control cells were transfected with control siRNA (in G) and siAR (in H). After 24 h, cells were plated in 96-well plates and incubated in the presence of DHT. (I) Enhanced migration of CWR22Rv1 cells by MDC1 knockdown was impaired by AR knockdown. CWR22Rv1 with stably knockdown of MDC1 or control cells were transfected with control siRNA and siAR in the presence of DHT. After 48 h, cells were plated into transwell chambers and detected after 20 h.
We then ask whether AR is required for MDC1 functions in PCa, to this end, we turned to CWR22Rv1 PCa cells and performed cell proliferation and migration experiments after downregulation of AR by siRNA with or without MDC1 depletion. The silencing efficiency of AR was evaluated (Supplementary Figure S8). In the presence of DHT, MDC1 depletion promoted PCa cell proliferation and migration (Figure 5G and I left), downregulation of AR attenuated PCa cell proliferation promoted by MDC1 knockdown (Figure 5H). In addition, PCa cell migration increased by MDC1 depletion was almost totally impaired by AR silencing (Figure 5I). Taken together, these results suggested that suppression of growth and migration of PCa cells induced by MDC1 is related to AR.
MDC1 participates in positive modulation of AR-induced target genes transcription
The target genes of AR are involved in diverse cellular processes including cell cycle progression, tumor suppression, differentiation, metastasis and so on. To further investigate the potential contribution of MDC1 in regulation of AR target genes, we turned to LNCaP and CWR22Rv1 cell lines in which the AR is constitutively expressed and analyzed the impact of MDC1 on androgen-induced expression of 17 putative AR target genes with different biological functions. We identified nine genes that were significantly decreased in LNCaP cells and eight genes in CWR22Rv1 cells by MDC1 knockdown. MDC1 knockdown significantly suppressed androgen-induced mRNA expression of AR target genes, including Vinculin, p21, NKX3.1, PMEPA1, FKBP5, TMPRSS2, PSA, SLC45H31 and JAG1, while not affecting HUS1, KRT18, FASN, KLK4, ITGAV, BMPRIB, B4GALT1 and ALDH1A3, in LNCaP cells (Figure 6A, upper). And induction folds of Vinculin, p21, NKX3.1, KRT18, TMPRSS2, PSA, SLC45H31 and JAG1 mRNA expression by DHT were significantly reduced in CWR22Rv1 cells with MDC1 knockdown, while HUS1, PMEPA1, FKBP5, FASN, KLK4, ITGAV, BMPRIB, B4GALT1 and ALDH1A3 were not affected (Figure 6A, lower). Two of above AR target genes, p21 that inhibits cell growth and Vinculin that promotes epithelial differentiation and inhibits metastasis, were further confirmed at protein level. Western blot analysis of the endogenous protein expression in CWR22Rv1 cells confirmed that DHT-AR was able to induce the expression of p21 and Vinculin (Figure 6B). Meanwhile, shMDC1, which led to an 80% reduction of MDC1 protein levels, decreased the expression of p21 or Vinculin enhanced by DHT treatment. In good agreement, ectopic expression of MDC1 in CWR22Rv1 cells was able to enhance the expression of p21 and Vinculin in a dose-dependent manner (Supplementary Figure S9). In contrast, the expression of p21 or Vinculin has not been reduced by shMDC1 in AR-negative PCa cell lines, including DU145 cells and PC3 cells (Figure 6B). These results suggested that MDC1 is selectively involved in a subset of endogenous androgen-regulated genes transactivation in vivo.
Figure 6.
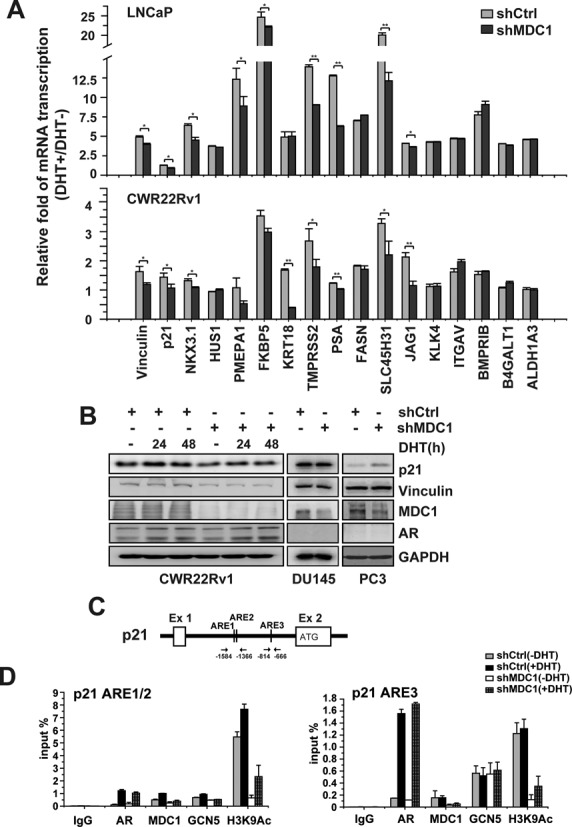
Knockdown MDC1 inhibits androgen induction of AR target genes. (A) Real-time PCR analysis showing the effect of MDC1 knockdown on DHT-dependent activation of 17 AR target genes. LNCaP or CWR22Rv1 cells were infected with either control shRNA lentivirus (shCtrl) or shRNA against MDC1 lentivirus (shMDC1), and treated with or without ligand (DHT). The cells were collected for RNA isolation after DHT treatment for 24 h. Induction of mRNA expression was expressed as the ratio of target gene mRNA levels normalized to β-Actin levels between cells treated and untreated DHT. *P < 0.05; **P < 0.01. (B) MDC1 knockdown diminishes DHT induced p21 and Vinculin expression in CWR22Rv1 cells. Western blot analysis in the whole cell lysates of CWR22RV1, DU145 or PC3 cells with shMDC1 or control shCtrl as indicated. (C) Schemetic representation of putative AREs in p21. (D) MDC1 affects the epigenetic changes and GCN5 recruitment at putative AREs of p21. CWR22Rv1 cells with knockdown of MDC1 by shRNA were incubated with or without DHT for 4 h and were subject to ChIP assay with antibodies indicated. The precipitated chromatin was normalized to input DNA signal as a percentage. Data represents the mean values (±SD) of triplicate real-time PCR. The precipitated chromatin was amplified by real-time qPCR using primers flanking p21 ARE1/2 (−1584 to −1366 bp) and p21 ARE3 (−814 to −666 bp).
To assess the epigenetic changes and GCN5 recruitment upon MDC1 manipulation at endogenous target genes of AR, we focused on p21. According to the previous report (41,42), bioinformatics analysis showed that there are three putative AR binding sites in Intron1–2 of p21, which are consisting of a 6 bp sequence resembling the consensus half site of an ARE (5′-TGTYCT-3′) or the same sequence as a half ARE (5′-TGTYCT-3′). We named them p21 ARE1 (at position −1512 to −1507 upstream from start codon, 5′-TGTCCC-3′), p21 ARE2 (at position −1444 to −1439, 5′-TGTCCC-3′) and p21 ARE3 (at position −725 to −720, 5′-TGTTCT-3′) (Figure 6C). We then assessed the recruitment of AR, MDC1, GCN5 and the epigenetic changes at chromatin encompassing the loci. As expected, AR, MDC1 and GCN5 were recruited at both p21 ARE1/2 and p21 ARE3. MDC1 knock down decreased of H3K9Ac level and GCN5 recruitment at p21 ARE1/2 (Figure 6D, left panel). At p21 ARE3, MDC1 knockdown decreased H3K9Ac level, but not GCN5 recruitment (Figure 6D, right panel), suggesting that other mechanisms may be involved.
These results indicated that MDC1 is involved in regulation of a subset of androgen-regulated genes and both the epigenetic changes and GCN5 recruitment at p21 ARE1/2 are affected by MDC1. Together with the effect of MDC1 on AR-positive and AR-negative PCa cells, these data suggested that suppression of PCa proliferation and migration by MDC1 may be mediated, at least in part, through regulation of AR target genes.
The expression of MDC1 in clinical prostate biopsies
As MDC1 has a role in suppression of growth and migration of PCa cells, we next examined the expression of MDC1 in clinical prostate biopsies including 98 cases of PCa and 33 cases of benign prostatic hyperplasia (BPH). We compared the expression levels of MDC1 among BPH, well and moderately differentiated PCa (Gleason score 2–6) and poorly differentiated PCa (Gleason score 7–10). The representative images were shown in Figure 7A. Contrary to the staining of BPH tissues, immunohistochemistry using a well characterized antibody against MDC1 showed positive staining in well and moderately differentiated PCa tissues, but the poorly differentiated PCa tissues showed moderately decreased staining (Figure 7B). And the expressions of AR target gene p21 are similar to MDC1 (Figure 7C). These results suggested that MDC1 might be activated in early carcinomas as a barrier of tumorigenesis and plays a role in suppression of more advanced stages of PCa.
Figure 7.
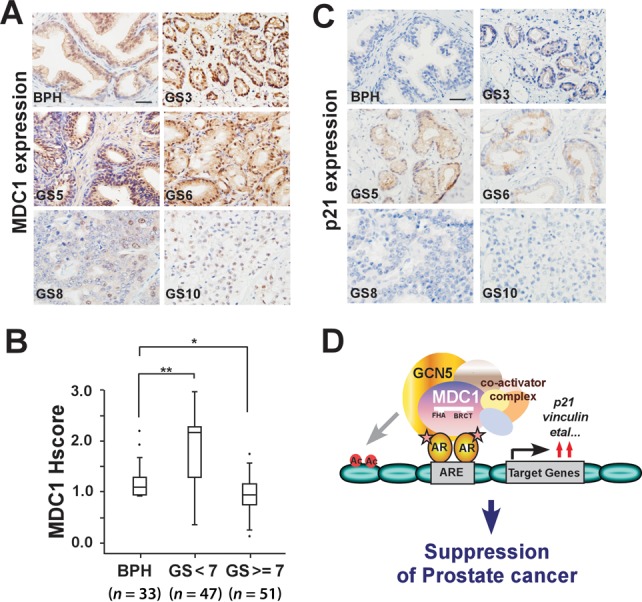
The expression of MDC1 in clinical specimens. (A) Representative images of MDC1 immunohistochemical staining in PCa (Gleason score [GS] 3, 5, 6, 8 and 10) compared with benign prostate hyperplasia (BPH). Scale bar, 25 μm. (B) MDC1 expression in PCa (GS < 7, GS ≥ 7) compared with BPH. Mann–Whitney U test was used for statistical significance. The MDC1 expression scores were shown as box plots, with the horizontal lines representing the median; the bottom and top of the boxes representing the 25th and 75th percentiles, respectively; and the vertical bars representing the range of data. Extreme cases were marked with a dot. *P < 0.05; **P < 0.01. (C) Representative images of p21 immunohistochemical staining in PCa. Scale bar, 25 μm. (D) Schematic representation of MDC1 co-activator functions on AR-induced transactivation and suppression of prostate cancer.
DISCUSSION
MDC1 with a crucial role in the DNA damage response also participates in regulating meiotic silencing (43). However, the role of MDC1 in modulating gene transcription is not clearly known. In this study, we have functionally identified MDC1 using a screening designed to isolate proteins that modulate AR action in a Drosophila experimental system (8,28). We show that MDC1 increases AR-induced transactivation in both Drosophila and mammalian cells. Importantly, we demonstrate that MDC1 acts as a modifier of AR-PEV in Drosophila, suggesting that MDC1 might be involved in chromatin remodeling. Furthermore, we show that MDC1 facilitates the association between AR and GCN5, thereby increasing histone H3 acetylation level on cis-regulatory elements of AR target genes for active gene transcription. Finally, we demonstrate that MDC1 participates in suppression of PCa cell growth and migration.
MDC1, a multifaceted nuclear protein that facilitates AR–HAT interactions
We demonstrate here that MDC1 associates with AR in mammalian cells or tissues (Figure 2), and acts as a co-activator of AR (Figures 1 and 3). This finding is in agreement with a recent study indicating that MDC1 regulates gene transcription in the absence of DNA damage response (44) and a previous report which showed residues 508–995 of MDC1 possesses transactivation activity (45). The N-terminus of MDC1 contains a forkhead-associated (FHA) domain that interacts with ATM, Chk2 and components of the Mre11/Rad50/Nbs1 (MRN) complex, while tandem BRCA1 C-terminal (BRCT) domains of MDC1 interact with gH2AX (46). In this study, we demonstrate that MDC1 interacts with AR mainly through its C-terminal 1699–2089 aa fragment or N-terminal 1–133 aa fragment (Figure 2G). It may attribute to the spatial structure and reflect different partners that MDC1 interacts with in the different contexts.
P/CAF, which belongs to the same family as GCN5, has previously been shown to directly interact with 505–676 aa of AR (47,48). Our results showed that GCN5 also directly interacts with AR at its 254–532 aa region (Supplementary Figure S7). Given its multiple domain-mediated interactions, MDC1 has the potential to act as a scaffolding protein to bring proteins together. Supporting this view, as MDC1 also directly interacts with AR, our findings suggested that MDC1 may facilitate AR and GCN5 interaction (Figure 4 and Supplementary Figure S5). Recruitment of GCN5 to promoters has emerged as a general mechanism of transcription activation of target genes. For example, GCN5 as a component of the c-Myc interacting protein TRRAP/GCN5 complex or TFTC/STAGA complex acts as a positive co-factor for activation by NRs (8,36). Overall, several protein-interaction domains of MDC1 allows it to engage in multiple interactions with HAT as well as AR.
MDC1 functions as a co-activator of AR in prostate cancer
AR belongs to the NR superfamily and is responsible for mediating all the biological actions of androgen in the target tissues, plays a central role in the development of PCa and castration-resistant PCa (CRPC) (49,50). Recent studies suggest that AR and its target genes play both suppressive and proliferative roles in PCa growth (4,5,51), but androgen deprivation therapy (52,53) or targeting PCa AR with siRNA (54) could promote PCa cell migration/invasion. So understanding the mechanism underlying the regulation of AR activity is important for the selectively control of AR in PCa and CRPC treatment. Our results obtained in both AR-dependent PEV Drosophila model and mammalian cells suggest that MDC1 acts as a co-activator of AR (Figures 1 and 3). This raises the following questions: how does the MDC1-coactivated AR contributing to PCa?
A growing body of evidence suggests that some AR co-activators with reduced expression in PCa participate in tumor suppression, such as ART-27, ARA70, BRCA1, p44 and TBLR1 (4,15–18). Our results suggest that MDC1, which is a new co-activator of AR, inhibits both growth and migration of PCa (Figure 5). Knockdown of AR attenuated the enhancement of growth stimulation and cell migration by MDC1 depletion (Figure 5G-5I), together with the effect of MDC1 on the suppression of cell growth but not migration in AR-negative DU145 cells (Figure 5C and F), these results support the conclusion: the suppression of cell growth and migration of PCa induced by MDC1 is associated with AR. Some AR co-activators participating PCa suppression are likely to regulate a subset of AR target genes important to prostate growth suppression and differentiation (15,16,18). Our results showed that MDC1 indeed selectively regulates a subset of AR target genes (Figure 6A). Unexpectedly, among the AR target genes regulated by MDC1, there were both tumor suppressors (e.g. Vinculin, p21, NKX3.1) and tumor promoters (e.g. TMPRSS2, PSA, SLC45H31). So we presume that MDC1-mediated suppression of PCa might be an overall effect of the target genes. Since MDC1 increases AR-dependent transcription of p21 or Vinculin, which plays the important suppressive roles in cancer, the effect of MDC1 on PCa is mediated, at least in part, through the regulation of distinct AR target genes. AR variants are more abundantly and frequently expressed in CRPC (55–57). In addition to the modulation of full length AR function, we also found that MDC1 interacts with the endogenous AR variants in CWR22Rv1 cells (Figure 2C) and enhances AR-V7-induced transactivation (Supplementary Figure S4). These data suggested that MDC1 is involved in modulating the functions of AR variants.
Our findings, together with those from the previous reports, significantly expand our understanding of the physiological roles of MDC1 in tumor progression by revealing its two integrated functions. First, MDC1 acts as a DNA damage checkpoint factor in the cellular response to double-strand breaks, regulating G2/M transition. Meanwhile, genomic instability caused by the loss of MDC1 does contribute to tumorigenesis, implying its role in tumor suppression. Second, MDC1 serves as a co-activator of AR for enhancing AR-induced target gene transcription. This line of evidence establishes MDC1 as a crucial regulator of tumor development by virtue of its coordination of DNA damage checkpoint, maintenance of genomic stability and gene transcription through both DNA damage responsive and transcriptional co-factor functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We appreciate Mr Yunlong Huo, Dr Zhongbo Yin and Ms Lin Lin for helpful technique support. The anti-HP1 polyclonal antibody (C1A9) developed by investigators was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology Science, Iowa City, IA 52242.
FUNDING
973 Program Grant from the Ministry of Science and Technology of China [2013CB945201]; National Natural Science Foundation of China [30871390, 31171259, 31271364 31401115]. Ministry of Education fund innovation team [IRT 13101]; Ministry of Education Science and technology research [213008A]. Funding for open access charge: 973 Program Grant from the Ministry of Science and Technology of China [2013CB945201]; National Natural Science Foundation of China [30871390, 31171259, 31271364 31401115]. Ministry of Education fund innovation team [IRT 13101]; Ministry of Education Science and technology research [213008A].
Conflict of interest statement. None declared.
REFERENCES
- 1.Xu K., Wu Z.J., Groner A.C., He H.H., Cai C., Lis R.T., Wu X., Stack E.C., Loda M., Liu T., et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J., Mani R.S., Cao Q., Brenner C.J., Cao X., Wang X., Wu L., Li J., Hu M., Gong Y., et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh S., Hu Y.C., Rahman M., Lin H.K., Hsu C.L., Ting H.J., Kang H.Y., Chang C. Increase of androgen-induced cell death and androgen receptor transactivation by BRCA1 in prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11256–11261. doi: 10.1073/pnas.190353897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chng K.R., Chang C.W., Tan S.K., Yang C., Hong S.Z., Sng N.Y., Cheung E. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31:2810–2823. doi: 10.1038/emboj.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perissi V., Aggarwal A., Glass C.K., Rose D.W., Rosenfeld M.G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Bassets I., Kwon Y.S., Telese F., Prefontaine G.G., Hutt K.R., Cheng C.S., Ju B.G., Ohgi K.A., Wang J., Escoubet-Lozach L., et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A., et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 10.Clark E.L., Coulson A., Dalgliesh C., Rajan P., Nicol S.M., Fleming S., Heer R., Gaughan L., Leung H.Y., Elliott D.J., et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu K., Shimelis H., Linn D.E., Jiang R., Yang X., Sun F., Guo Z., Chen H., Li W., Kong X., et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell. 2009;15:270–282. doi: 10.1016/j.ccr.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang Y., Zhu Z., Han G., Ye X., Xu B., Peng Z., Ma Y., Yu Y., Lin H., Chen A.P., et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Guo J., Li Y., Bavarva J.H., Qian C., Brahimi-Horn M.C., Tan D., Liu W. Inactivation of androgen-induced regulator ARD1 inhibits androgen receptor acetylation and prostate tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3053–3058. doi: 10.1073/pnas.1113356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahl P., Gullotti L., Heukamp L.C., Wolf S., Friedrichs N., Vorreuther R., Solleder G., Bastian P.J., Ellinger J., Metzger E., et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 15.Peng Y., Chen F., Melamed J., Chiriboga L., Wei J., Kong X., McLeod M., Li Y., Li C.X., Feng A., et al. Distinct nuclear and cytoplasmic functions of androgen receptor cofactor p44 and association with androgen-independent prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5236–5241. doi: 10.1073/pnas.0712262105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taneja S.S., Ha S., Swenson N.K., Torra I.P., Rome S., Walden P.D., Huang H.Y., Shapiro E., Garabedian M.J., Logan S.K. ART-27, an androgen receptor coactivator regulated in prostate development and cancer. J. Biol. Chem. 2004;279:13944–13952. doi: 10.1074/jbc.M306576200. [DOI] [PubMed] [Google Scholar]
- 17.Ligr M., Li Y., Zou X., Daniels G., Melamed J., Peng Y., Wang W., Wang J., Ostrer H., Pagano M., et al. tumor suppressor function of androgen receptor coactivator ARA70alpha in prostate cancer. Am. J. Pathol. 2010;176:1891–1900. doi: 10.2353/ajpath.2010.090293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels G., Li Y., Gellert L.L., Zhou A., Melamed J., Wu X., Zhang X., Zhang D., Meruelo D., Logan S.K., et al. TBLR1 as an androgen receptor (AR) coactivator selectively activates AR target genes to inhibit prostate cancer growth. Endocr. Relat. Cancer. 2014;21:127–142. doi: 10.1530/ERC-13-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung C., Kim R.S., Zhang H.J., Lee S.J., Jeng M.H. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 20.Wu K., Katiyar S., Witkiewicz A., Li A., McCue P., Song L.N., Tian L., Jin M., Pestell R.G. The cell fate determination factor dachshund inhibits androgen receptor signaling and prostate cancer cellular growth. Cancer Res. 2009;69:3347–3355. doi: 10.1158/0008-5472.CAN-08-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazumdar A., Wang R.A., Mishra S.K., Adam L., Bagheri-Yarmand R., Mandal M., Vadlamudi R.K., Kumar R. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat. Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 22.Lakshmikanthan V., Zou L., Kim J.I., Michal A., Nie Z., Messias N.C., Benovic J.L., Daaka Y. Identification of betaArrestin2 as a corepressor of androgen receptor signaling in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9379–9384. doi: 10.1073/pnas.0900258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S.P., Bartek J., Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg M., Stucki M., Falck J., D'Amours D., Rahman D., Pappin D., Bartek J., Jackson S.P. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 25.Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi M., Ozaki T., Yamamoto H., Hanamoto T., Kikuchi H., Furuya K., Asaka M., Delia D., Nakagawara A. NFBD1/MDC1 associates with p53 and regulates its function at the crossroad between cell survival and death in response to DNA damage. J. Biol. Chem. 2007;282:22993–23004. doi: 10.1074/jbc.M611412200. [DOI] [PubMed] [Google Scholar]
- 27.Minter-Dykhouse K., Ward I., Huen M.S., Chen J., Lou Z. Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J. Cell Biol. 2008;181:727–735. doi: 10.1083/jcb.200801083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Takeyama K., Sawatsubashi S., Ito S., Suzuki E., Yamagata K., Tanabe M., Kimura S., Fujiyama S., Ueda T., et al. Corepressive action of CBP on androgen receptor transactivation in pericentric heterochromatin in a Drosophila experimental model system. Mol. Cell. Biol. 2009;29:1017–1034. doi: 10.1128/MCB.02123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito S., Fujiyama-Nakamura S., Kimura S., Lim J., Kamoshida Y., Shiozaki-Sato Y., Sawatsubashi S., Suzuki E., Tanabe M., Ueda T., et al. Epigenetic silencing of core histone genes by HERS in Drosophila. Mol. Cell. 2012;45:494–504. doi: 10.1016/j.molcel.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 30.Dronamraju R., Mason J.M. Recognition of double strand breaks by a mutator protein (MU2) in Drosophila melanogaster. PLoS Genet. 2009;5:e1000473. doi: 10.1371/journal.pgen.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y., Goto K., Saitoh M., Yanase T., Nomura M., Okabe T., Takayanagi R., Nawata H. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particle-binding protein as a coactivator for the receptor. J. Biol. Chem. 2002;277:30031–30039. doi: 10.1074/jbc.M203811200. [DOI] [PubMed] [Google Scholar]
- 32.Chapman J.R., Jackson S.P. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agoulnik I.U., Vaid A., Bingman W.E. 3rd, Erdeme H., Frolov A., Smith C.L., Ayala G., Ittmann M.M., Weigel N.L. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 34.Wang C., Li Y., Zhang H., Liu F., Cheng Z., Wang D., Wang G., Xu H., Zhao Y., Cao L., et al. Oncogenic PAK4 regulates Smad2/3 axis involving gastric tumorigenesis. Oncogene. 2014;33:3473–3484. doi: 10.1038/onc.2013.300. [DOI] [PubMed] [Google Scholar]
- 35.Fujiki R., Kim M.S., Sasaki Y., Yoshimura K., Kitagawa H., Kato S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005;24:3881–3894. doi: 10.1038/sj.emboj.7600853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Yanagisawa J., Kitagawa H., Yanagida M., Wada O., Ogawa S., Nakagomi M., Oishi H., Yamamoto Y., Nagasawa H., McMahon S.B., et al. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell. 2002;9:553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- 37.Shao W., Halachmi S., Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol. Cell. Biol. 2002;22:3358–3372. doi: 10.1128/MCB.22.10.3358-3372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohgaki T., Bohgaki M., Cardoso R., Panier S., Zeegers D., Li L., Stewart G.S., Sanchez O., Hande M.P., Durocher D., et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 2011;7:e1001381. doi: 10.1371/journal.pgen.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Corsa C.A., Pan P.W., Wu L., Ferguson D., Yu X., Min J., Dou Y. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol. Cell. Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haelens A., Verrijdt G., Schoenmakers E., Alen P., Peeters B., Rombauts W., Claessens F. The first exon of the human sc gene contains an androgen responsive unit and an interferon regulatory factor element. Mol. Cell. Endocrinol. 1999;153:91–102. doi: 10.1016/s0303-7207(99)00079-9. [DOI] [PubMed] [Google Scholar]
- 42.Roche P.J., Hoare S.A., Parker M.G. A consensus DNA-binding site for the androgen receptor. Mol. Endocrinol. 1992;6:2229–2235. doi: 10.1210/mend.6.12.1491700. [DOI] [PubMed] [Google Scholar]
- 43.Ichijima Y., Ichijima M., Lou Z., Nussenzweig A., Camerini-Otero R.D., Chen J., Andreassen P.R., Namekawa S.H. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 2011;25:959–971. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson K.A., Colavito S.A., Schulz V., Wakefield P.H., Sessa W., Tuck D., Stern D.F. NFBD1/MDC1 regulates Cav1 and Cav2 independently of DNA damage and p53. Mol. Cancer Res. 2011;9:766–781. doi: 10.1158/1541-7786.MCR-10-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozaki T., Nagase T., Ichimiya S., Seki N., Ohiri M., Nomura N., Takada N., Sakiyama S., Weber B.L., Nakagawara A. NFBD1/KIAA0170 is a novel nuclear transcriptional transactivator with BRCT domain. DNA Cell Biol. 2000;19:475–485. doi: 10.1089/10445490050128403. [DOI] [PubMed] [Google Scholar]
- 46.Kim J.E., Minter-Dykhouse K., Chen J. Signaling networks controlled by the MRN complex and MDC1 during early DNA damage responses. Mol. Carcinog. 2006;45:403–408. doi: 10.1002/mc.20221. [DOI] [PubMed] [Google Scholar]
- 47.Fu M., Wang C., Reutens A.T., Wang J., Angeletti R.H., Siconolfi-Baez L., Ogryzko V., Avantaggiati M.L., Pestell R.G. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 48.Reutens A.T., Fu M., Wang C., Albanese C., McPhaul M.J., Sun Z., Balk S.P., Janne O.A., Palvimo J.J., Pestell R.G. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol. Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 49.Qi J., Tripathi M., Mishra R., Sahgal N., Fazli L., Ettinger S., Placzek W.J., Claps G., Chung L.W., Bowtell D., et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23:332–346. doi: 10.1016/j.ccr.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman B.J., Feldman D. The development of androgen-independent prostate cancer. Nat. Rev. Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 51.Niu Y., Altuwaijri S., Lai K.P., Wu C.T., Ricke W.A., Messing E.M., Yao J., Yeh S., Chang C. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12182–12187. doi: 10.1073/pnas.0804700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paez-Ribes M., Allen E., Hudock J., Takeda T., Okuyama H., Vinals F., Inoue M., Bergers G., Hanahan D., Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin T.H., Lee S.O., Niu Y., Xu D., Liang L., Li L., Yeh S.D., Fujimoto N., Yeh S., Chang C. Differential androgen deprivation therapies with anti-androgens casodex/bicalutamide or MDV3100/Enzalutamide versus anti-androgen receptor ASC-J9(R) Lead to promotion versus suppression of prostate cancer metastasis. J. Biol. Chem. 2013;288:19359–19369. doi: 10.1074/jbc.M113.477216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izumi K., Fang L.Y., Mizokami A., Namiki M., Li L., Lin W.J., Chang C. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol. Med. 2013;5:1383–1401. doi: 10.1002/emmm.201202367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo Z., Yang X., Sun F., Jiang R., Linn D.E., Chen H., Kong X., Melamed J., Tepper C.G., Kung H.J., et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun S., Sprenger C.C., Vessella R.L., Haugk K., Soriano K., Mostaghel E.A., Page S.T., Coleman I.M., Nguyen H.M., Sun H., et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao B., Qi Y., Zhang G., Xu D., Zhan Y., Alvarez X., Guo Z., Fu X., Plymate S.R., Sartor O., et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


