Abstract
Circadian rhythm, or daily oscillation, of behaviors and biological processes is a fundamental feature of mammalian physiology that has developed over hundreds of thousands of years under the continuous evolutionary pressure of energy conservation and efficiency. Evolution has fine-tuned the body's clock to anticipate and respond to numerous environmental cues in order to maintain homeostatic balance and promote survival. However, we now live in a society in which these classic circadian entrainment stimuli have been dramatically altered from the conditions under which the clock machinery was originally set. A bombardment of artificial lighting, heating, and cooling systems that maintain constant ambient temperature; sedentary lifestyle; and the availability of inexpensive, high-calorie foods has threatened even the most powerful and ancient circadian programming mechanisms. Such environmental changes have contributed to the recent staggering elevation in lifestyle-influenced pathologies, including cancer, cardiovascular disease, depression, obesity, and diabetes. This review scrutinizes the role of the body's internal clocks in the hard-wiring of circadian networks that have evolved to achieve energetic balance and adaptability, and it discusses potential therapeutic strategies to reset clock metabolic control to modern time for the benefit of human health.
-
Introduction
Circadian clock machinery
Evolution of the CRY proteins
Interplay between the clock and energy metabolism
Clock metabolic control in modern societies: programming in need of an upgrade
-
Tissue-Specific Contributions to Circadian Physiology
Brain: the command center
Brown adipose tissue: the thermogenic center
White adipose tissue: the storage center
Liver: the fuel management center
Heart: the distribution center
Pancreas: the glycemic control center
Skeletal muscle: the motion center
Gut microbiota: the food-processing center
Conclusion
I. Introduction
The evolutionary biologist Theodosius Dobzhansky posited that “nothing in biology makes sense except in the light of evolution” (1). This doctrine applies well to the observation of endogenous biological rhythms whose phase is approximately the length of a day on earth, which are referred to as circadian rhythm. These measurable systemic outputs are the end-product of a vast, coordinated circuitry, central to which are a fundamental molecular mechanism referred to as the core clock. Most organisms on the planet have such a clock, which is entrained by light and anticipates as well as adapts to external demands to promote organismal fitness. Hundreds of thousands of years of environmental (eg, the 24-hour period of the earth's rotation about its axis) and nutrient-derived (eg, availability of food) inputs have shaped the rhythmic programming of biological and behavioral output, conferring the presumed benefit of selective advantage.
A. Circadian clock machinery
One of the simplest biological clocks is present in cyanobacteria and consists of three proteins (KaiA, KaiB, and KaiC) that oscillate in a self-sustained phosphorylation/dephosphorylation cycle. This clock prevents DNA replication during daylight under the harmful UV rays of the sun and increases organismal survival and reproductive fitness (2–4). However, the complexity of higher-order metazoans demands a more advanced, multifaceted system.
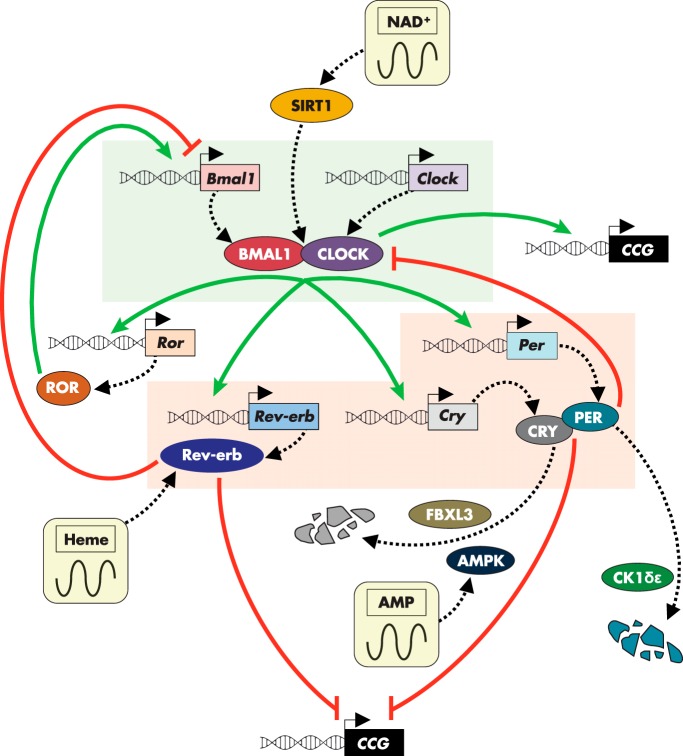
In mammals, the core molecular clock that is present in each cell is comprised of an autoregulatory transcriptional/translational feedback loop (5–7) that is reset daily by the master clock located in the hypothalamic suprachiasmatic nucleus (SCN) (8, 9). As detailed in Figure 1, two bHLH transcriptional activators, circadian locomotor output cycles kaput (CLOCK) (10, 11) and brain and muscle ARNT-like 1 (BMAL1 a.k.a. ARNTL) (12, 13), heterodimerize and bind to promoter E-box elements to induce expression of the repressive arm of the clock, which include cryptochromes 1 and 2 (CRY1/2) (14, 15) and periods 1, 2, and 3 (PER1/2/3) (16) and the nuclear receptors, Rev-erbα and β (17–19). CRY and PER form a regulatory complex, which directly inhibits CLOCK-BMAL1 transactivating function, whereas Rev-erbα and -β repress Bmal1 transcription (20) through recruitment of histone deacetylase 3 (21). Another set of nuclear receptors, RAR-orphan receptor α and γ (RORα and -γ), are induced by CLOCK-BMAL1 and function as transcriptional circadian activators (22) to positively feedback on Bmal1 expression (23) in competition with Rev-erbα and -β (24). Additionally, casein kinase 1δ and -ϵ (CK1δ/ϵ) contribute to clock function through the phosphorylation and destabilization of PER proteins (25–27) while FBXL3 directs the E3 ubiquitin ligase-mediated degradation of CRY proteins (28–30).
Figure 1.
Interplay between the cell-autonomous clock and metabolism. The core biological clock, present in every cell in the body, consists of an autoregulatory transcriptional feedback loop. The activating arm (highlighted in green) is comprised of the transcriptional regulators BMAL1 and CLOCK, which heterodimerize and induce the expression of the inhibitory arm (highlighted in red). The inhibitory arm contains Rev-erb, CRY, and PER factors. Rev-erb acts to suppress transcription of Bmal1, whereas CRY and PER directly inhibit the activating function of BMAL1/CLOCK heterodimer complexes. Members of both activating and inhibitory arms of the clock coordinate metabolic programming through the transcriptional control of clock-controlled genes (CCG) involved in a wide array of bioenergetic networks. The metabolic output resulting from this regulation feeds back on individual clock components, linking the energetic fitness of the cell with circadian functionality. Various nodes in this sustained molecular loop are subject to further control by intracellular oscillations of key metabolites, including heme, NAD+, and AMP.
B. Evolution of the CRY proteins
Despite the dramatic variation in the complexity of metazoans, there is significant conservation of the core components of this evolutionary clock toolset. Instead of synthesizing entirely new parts de novo, there appears to be a repurposing of master regulatory factors to accommodate the selective pressures of the individual organism. This concept is exemplified in the CRY proteins. This core clock component possesses strong sequence homology reminiscent of photolyases (31, 32), which are light-activated DNA repair enzymes, suggesting that the ancient ancestor of this timekeeper tightly linked the active maintenance of genomic integrity with exposure to the sun to counteract the damaging effects of the UV-induced pyrimidine dimerization.
In Drosophila and plants, CRY homologs function as blue light photoreceptors and directly connect light exposure to regulation of the molecular clock (33–35). Interestingly, both these photolyase and photoreceptor properties appear to have been lost in mammals (36), most likely because the specific selective pressure that faced simpler eukaryotes was superseded by a more pressing demand or was circumvented by an alternate mechanism.
C. Interplay between the clock and energy metabolism
The circadian clock machinery is also responsible for modulating the expression of numerous tissue-specific, bioenergetic programs to orchestrate systemic metabolic rhythms (37–39). This communication between the clock and physiological function is of chief importance when considering evolutionary influence on circadian networks. However, binding of these circadian transcriptional regulators to obligate sequences in target promoters does not solely explain the rhythmicity of all gene products (40). To ensure appropriate coordination of complex metabolic biologies, such as those found in mammals, clock regulators employ an array of accessory activators and repressors to coordinate multiple, distinct transcriptional phases throughout the day (41). Moreover, a significant amount of post-transcriptional processing and translational control, including rhythmic RNA-binding proteins (42), contributes additional layers of regulation in defining the circadian landscape (43–46).
The deeply rooted interplay between circadian rhythm and evolutionary homeostatic pressure is evidenced by the numerous mechanisms through which the clock is directly wired into the energetic state of the cell (47) (summarized in Figure 1). Rhythmic genomic binding of CLOCK-BMAL1 heterodimers is profoundly influenced by NAD+/NADH redox status (48). Circadian oscillation of the NAD+ biosynthetic enzyme, nicotinamide phosphoribosyltransferase, drives rhythmic levels of NAD+, which activate the NAD+-dependent deacetylases, sirtuin 1 (SIRT1) in the cytosol and nucleus (49–51) and SIRT3 in the mitochondria (52), to direct metabolic output. Heme acts as a ligand for Rev-erbα, thereby conferring an energy-sensing capacity to the repressive arm of the clock (53, 54). Additionally, the circadian clock is sensitive to changes in the AMP/ATP ratio through AMP-activated protein kinase (AMPK)-mediated phosphorylation and subsequent degradation of CRY (55). Perhaps the oldest and most conserved example of this integral pairing of circadian rhythm and selective evolutionary pressure is in the transcription-independent cycling of peroxiredoxin enzymes, which protect against the daily rhythmic increase in reactive oxygen species and is found across all domains of life from bacteria to eukaryota (56–58). However, the physiological readout of this circadian rhythm remains to be established (59).
D. Clock metabolic control in modern societies: programming in need of an upgrade
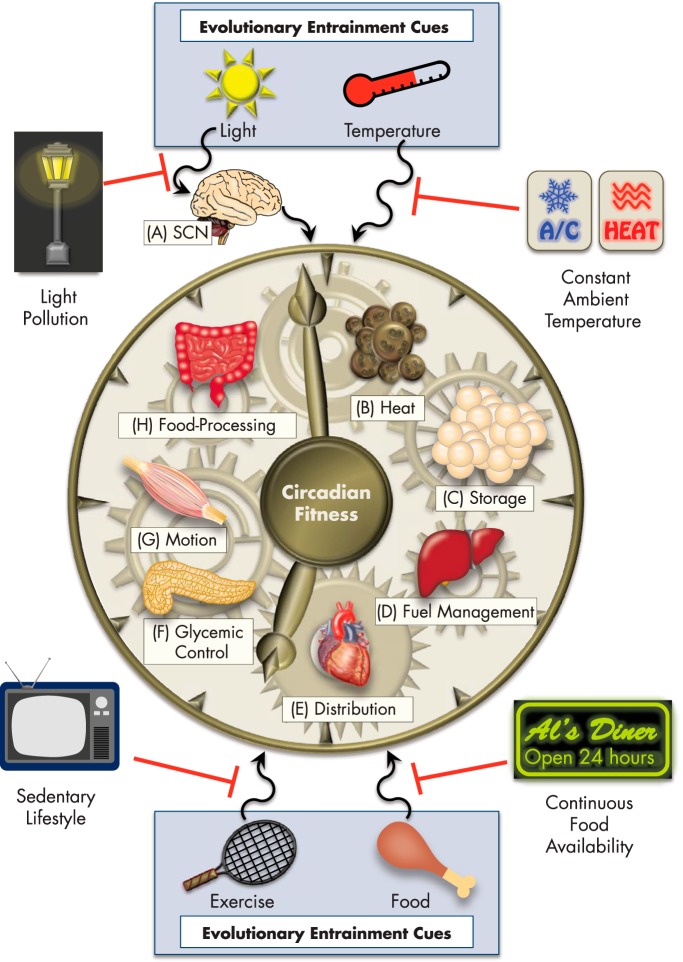
Evolution has instated this molecular framework of circadian control to adapt and thrive under the selective pressures of food scarcity, seasonal changes in sunlight availability, and variable range of temperature exposure. Although this fine-tuned system was sufficient for many thousands of years, we have rapidly developed societal conditions, which seem to exceed the adaptive limitations of our circadian programming (Figure 2). With the introduction of 24-hour fast food restaurants and the help of multibillion dollar snack and beverage industries, cheap and calorically dense meals are accessible at any time of day. This trend is even more detrimental to human health combined with a growing population of shift workers, more regular travel across time zones, social jet lag, and nearly constant exposure to artificial light pollution. Making matters worse, most people work, recreate, and reside in indoor conditions where heating and cooling advances maintain a continuous ambient environment. Coupling this to a largely sedentary lifestyle creates a situation requiring little or no energy expenditure to defend our body temperature or exert physical activity, in stark contrast to the realities with which our ancestors were frequently faced.
Figure 2.
Impact of modern environments on evolutionarily programmed circadian functions. Sunlight, temperature, physical activity, and food intake serve as basic entraining cues, or zeitgebers, that coordinate tissue-specific circadian processes to cumulatively define whole organismal physiology. Among these zeitgebers, light is the chief synchronization cue and acts to reset the master clock (A) in the hypothalamic SCN each day, which then relays signaling to peripheral tissues. Evolutionarily fine-tuned, tissue-specific circadian processes discussed in this review are depicted, including: B, heat production by brown adipose; C, energy storage by white adipose; D, fuel source management between carbohydrate and lipid substrates in the liver; E, distribution or circulation of blood-borne factors, hormones, and metabolites by the heart; F, control of blood glucose levels by the pancreas; G, capacity for movement and activity by skeletal muscle; and H, food processing and nutrient extraction by the gut microbiome. A combination of light pollution from artificial light sources, sedentary lifestyles largely lacking physical activity, continuous access to high-calorie foods, and living conditions maintained at constant ambient temperature have all contributed to the disruption of circadian fitness.
It is likely not a coincidence that radical adjustments in these environmental parameters of nutrition, light, and temperature are correlated with dramatic increases in the rates of obesity, diabetes, cardiovascular disease, depression, and many types of cancer (60–71). To this end, we will now explore tissue-specific circadian biology with an evolutionary perspective and examine how these functions have or have not become outdated or broken down in the face of modern societal conditions. Ultimately, we hope that these insights will identify areas of circadian metabolic control that can be targeted for novel therapeutic strategies to rewire specific clock-regulated pathways with minimal or no detriment to the core machinery.
II. Tissue-Specific Contributions to Circadian Physiology
Mammalian integrative physiology requires that a network of specialized cells, organs, and tissues communicate with one another via the nervous and endocrine systems. This system is inherently interactive, with certain organs acting as nodes, or centers, that play disproportionate roles in behavior, energy consumption and storage, and fuel selection, as detailed in this section. However, an important consideration when assessing tissue-specific circadian metabolic control is the contribution of central regulation by the master clock vs the cell-autonomous clocks. This distinction is best addressed using conditional, tissue-specific deletion and will be noted, where possible, throughout the section.
A. Brain: the command center
The SCN is the master circadian regulator that synchronizes the rest of the cellular clocks. Light, one of the chief zeitgebers, is sensed by a specific set of photoreceptors located in the retina (72, 73), and signals are relayed to the hypothalamic SCN (Figure 2A). The SCN in turn communicates with other neurons in the central nervous system to coordinate organismal rhythmicity in a highly complex network that has been more extensively reviewed elsewhere (9, 74). The absolute requirement of the SCN in systemic circadian control was demonstrated by specifically introducing lesions in the hypothalamus. Early, pioneering studies found that lesions that destroyed or disrupted the SCN ablated rhythmicity in both drinking and physical activity (75) as well as corticosterone levels (76) in rats. Similar experiments further demonstrated that the SCN was also required for the daily pattern of blood glucose concentration (77, 78). Finally, SCN grafts into animals with disrupted circadian biology were able to rescue and restore organismal rhythmicity (79).
Wiring the synchronization of all the individual peripheral clocks through the light-controlled SCN provides a number of evolutionary advantages. Most importantly, one master regulator more accurately ensures appropriate phase alignment in the diverse organ systems. Coupling this master controller to an entrainment as continuous and as regular as light allows the organism to respond not only to daily changes but also to seasonal changes. Moreover, one of the first functions of biological clocks may have been to provide a mechanism to taper mutation-sensitive cellular processes like genome replication during daylight exposure to UV radiation (2). However, the central importance of the SCN in whole-animal circadian control also renders it an unlikely point of therapeutic intervention for correcting outdated evolution-imposed circuitry. Instead, restoring clock function and circadian robustness in neuronal networks may be a more beneficial pharmacological approach to address deficits in the central nervous system clock. This strategy has been promising in ameliorating anxiety-like behavior (80). Corrective pharmacological approaches could have a profound impact on societal health, given the numerous genetic variants or mutations in clock genes that have been identified in the population (81–85).
B. Brown adipose tissue: the thermogenic center
Brown adipose tissue (BAT) is a major source of mammalian facultative thermogenesis (ie, additional heat production above the basal metabolic rate) (86). The classic model of BAT heat production begins with an initial stimulating event, such as exposure to cold temperature that elicits brown adipose activity through blood-borne hormonal and nutrient prompts as well as direct neuronal input from the hypothalamus (86, 87). This function is accomplished through simultaneous consumption of lipid and carbohydrate fuel sources and rerouting of the mitochondrial proton gradient through the thermogenic effector, uncoupling protein 1 (86).
The unique, energy-dissipating activity of BAT is also controlled in a circadian manner (88–90) (Figure 2B) by the clock components, Rev-erbα (91), PER2 (92), and BMAL1 (93, 94). Rev-erbα levels peak while animals are sleeping and result in direct transcriptional repression of the BAT thermogenic program, including Ucp1 (91). Whole-animal genetic deletion of Rev-erbα largely abolishes the oscillation of BAT activity and whole-animal core temperature, suggesting that clock control of brown adipose is a major driver of the circadian rhythm of body temperature. Consistent with this model, daily increases and decreases in BAT thermogenesis, as measured by implanted telemetric thermometers, were found to precede corresponding changes in core temperature (95). To further underscore the potential impact and physiological relevance of BAT heat production, previous studies have demonstrated that even subtle changes in body temperature, well within the thermogenic range driven by Rev-erbα, can synchronize or reset peripheral clocks (96, 97).
Clock-mediated regulation of BAT thermogenic function also exemplifies the integral association between circadian rhythm and evolutionary demand. As ancient eutherians underwent the transition from ectotherms to endotherms, BAT adopted a more clearly defined role and conferred a selective advantage not only to survive but also to thrive in adverse environmental climates (98). However, given the scarcity of food with which our ancestors were faced for thousands of years, the energy-dissipating nature of BAT would also prove highly unfavorable if left unchecked. In this regard, we hypothesize that the circadian clock was evolutionarily programmed to “turn off” BAT when its function was not needed, such as during sleep when most animals are sheltered in ambient environments. BAT activity begins increasing once animals awaken to provide the necessary thermal protection for hunting and gathering food in harsh environments. This network has even been further equipped with a fail-safe in the event that an animal is suddenly exposed to cold temperatures while sleeping. The chief circadian BAT regulator, Rev-erbα, is rapidly and significantly reduced, thereby facilitating full induction of the thermogenic response (91).
This rhythmic mechanism of decreasing BAT activity during sleep is perfectly in line with evolutionary principles but is now more of a hindrance to metabolic health in the face of late night eating and calorie-rich, Western diets. Targeting this brake on BAT function could provide a way to increase the daily BAT consumption of glucose and fat stores in a safer and more specific manner than through the more classic, ubiquitous pathway of adrenergic signaling. Furthermore, preventing the BAT clock from actively suppressing calorie-burning at night could potentially increase BAT output without an uncomfortable or dangerous elevation in body temperature that is perhaps more likely with strategies that enhance BAT activity beyond its endogenous maximal capacity. However, whether or not the difference in energy balance from unlocking the circadian inhibition of BAT would be significant enough to mitigate the effects of metabolic syndrome is as yet undetermined (91). Given that the circadian rhythm of BAT function is also correlated with food intake and BAT is believed to be a contributor to diet-induced thermogenesis (99), further experiments must also be carried out to tease apart the mechanistic contributions from central and dietary control of the oscillation of BAT activity.
C. White adipose tissue: the storage center
The body's chief reservoir for diet-derived energy is white adipose tissue, which accumulates triglycerides. With a potential energy of 9 kcal/g, triglycerides are the most efficient storage form of biological energy. During periods of fasting, white adipose mobilizes these lipid stores for use as oxidative substrates in peripheral tissue such as skeletal muscle. In this manner, evolution has programmed the circadian clock to increase lipolytic pathways during sleep to compensate for the absence of dietary energy sources in mouse (100, 101) and human (102, 103). Conversely, while animals are awake and feeding, white adipose builds up triglyceride stores with liver-produced lipid (Figure 2C).
Circadian control of lipid mobilization is mediated, at least in part, through CLOCK and BMAL1-induced rhythm of adipose triglyceride lipase (Atgl) and hormone sensitive lipase (Hsl) gene expression (100). Indeed, genetic disruption of Bmal1 or use of the dominant negative ClockΔ19 (10, 104) mutant results in arrhythmic levels of serum free fatty acids and decreased lipolysis rates (100, 101), which contributes to the development of obesity. Additionally, adipose-specific Bmal1 deletion also seems to influence adipose-hypothalamic cross talk through alterations in the level of polyunsaturated lipid species that are released into the blood (101). The repressing arm of the clock modulates lipid metabolism through PER2-dependent suppression of the highly adipogenic and insulin-sensitizing nuclear receptor peroxisome proliferator-activated receptor-γ (105) and Rev-erbα-dependent transcriptional repression of lipoprotein lipase (106).
Adipose tissue is also a key contributor to endocrine signaling (107) through the production of adipokines, including leptin (108), resistin (109), and adiponectin (110), whose circulating levels are robustly circadian (111, 112). The satiety adipokine, leptin, reaches peak serum levels in the early period of the inactive phase in both diurnal (eg, monkey and human) (113–116) and nocturnal (eg, mouse and rat) (117, 118) animals. This is consistent with a suppression of appetite in anticipation of sleep. Additionally, adiponectin and resistin exhibit rhythmic expression patterns, although the physiological function of this oscillation remains unknown (111, 119, 120).
The conserved circadian functions of white adipose tissue serve to modulate systemic lipid homeostasis through storage of triglyceride during feeding and provision of peripheral tissues with fatty acid substrate during fasting. However, current lifestyle patterns characterized by frequent and high-caloric feeding with shorter periods of fasting as well as disruptive sleep events, such as jet lag and shift work, have negatively tilted white adipose metabolism toward storage and accumulation. A trait once selectively desirable for organismal survival when food was scarce now contributes to ever-expanding waistlines and deleterious metabolic diseases.
Therapeutic intervention to correct circadian dysregulation of white adipose metabolism would likely have to be coupled to a method of increasing fat oxidation in peripheral tissues. Simply limiting the triglyceride accumulation in adipose depots could result in a rerouting and deposition of the lipid in other tissues, such as skeletal muscle and liver, potentially with even more detrimental consequences. On the other hand, further elucidation of the circadian endocrine contributions of white adipose could prove beneficial in developing novel strategies to synchronize systemic energy homeostasis.
D. Liver: the fuel management center
The liver is an organ that has been evolutionarily tasked with the responsibility of maintaining systemic energy homeostasis through the coordinated synthesis and storage of lipid and carbohydrate fuel sources (Figure 2D). To most effectively achieve this balance, the liver receives cues from the master clock in the SCN as well as independent signals from the animal's feeding and fasting behavior (121–124). However, these neuronal and nutrient-mediated contributors to hepatic metabolic oscillation do not function in mutual exclusivity. For example, the fasting-induced hepatokine, fibroblast growth factor 21, directly influences central circadian networks through binding to its obligate coreceptor, β-KLOTHO, in the SCN (125). Additionally, adrenally produced glucocorticoid hormones antagonize the capacity of altered feeding times to uncouple central and peripheral control over liver metabolic rhythms (126).
Circadian coordination of hepatic energy homeostasis employs both activating and repressing arms of the core clock (127, 128). CLOCK and BMAL1 exert a profound influence on gluconeogenesis and systemic oscillation of blood glucose (129–132). CLOCK plays a role in the glycogen synthesis pathway by promoting Gys2 expression (133). Evidence from whole-animal knockout models demonstrates that PER2 also stimulates glycogen levels during refeeding through increases in Gys2 (134). Conversely, glucose production is negatively regulated by the CRY proteins, which attenuate glucagon-mediated increases in cAMP levels and diminish gluconeogenic output (135). CRY1 and -2 also interact with glucocorticoid receptor in a hormone-dependent manner to directly modulate the expression of the gluconeogenic effector enzyme, phosphoenolpyruvate carboxykinase 1 (Pck1 or Pepck) (136).
The anticipatory induction in hepatic lipogenic genes prior to an animal waking and feeding is almost exclusively mediated by derepression of Rev-erbα/β and histone deacetylase 3 target genes (21, 137, 138). This phenomenon is observed in both whole-animal and tissue-specific models of Rev-erbα disruption, thereby indicating significant contributions by the peripheral liver clock. Rev-erbα also modulates the rhythmicity of cholesterol metabolism through the transcription factor, sterol regulatory element-binding protein, and bile acid synthesis through regulation of Cyp7a1 (139, 140). Oscillations in NAD+ biosynthesis drive SIRT3 activation in mitochondria, which leads to circadian increases in liver fatty acid utilization during the sleep phase (52). Daily NAD+ rhythm in the cytosol and nucleus coordinates SIRT1-mediated deacetylation of histones and core clock components to orchestrate gene expression (49–51). In addition to SIRT1-dependent histone deacetylation, circadian genome-wide control of hepatic metabolic programming is elicited in an intricate network of transcriptional phases driven by individual clock factors (41, 43, 141).
Cumulatively, these mechanisms define how the liver clock is hardwired to maintain blood glucose homeostasis. Liver glycogen stores are depleted while the animal is asleep and in a fasted state and then replenished throughout the waking period when the animal maintains blood glucose through dietary means (133). The increases in the hepatic lipogenic program that proceed the animal's first feeding likely served an evolutionary advantage in maximizing the amount of energy that could be extracted from a meal and deposited in adipose depots. Interestingly, the liver has been shown to mediate intertissue cross talk through the rhythmic production phosphatidylcholine 18:0/18:1 to influence circadian lipid utilization in skeletal muscle (142).
The liver's circadian programming, which for thousands of years was advantageous due to its coordination with nutrient availability, eating patterns, and lifestyle habits, is challenged by our modern society characterized by caloric excess and increasing prevalence of sleep disruptions. Indeed, an ad libitum high-fat diet (HFD) impairs liver circadian function (143), and shift workers exhibit significantly higher rates of fatty liver (144). Moreover, studies examining time-restricted feeding in mice suggest that the old adage “you are what you eat” should be amended to also include “you are WHEN you eat.” When animals were forced to eat during the light period when they normally sleep, metabolic rhythms in energy expenditure and body temperature rhythms were completely reversed (145). However, restricting the time of food consumption to the normal murine feeding period at night made it possible to synchronize liver metabolic pathways, correct dysfunction in the diabetic db/db model (146), and prevent the development of HFD-induced liver pathology, without reduction in total daily caloric intake (147). Strikingly, even time-restricting the HFD only during the week and allowing mice ad libitum access during the weekend to simulate a more recreational human lifestyle was sufficient to retain the metabolic benefits observed under constitutive time-restricted feeding (148).
The preponderance of data would therefore suggest that the most effective “treatment” would be simply adopting a lifestyle in which eating was restricted to approximately the same time and within daylight hours. However, this is an unachievable goal for the growing percentage of the population working at night or repeatedly navigating between time zones. Given that genetic and dietary disruption of the mouse liver clock leads to aberrantly overactive lipogenesis and, consequently, steatosis, negatively targeting hepatic lipid production within the circadian window in which lipogenesis is normally suppressed could serve as a corrective measure. This concept has already been proposed for circadian control of glucose homeostasis. Ectopic induction of CRY1 suppressed gluconeogenesis and improved insulin sensitivity in a diabetic mouse model (135).
E. Heart: the distribution center
The heart is responsible for ensuring the continuous circulation of blood-borne oxygen, metabolites, and hormones required for coordinating appropriate organismal homeostasis. This workload on the heart increases significantly when animals are active, and therefore the clock has been configured to accordingly modulate cardiac metabolism throughout the day (149, 150) (Figure 2E). The circadian rhythm of human heart rate and blood pressure reaches its nadir, or lowest point, in the early hours of the morning corresponding to the point of deepest sleep and lowest core body temperature (151). However, before waking, both heart rate and blood pressure spike (151). The manner in which these physiological events precede the act of arousal and prepare the heart for a heightened workload exemplifies the anticipatory nature of circadian evolutionary programming.
This daily oscillation is facilitated metabolically by a transition between fuel sources. Cardiomyocytes undergo circadian-driven increases in glucose uptake and glycolysis as well as elevation in triglyceride synthesis immediately before and during the active waking hours (152–154). Similar results were seen in rats where hearts isolated during the dark phase, when rodent activity peaks, demonstrated the highest levels of contractile performance, carbohydrate oxidation, and oxygen consumption (155). The rhythm of these processes was largely ablated in cardiomyocyte-specific models of ClockΔ19 mutant expression or Bmal1 deletion (153, 156, 157), indicating a functional dependence on the heart clock itself and a direct role in cardiac disease. Downstream of the clock, the transcriptional regulator, Krüppel-like factor 15, contributes to the circadian control of cardiac repolarization, deficits of which play a causative role in the development of cardiac arrhythmias (158). Furthermore, the daily decrease or “dip” in blood pressure is essential, and individuals who maintain a constitutively elevated cardiac workload are at significantly higher risk for damage to peripheral organs and vasculature (159–161).
The evolutionary structuring of the heart clock to increase carbohydrate utilization and workload before arousal demonstrates a clear selective advantage in preparing animals for acute bursts of activity necessary for securing food or safety (162). However, in the context of modern society, that same circadian program has profoundly impacted the health of individuals undergoing acute or chronic sleep disruption, particularly in the form of shift work (163–166). Therapeutic intervention into the cardiac clock presents a risky proposition given the heart's critical role. To this end, perhaps the most viable option in improving cardiovascular circadian health may simply be to restore the robustness of the amplitude of the heart clock in individuals suffering from genetic or environmental circadian disturbances. This improvement could be exacted through small-molecule activation of the function of clock components. This functional activation would likely have to occur independent of protein stabilization because increased clock factor half-life could negatively impact periodicity.
F. Pancreas: the glycemic control center
Continuous maintenance of blood glucose levels within a narrow concentration range is absolutely critical for neuronal function and survival. Glycemic homeostasis is monitored and sustained by the coordinated release of insulin or glucagon from the pancreas to lower or raise blood glucose, respectively (Figure 2F). Given the sleep/wake, feeding/fasting, and active/inactive patterns that mammals exhibit, the biological clock has wired the pancreas to anticipate daily periods of glucose surges or drops (167–169). Indeed, pancreatic genes involved in the insulin production and secretion pathway oscillate (170), and circulating insulin levels have a circadian rhythm in both rodents (171) and humans (172, 173). In humans, this cycling reaches its lowest point during the early hours of the morning and peaks in the mid-to-late afternoon (172, 173). Although daily feeding patterns likely play an integral role in coordinating circadian insulin secretion, isolated rat islets exhibit a cell autonomous rhythm of insulin release, underscoring the importance of the intrinsic pancreatic clock (174).
Whole-body Bmal1 knockout and dominant negative ClockΔ19 mouse models demonstrate defects in β-cell proliferation and function and glucose-stimulated insulin secretion resulting in whole-animal glucose intolerance (175–177). One route through which BMAL1 is protective of β-cell function appears to be through preventing reactive oxygen species buildup via transcriptional induction in the nuclear factor erythroid 2-related factor 2 (Nrf2) master antioxidant regulatory factor (175, 177). The repressive arm of the clock also plays a role in the endocrine signaling from the pancreas. Disruption of Rev-erbα in β-cells and α-cells significantly attenuated glucose-stimulated insulin release (178) and low glucose-stimulated glucagon secretion (179), respectively. Conversely, glucose was more efficacious in stimulating insulin secretion in mice lacking PER2(180). Understanding how and when the clock most significantly influences insulin and glucagon secretion would be potentially informative in modulating appropriate dose concentrations throughout the day. This is particularly important given the advent of insulin analogs that are more long-lived than endogenous pancreatic insulin.
G. Skeletal muscle: the motion center
Skeletal muscle comprises the largest tissue mass in the human body and accounts for a majority of postprandial blood glucose disposal (181). Through contractile forces, skeletal muscle provides the essential means by which animals achieve movement, including the fundamental acts of survival such as seeking sustenance and shelter and avoiding predators and environmental hazards (Figure 2G). Skeletal muscle burns different fuels depending on the metabolic demand, relying more exclusively on glucose during fed conditions and resistance training but switching primarily to lipid utilization during fasting and prolonged endurance exercise (182).
Given the close relationship between physical activity and sleep/wake patterns, it is not surprising that numerous aspects of skeletal muscle bioenergetic programming are under the control of the clock in both mice (183, 184) and humans (185). Although earlier estimates identified 215 circadian genes in mouse skeletal muscle (186), more recent analyses have indicated that over 800 genes exhibit a circadian rhythm in the tissue (187). Genetic knockout models have been highly informative in uncovering the specific regulatory roles of the individual clock components in skeletal muscle metabolism.
Muscle-specific inducible deletion of BMAL1 revealed a striking deficiency in insulin-stimulated glucose uptake (188). These studies suggested that the activating arm of the clock is critical in orchestrating the switch in fuel source from lipid to glucose that occurs during the sleep-to-wake transition. Additionally, rescuing BMAL1 skeletal muscle expression in the context of a whole-animal knockout partially prevented decreases in overall activity and body weight and indicated that functional muscle rhythm has a profound impact on systemic energy homeostasis (189). Further underscoring the importance of clock control in muscle physiology is the finding that the key muscle differentiation transcription factor, MyoD, is circadian, and disruption of its normal expression pattern due to either ClockΔ19 mutation or Bmal1 deletion results in altered muscle structure and function (190).
The repressive arm of the clock also contributes to skeletal muscle circadian programming. Rev-erbα controls skeletal muscle lipid utilization, at least in part, through the direct transcriptional regulation of lipoprotein lipase as evidenced by whole-animal Rev-erbα deletion (106). Rev-erbα was also found to positively impact exercise capacity and oxidative metabolism by impinging on the LKB1-AMPK-SIRT1-PGC-1α axis (191). Interestingly, the transcriptional coactivator, PGC-1α, has previously been shown to induce the expression of both Bmal1 and Rev-erbα in skeletal muscle through the ROR nuclear receptors (192), ideally illustrating how the cross talk between the clock and metabolic networks goes in both directions.
Similar to other tissues such as brown adipose, the circadian control of skeletal muscle metabolism, particularly energy substrate selection, was likely evolved as a means of conservation. Carbohydrate utilization is lowered during the resting period, in which the animal is fasting, and lipid becomes the predominant energy source to preserve blood glucose levels for appropriate brain function. Before waking, the clock again signals an anticipatory transition that reinstates glucose as the chief fuel source readying the animal for physical activity (eg, food searching). Interestingly, the skeletal muscle peripheral clock not only dictates a rhythm of functional capacity but is itself subject to influence and synchronization by physical activity (193). This helps to explain how the sedentary lifestyle of modern man has dramatically impacted evolutionary-instated mechanisms of entrainment. The influence of physical activity on the skeletal muscle clock also indicates that exercise at different times of day may be more or less beneficial and therefore has significant implications for human health. Additionally, understanding the molecular mechanism of circadian skeletal muscle control may provide potent therapeutic avenues for metabolic disease. Determining the downstream machinery through which the skeletal muscle clock switches between fuel sources or modulates glucose uptake could provide potent, tissue-specific targets to release the evolutionary brakes to counteract diabetic hyperglycemia or elevate fat-oxidizing capacity.
H. Gut microbiota: the food-processing center
The human intestine is home to an enormous and highly diverse population of microorganisms, known as the gut microbiota, that has coevolved with its host to profoundly influence organismal energy homeostasis (194–196). The unique composition of the bacterial milieu confers different properties to the host, including influence over which nutrients can be extracted from foodstuffs (Figure 2H). Disruption of this delicate host/microbe balance can lead to severe pathophysiological consequences, such as cardiovascular disease (197) and obesity (198). Therefore, the intestinal/gut microbial axis is critical for how the host “sees” a meal. Given the circadian nature of food intake, it is not surprising that this dynamic is subject to regulation by the clock.
Indeed, the counterregulatory activities of clock components, RORα, and Rev-erbα contribute to circadian cross talk between the intestinal epithelial cells and the gut microbiota through the transcriptional regulation of toll-like receptors (199). The core clock also impacts the oscillation of bacterial species comprising the microbiota. Whole-body genetic disruption of the host clock through Per1 and -2 deletion resulted in loss of bacterial rhythm (200). Moreover, environmental disruption of mouse and human circadian rhythm by phase-shifting light:dark cycles and jet lag across time zones, respectively, significantly altered 24-hour bacterial patterning (200). Circadian rhythm of the gut microbiota is similarly influenced by dietary composition. HFD negatively impacts the diurnal cycling of the gut microbiota; however, time-restricting HFD to a portion of the dark, active period partially rescues normal oscillation (201). Combining multiple environmental disruptions and phase-shifting the light:dark cycles of mice fed a high-fat/high-sugar diet further compounded the detrimental effects on the gut microbiome (202).
From a therapeutic perspective, the gut microbiome offers unique avenues for developing novel treatment strategies (203). Proof-of-concept studies have shown that bacteria that were genetically engineered to produce a beneficial metabolite were capable of reducing adiposity, insulin resistance, and hepatic steatosis in obese mice (204). Similar probiotic approaches could be used to correct, re-establish, or maintain appropriate rhythm in the gut flora population. The gut microbiome and its circadian patterning likely coevolved with the host to maximize calorie extraction, given the relative scarcity of food availability for thousands of years. Additional investigation of the microbiome composition at different times of day (200) could reveal specific oscillating bacterial subtypes responsible for diurnal variation in the capacity for processing foodstuffs.
III. Conclusion
A central thesis of this review is that evolutionarily selected circadian clock mechanisms are impacted by modern lifestyles. This new challenge raises the possibility of circadian-based therapeutic strategies that modulate the activities of core clock components. For example, Rev-erbα agonists have been suggested to increase energy expenditure and reduce adiposity in diet-induced obese mouse models (205), enhance exercise capacity (206), and suppress anxiety-like behavior (80). Nevertheless, despite these potential beneficial effects, targeting core clock components carries a risk, given their functional presence in every cell of the body. The diverse metabolic roles of clock factors in various tissues further complicate the systemic use of ubiquitous agonists or antagonists.
To this end, delving deeper into the tissue-specific mechanisms through which the clock controls key metabolic pathways offers a selective advantage with lower risk for adverse side effects. Importantly, taking the evolutionary purpose of various clock-controlled functions into account provides a valuable filter through which to pinpoint the networks that might be most clinically efficacious. For example, can we develop targeted therapeutic strategies to stop clock-mediated down-regulation of brown fat heat production, increase muscle glucose sensitivity throughout the day, or attenuate aberrant hepatic lipogenesis in individuals with sleep disruption? Unbiased small-molecule and small interfering RNA screening could be used to identify key, tissue-specific surface receptors and intracellular factors that mediate circadian metabolic control in a similar manner as has been previously employed for more general oscillatory modulators (207–210). This strategy has the potential to yield highly potent and selective novel therapies without global detriment to the core clock.
Not only is this concept feasible, but the likelihood is that many commercially available drugs are already functioning in this very manner. In fact, more than 50% of the best-selling drugs in the United States target a circadian gene product (211). This finding demonstrates the profound importance of determining the most appropriate temporal window of therapeutic opportunity for each for drug target. Indeed, reevaluation of dosage and time of delivery for existing drugs that target circadian metabolic programs may improve efficacy or reduce negative side effects.
Evolution has installed in us a powerful internal clock programmed to anticipate physiological events based on a battery of entrainment cues, including light, temperature, food, and physical activity. Unfortunately, the infrastructure of today's society and our lifestyle choices have “thrown a wrench” into our biological clockwork. To this end, uncovering the molecular underpinnings through which each tissue's circadian metabolism is coordinated, we may be able to tinker with the evolutionary toolbox and bring the clock up to modern times.
Acknowledgments
We thank Dr Anne Bugge for critical reading of the manuscript.
Research on circadian rhythm in the authors' laboratories was supported by National Institutes of Health Grant R01 DK45586 (to M.A.L.), the JPB Foundation (to M.A.L.), National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant F-32 DK095563-02 (to Z.G.-H.), Novo Nordisk Foundation Project Grant (to Z.G.-H.), the Danish Council for Independent Research Sapere Aude Starting Grant 4002-00024B FSS (to Z.G.-H.), and the European Research Council Starting Grant 639382 aCROBAT (to Z.G.-H.).
Disclosure Summary: The authors have nothing to declare.
Footnotes
- AMPK
- AMP-activated protein kinase
- BAT
- brown adipose tissue
- BMAL1
- brain and muscle Arnt-like protein-1
- CLOCK
- circadian locomotor output cycles kaput
- CRY
- cryptochrome
- HFD
- high-fat diet
- PER
- period
- RORα and -γ
- RAR-orphan receptor α and γ
- SCN
- suprachiasmatic nucleus
- SIRT1
- sirtuin 1.
References
- 1. Dobzhansky T. Nothing in biology makes sense except in the light of evolution. Am Biol Teach. 1973;35(3):125–129. [Google Scholar]
- 2. Simons MJ. The evolution of the cyanobacterial posttranslational clock from a primitive “phoscillator”. J Biol Rhythms. 2009;24(3):175–182. [DOI] [PubMed] [Google Scholar]
- 3. Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14(16):1481–1486. [DOI] [PubMed] [Google Scholar]
- 4. Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shearman LP. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–1019. [DOI] [PubMed] [Google Scholar]
- 7. Robinson I, Reddy AB. Molecular mechanisms of the circadian clockwork in mammals. FEBS Lett. 2014;588(15):2477–2483. [DOI] [PubMed] [Google Scholar]
- 8. Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4(8):649–661. [DOI] [PubMed] [Google Scholar]
- 9. Moore RY. Organization and function of a central nervous system circadian oscillator: the suprachiasmatic hypothalamic nucleus. Fed Proc. 1983;42(11):2783–2789. [PubMed] [Google Scholar]
- 10. King DP, Zhao Y, Sangoram AM, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vitaterna MH, King DP, Chang AM, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hogenesch JB, Chan WK, Jackiw VH, et al. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272(13):8581–8593. [DOI] [PubMed] [Google Scholar]
- 13. Ikeda M, Nomura M. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem Biophys Res Commun. 1997;233(1):258–264. [DOI] [PubMed] [Google Scholar]
- 14. Todo T, Ryo H, Yamamoto K, et al. Similarity among the Drosophila (6–4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272(5258):109–112. [DOI] [PubMed] [Google Scholar]
- 15. Kobayashi K, Kanno S, Smit B, van der Horst GT, Takao M, Yasui A. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res. 1998;26(22):5086–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68(9):2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyajima N, Horiuchi R, Shibuya Y, et al. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell. 1989;57(1):31–39. [DOI] [PubMed] [Google Scholar]
- 18. Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA α transcriptional unit. Mol Cell Biol. 1989;9(3):1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dumas B, Harding HP, Choi HS, et al. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol Endocrinol. 1994;8(8):996–1005. [DOI] [PubMed] [Google Scholar]
- 20. Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. [DOI] [PubMed] [Google Scholar]
- 21. Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. [DOI] [PubMed] [Google Scholar]
- 23. Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12(5):441–448. [DOI] [PubMed] [Google Scholar]
- 24. Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20(5):391–403. [DOI] [PubMed] [Google Scholar]
- 25. Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIδ and CKIϵ in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106(50):21359–21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94(1):83–95. [DOI] [PubMed] [Google Scholar]
- 27. Kloss B, Price JL, Saez L, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iϵ. Cell. 1998;94(1):97–107. [DOI] [PubMed] [Google Scholar]
- 28. Siepka SM, Yoo SH, Park J, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129(5):1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoo SH, Mohawk JA, Siepka SM, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152(5):1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8(2):139–148. [DOI] [PubMed] [Google Scholar]
- 31. Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103(6):2203–2237. [DOI] [PubMed] [Google Scholar]
- 32. Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol. 2003;57(suppl 1):S286–S289. [DOI] [PubMed] [Google Scholar]
- 33. Emery P, Stanewsky R, Helfrich-Förster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26(2):493–504. [DOI] [PubMed] [Google Scholar]
- 34. Czarna A, Berndt A, Singh HR, et al. Structures of Drosophila cryptochrome and mouse cryptochrome 1 provide insight into circadian function. Cell. 2013;153(6):1394–1405. [DOI] [PubMed] [Google Scholar]
- 35. Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 2003;54:469–496. [DOI] [PubMed] [Google Scholar]
- 36. Chaves I, Yagita K, Barnhoorn S, Okamura H, van der Horst GT, Tamanini F. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol Cell Biol. 2006;26(5):1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. [DOI] [PubMed] [Google Scholar]
- 38. Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. [DOI] [PubMed] [Google Scholar]
- 39. Ko CH. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. [DOI] [PubMed] [Google Scholar]
- 40. Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang B, Everett LJ, Jager J, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159(5):1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morf J, Rey G, Schneider K, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338(6105):379–383. [DOI] [PubMed] [Google Scholar]
- 43. Koike N, Yoo SH, Huang HC, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reddy AB, Karp NA, Maywood ES, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. [DOI] [PubMed] [Google Scholar]
- 45. Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mauvoisin D, Dayon L, Gachon F, Kussmann M. Proteomics and circadian rhythms: it's all about signaling! Proteomics. 2015;15:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. [DOI] [PubMed] [Google Scholar]
- 48. Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293(5529):510–514. [DOI] [PubMed] [Google Scholar]
- 49. Ramsey KM, Yoshino J, Brace CS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. [DOI] [PubMed] [Google Scholar]
- 52. Peek CB, Affinati AH, Ramsey KM, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342(6158):1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol. 2007;14(12):1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yin L, Wu N, Curtin JC, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–1789. [DOI] [PubMed] [Google Scholar]
- 55. Lamia KA, Sachdeva UM, DiTacchio L, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Edgar RS, Green EW, Zhao Y, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Neill JS, van Ooijen G, Dixon LE, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469(7331):554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. O'Neill JS, Feeney KA. Circadian redox and metabolic oscillations in mammalian systems. Antioxid Redox Signal. 2014;20(18):2966–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9(12):886–896. [DOI] [PubMed] [Google Scholar]
- 61. Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121(6):2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113(3):103–112. [DOI] [PubMed] [Google Scholar]
- 63. Wang XS, Armstrong ME, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med (Lond). 2011;61(2):78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sack RL, Auckley D, Auger RR, et al. Circadian rhythm sleep disorders: part II, advanced sleep phase disorder, delayed sleep phase disorder, free-running disorder, and irregular sleep-wake rhythm. An American Academy of Sleep Medicine review. Sleep. 2007;30(11):1484–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sack RL, Auckley D, Auger RR, et al. Circadian rhythm sleep disorders: part I, basic principles, shift work and jet lag disorders. An American Academy of Sleep Medicine review. Sleep. 2007;30(11):1460–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med. 2011;8(12):e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suwazono Y, Dochi M, Oishi M, Tanaka K, Kobayashi E, Sakata K. Shiftwork and impaired glucose metabolism: a 14-year cohort study on 7104 male workers. Chronobiol Int. 2009;26(5):926–941. [DOI] [PubMed] [Google Scholar]
- 68. Roenneberg T, Allebrandt KV, Merrow M, Vetter C. Social jetlag and obesity. Curr Biol. 2012;22(10):939–943. [DOI] [PubMed] [Google Scholar]
- 69. Beihl DA, Liese AD, Haffner SM. Sleep duration as a risk factor for incident type 2 diabetes in a multiethnic cohort. Ann Epidemiol. 2009;19(5):351–357. [DOI] [PubMed] [Google Scholar]
- 70. Meisinger C, Heier M, Loewel H. Sleep disturbance as a predictor of type 2 diabetes mellitus in men and women from the general population. Diabetologia. 2005;48(2):235–241. [DOI] [PubMed] [Google Scholar]
- 71. Spiegel K, Leproult R, Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354(9188):1435–1439. [DOI] [PubMed] [Google Scholar]
- 72. Güler AD, Ecker JL, Lall GS, et al. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453(7191):102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: an opsin in melanophores, brain, and eye. Proc Natl Acad Sci USA. 1998;95(1):340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bartness TJ, Song CK, Demas GE. SCN efferents to peripheral tissues: implications for biological rhythms. J Biol Rhythms. 2001;16(3):196–204. [DOI] [PubMed] [Google Scholar]
- 75. Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA. 1972;69(6):1583–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42(1):201–206. [DOI] [PubMed] [Google Scholar]
- 77. Yamamoto H, Nagai K, Nakagawa H. Role of SCN in daily rhythms of plasma glucose, FFA, insulin and glucagon. Chronobiol Int. 1987;4(4):483–491. [DOI] [PubMed] [Google Scholar]
- 78. La Fleur SE, Kalsbeek A, Wortel J, Buijs RM. A suprachiasmatic nucleus generated rhythm in basal glucose concentrations. J Neuroendocrinol. 1999;11(8):643–652. [DOI] [PubMed] [Google Scholar]
- 79. Tousson E, Meissl H. Suprachiasmatic nuclei grafts restore the circadian rhythm in the paraventricular nucleus of the hypothalamus. J Neurosci. 2004;24(12):2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Banerjee S, Wang Y, Solt LA, et al. Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat Commun. 2014;5:5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dashti HS, Follis JL, Smith CE, et al. Habitual sleep duration is associated with BMI and macronutrient intake and may be modified by CLOCK genetic variants. Am J Clin Nutr. 2015;101(1):135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goumidi L, Grechez A, Dumont J, et al. Impact of REV-ERB α gene polymorphisms on obesity phenotypes in adult and adolescent samples. Int J Obes (Lond). 2013;37(5):666–672. [DOI] [PubMed] [Google Scholar]
- 83. Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond). 2008;32(4):658–662. [DOI] [PubMed] [Google Scholar]
- 84. Sookoian S, Gemma C, Gianotti TF, Burgueño A, Castaño G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87(6):1606–1615. [DOI] [PubMed] [Google Scholar]
- 85. Sookoian S, Castaño G, Gemma C, Gianotti TF, Pirola CJ. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol. 2007;13(31):4242–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. [DOI] [PubMed] [Google Scholar]
- 87. Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404(6778):652–660. [DOI] [PubMed] [Google Scholar]
- 88. van der Veen DR, Shao J, Chapman S, Leevy WM, Duffield GE. A diurnal rhythm in glucose uptake in brown adipose tissue revealed by in vivo PET-FDG imaging. Obesity (Silver Spring). 2012;20(7):1527–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zvonic S, Ptitsyn AA, Conrad SA, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55(4):962–970. [DOI] [PubMed] [Google Scholar]
- 90. Redlin U, Nuesslein B, Schmidt I. Circadian changes of brown adipose tissue thermogenesis in juvenile rats. Am J Physiol. 1992;262:R504–R508. [DOI] [PubMed] [Google Scholar]
- 91. Gerhart-Hines Z, Feng D, Emmett MJ, et al. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 2013;503(7476):410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chappuis S, Ripperger JA, Schnell A, et al. Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol Metab. 2013;2(3):184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nam D, Guo B, Chatterjee S, et al. The adipocyte clock controls brown adipogenesis via TGF-β/BMP signaling pathway [published online March 6, 2015]. J Cell Sci. doi:10.1242/jcs.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li S, Yu Q, Wang GX, Lin JD. The biological clock is regulated by adrenergic signaling in brown fat but is dispensable for cold-induced thermogenesis. PLoS One. 2013;8(8):e70109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yang YL, Shen ZL, Tang Y, Wang N, Sun B. Simultaneous telemetric analyzing of the temporal relationship for the changes of the circadian rhythms of brown adipose tissue thermogenesis and core temperature in the rat [in Chinese]. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2011;27(3):348–352. [PubMed] [Google Scholar]
- 96. Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26(6):567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Oelkrug R, Goetze N, Exner C, et al. Brown fat in a protoendothermic mammal fuels eutherian evolution. Nat Commun. 2013;4:2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281(5726):31–35. [DOI] [PubMed] [Google Scholar]
- 100. Shostak A, Meyer-Kovac J, Oster H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes. 2013;62(7):2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Paschos GK, Ibrahim S, Song WL, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18(12):1768–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hagström-Toft E, Bolinder J, Ungerstedt U, Arner P. A circadian rhythm in lipid mobilization which is altered in IDDM. Diabetologia. 1997;40(9):1070–1078. [DOI] [PubMed] [Google Scholar]
- 103. Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Grimaldi B, Bellet MM, Katada S, et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12(5):509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Delezie J, Dumont S, Dardente H, et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26(8):3321–3335. [DOI] [PubMed] [Google Scholar]
- 107. Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;89(6):2548–2556. [DOI] [PubMed] [Google Scholar]
- 108. Halaas JL, Gajiwala KS, Maffei M, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269(5223):543–546. [DOI] [PubMed] [Google Scholar]
- 109. Steppan CM, Bailey ST, Bhat S, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409(6818):307–312. [DOI] [PubMed] [Google Scholar]
- 110. Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270(45):26746–26749. [DOI] [PubMed] [Google Scholar]
- 111. Ando H, Yanagihara H, Hayashi Y, et al. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146(12):5631–5636. [DOI] [PubMed] [Google Scholar]
- 112. van der Spek R, Kreier F, Fliers E, Kalsbeek A. Circadian rhythms in white adipose tissue. Prog Brain Res. 2012;199:183–201. [DOI] [PubMed] [Google Scholar]
- 113. Downs JL, Urbanski HF. Aging-related sex-dependent loss of the circulating leptin 24-h rhythm in the rhesus monkey. J Endocrinol. 2006;190(1):117–127. [DOI] [PubMed] [Google Scholar]
- 114. Kalra SP, Bagnasco M, Otukonyong EE, Dube MG, Kalra PS. Rhythmic, reciprocal ghrelin and leptin signaling: new insight in the development of obesity. Regul Pept. 2003;111:1–11. [DOI] [PubMed] [Google Scholar]
- 115. Heptulla R, Smitten A, Teague B, Tamborlane WV, Ma YZ, Caprio S. Temporal patterns of circulating leptin levels in lean and obese adolescents: relationships to insulin, growth hormone, and free fatty acids rhythmicity. J Clin Endocrinol Metab. 2001;86(1):90–96. [DOI] [PubMed] [Google Scholar]
- 116. Sinha MK, Ohannesian JP, Heiman ML, et al. Nocturnal rise of leptin in lean, obese, and non-insulin-dependent diabetes mellitus subjects. J Clin Invest. 1996;97(5):1344–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kalsbeek A, Fliers E, Romijn JA, et al. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology. 2001;142(6):2677–2685. [DOI] [PubMed] [Google Scholar]
- 118. Ahima RS, Prabakaran D, Flier JS. Postnatal leptin surge and regulation of circadian rhythm of leptin by feeding. Implications for energy homeostasis and neuroendocrine function. J Clin Invest. 1998;101(5):1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gavrila A, Peng CK, Chan JL, Mietus JE, Goldberger AL, Mantzoros CS. Diurnal and ultradian dynamics of serum adiponectin in healthy men: comparison with leptin, circulating soluble leptin receptor, and cortisol patterns. J Clin Endocrinol Metab. 2003;88(6):2838–2843. [DOI] [PubMed] [Google Scholar]
- 120. Scheer FA, Chan JL, Fargnoli J, et al. Day/night variations of high-molecular-weight adiponectin and lipocalin-2 in healthy men studied under fed and fasted conditions. Diabetologia. 2010;53(11):2401–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hara R, Wan K, Wakamatsu H, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6(3):269–278. [DOI] [PubMed] [Google Scholar]
- 122. Damiola F. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–493. [DOI] [PubMed] [Google Scholar]
- 124. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bookout AL, de Groot MH, Owen BM, et al. FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat Med. 2013;19(9):1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Balsalobre A, Brown SA, Marcacci L, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–2347. [DOI] [PubMed] [Google Scholar]
- 127. Kida K, Nishio T, Yokozawa T, Nagai K, Matsuda H, Nakagawa H. The circadian change of gluconeogenesis in the liver in vivo in fed rats. J Biochem (Tokyo). 1980;88(4):1009–1013. [DOI] [PubMed] [Google Scholar]
- 128. Kudo T, Tamagawa T, Kawashima M, Mito N, Shibata S. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms. 2007;22(4):312–323. [DOI] [PubMed] [Google Scholar]
- 129. Rudic RD, McNamara P, Curtis AM, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105(39):15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kennaway DJ, Owens JA, Voultsios A, Boden MJ, Varcoe TJ. Metabolic homeostasis in mice with disrupted Clock gene expression in peripheral tissues. Am J Physiol Regul Integr Comp Physiol. 2007;293(4):R1528–R1537. [DOI] [PubMed] [Google Scholar]
- 132. Kennaway DJ, Varcoe TJ, Voultsios A, Boden MJ. Global loss of bmal1 expression alters adipose tissue hormones, gene expression and glucose metabolism. PloS One. 2013;8(6):e65255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Doi R, Oishi K, Ishida N. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J Biol Chem. 2010;285(29):22114–22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zani F, Breasson L, Becattini B, et al. PER2 promotes glucose storage to liver glycogen during feeding and acute fasting by inducing Gys2 PTG and G L expression. Mol Metab. 2013;2(3):292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhang EE, Liu Y, Dentin R, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16(10):1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lamia KA, Papp SJ, Yu RT, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bugge A, Feng D, Everett LJ, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485(7396):123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Duez H, van der Veen JN, Duhem C, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology. 2008;135(2):689–698. [DOI] [PubMed] [Google Scholar]
- 140. Le Martelot G, Claudel T, Gatfield D, et al. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9(2):e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Liu S, Brown JD, Stanya KJ, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502(7472):550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kohsaka A, Laposky AD, Ramsey KM, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. [DOI] [PubMed] [Google Scholar]
- 144. Brunt EM. Pathology of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7(4):195–203. [DOI] [PubMed] [Google Scholar]
- 145. Satoh Y. Time-restricted feeding entrains daily rhythms of energy metabolism in mice. Am J Physiol Regul Integr Comp Physiol. 2006;290(5):R1276–R1283. [DOI] [PubMed] [Google Scholar]
- 146. Kudo T, Akiyama M, Kuriyama K, Sudo M, Moriya T, Shibata S. Night-time restricted feeding normalises clock genes and Pai-1 gene expression in the db/db mouse liver. Diabetologia. 2004;47(8):1425–1436. [DOI] [PubMed] [Google Scholar]
- 147. Hatori M, Vollmers C, Zarrinpar A, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Chaix A, Zarrinpar A, Miu P, Panda S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20(6):991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bray MS, Young ME. Diurnal variations in myocardial metabolism. Cardiovasc Res. 2008;79(2):228–237. [DOI] [PubMed] [Google Scholar]
- 150. Young ME. The circadian clock within the heart: potential influence on myocardial gene expression, metabolism, and function. Am J Physiol Heart Circ Physiol. 2006;290(1):H1–H16. [DOI] [PubMed] [Google Scholar]
- 151. Degaute JP, van de Borne P, Linkowski P, Van Cauter E. Quantitative analysis of the 24-hour blood pressure and heart rate patterns in young men. Hypertension. 1991;18(2):199–210. [DOI] [PubMed] [Google Scholar]
- 152. Durgan DJ, Moore MW, Ha NP, et al. Circadian rhythms in myocardial metabolism and contractile function: influence of workload and oleate. Am J Physiol Heart Circ Physiol. 2007;293(4):H2385–H2393. [DOI] [PubMed] [Google Scholar]
- 153. Durgan DJ, Pat BM, Laczy B, et al. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem. 2011;286(52):44606–44619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tsai JY, Kienesberger PC, Pulinilkunnil T, et al. Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem. 2010;285(5):2918–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Portman MA. Molecular clock mechanisms and circadian rhythms intrinsic to the heart. Circ Res. 2001;89(12):1084–1086. [PubMed] [Google Scholar]
- 156. Bray MS, Shaw CA, Moore MW, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008;294(2):H1036–H1047. [DOI] [PubMed] [Google Scholar]
- 157. Durgan DJ, Tsai JY, Grenett MH, et al. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int. 2011;28(3):187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Jeyaraj D, Haldar SM, Wan X, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483(7387):96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Giles TD. Circadian rhythm of blood pressure and the relation to cardiovascular events. J Hypertens Suppl. 2006;24(2):S11–S16. [DOI] [PubMed] [Google Scholar]
- 160. White WB. Importance of blood pressure control over a 24-hour period. J Manag Care Pharm. 2007;13(8 suppl B):34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Izzedine H, Launay-Vacher V, Deray G. Abnormal blood pressure circadian rhythm: a target organ damage? Int J Cardiol. 2006;107(3):343–349. [DOI] [PubMed] [Google Scholar]
- 162. Chatham JC, Young ME. Regulation of myocardial metabolism by the cardiomyocyte circadian clock. J Mol Cell Cardiol. 2013;55:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Souza BB, Monteze NM, de Oliveira FL, et al. Lifetime shift work exposure: association with anthropometry, body composition, blood pressure, glucose and heart rate variability. Occup Environ Med. 2015;72:208–215. [DOI] [PubMed] [Google Scholar]
- 164. Ito H, Nozaki M, Maruyama T, Kaji Y, Tsuda Y. Shift work modifies the circadian patterns of heart rate variability in nurses. Int J Cardiol. 2001;79:231–236. [DOI] [PubMed] [Google Scholar]
- 165. Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106(11):4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kroenke CH, Spiegelman D, Manson J, Schernhammer ES, Colditz GA, Kawachi I. Work characteristics and incidence of type 2 diabetes in women. Am J Epidemiol. 2007;165(2):175–183. [DOI] [PubMed] [Google Scholar]
- 167. Lamia KA, Evans RM. Metabolism: tick, tock, a β-cell clock. Nature. 2010;466(7306):571–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Marcheva B, Ramsey KM, Bass J. Circadian genes and insulin exocytosis. Cell Logist. 2011;1(1):32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Mühlbauer E, Wolgast S, Finckh U, Peschke D, Peschke E. Indication of circadian oscillations in the rat pancreas. FEBS Lett. 2004;564:91–96. [DOI] [PubMed] [Google Scholar]
- 170. Allaman-Pillet N, Roduit R, Oberson A, et al. Circadian regulation of islet genes involved in insulin production and secretion. Mol Cell Endocrinol. 2004;226:59–66. [DOI] [PubMed] [Google Scholar]
- 171. Ahlersová E, Ahlers I, Milárová R, Datelinka I, Toropila M. Circadian oscillations of thyroid hormones, insulin and glucagon in the blood of laboratory rats in the course of the year. Physiol Bohemoslov. 1984;33(4):309–319. [PubMed] [Google Scholar]
- 172. Boden G, Ruiz J, Urbain JL, Chen X. Evidence for a circadian rhythm of insulin secretion. Am J Physiol. 1996;271:E246–E252. [DOI] [PubMed] [Google Scholar]
- 173. Boden G, Chen X, Urbain JL. Evidence for a circadian rhythm of insulin sensitivity in patients with NIDDM caused by cyclic changes in hepatic glucose production. Diabetes. 1996;45(8):1044–1050. [DOI] [PubMed] [Google Scholar]
- 174. Peschke E, Peschke D. Evidence for a circadian rhythm of insulin release from perifused rat pancreatic islets. Diabetologia. 1998;41(9):1085–1092. [DOI] [PubMed] [Google Scholar]
- 175. Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54(1):120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lee J, Moulik M, Fang Z, et al. Bmal1 and β-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced β-cell failure in mice. Mol Cell Biol. 2013;33(11):2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Vieira E, Marroquí L, Batista TM, et al. The clock gene Rev-erbα regulates pancreatic β-cell function: modulation by leptin and high-fat diet. Endocrinology. 2012;153(2):592–601. [DOI] [PubMed] [Google Scholar]
- 179. Vieira E, Marroquí L, Figueroa AL, et al. Involvement of the clock gene Rev-erb α in the regulation of glucagon secretion in pancreatic α-cells. PLoS One. 2013;8(7):e69939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhao Y, Zhang Y, Zhou M, Wang S, Hua Z, Zhang J. Loss of mPer2 increases plasma insulin levels by enhanced glucose-stimulated insulin secretion and impaired insulin clearance in mice. FEBS Lett. 2012;586(9):1306–1311. [DOI] [PubMed] [Google Scholar]
- 181. Thiebaud D, Jacot E, DeFronzo RA, Maeder E, Jequier E, Felber JP. The effect of graded doses of insulin on total glucose uptake, glucose oxidation, and glucose storage in man. Diabetes. 1982;31(11):957–963. [DOI] [PubMed] [Google Scholar]
- 182. Kelley DE, Goodpaster BH, Storlien L. Muscle triglyceride and insulin resistance. Annu Rev Nutr. 2002;22:325–346. [DOI] [PubMed] [Google Scholar]
- 183. Harfmann BD, Schroder EA, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. J Biol Rhythms. 2015;30:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Lefta M, Wolff G, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. Curr Top Dev Biol. 2011;96:231–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Zambon AC, McDearmon EL, Salomonis N, et al. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003;4(10):R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. McCarthy JJ, Andrews JL, McDearmon EL, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31(1):86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–D1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Dyar KA, Ciciliot S, Wright LE, et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab. 2014;3(1):29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. McDearmon EL, Patel KN, Ko CH, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314(5803):1304–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Andrews JL, Zhang X, McCarthy JJ, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA. 2010;107(44):19090–19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Woldt E, Sebti Y, Solt LA, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. [DOI] [PubMed] [Google Scholar]
- 193. Schroder EA, Esser KA. Circadian rhythms, skeletal muscle molecular clocks, and exercise. Exerc Sport Sci Rev. 2013;41(4):224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. [DOI] [PubMed] [Google Scholar]
- 195. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. [DOI] [PubMed] [Google Scholar]
- 196. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. [DOI] [PubMed] [Google Scholar]
- 199. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–827. [DOI] [PubMed] [Google Scholar]
- 200. Thaiss CA, Zeevi D, Levy M, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–529. [DOI] [PubMed] [Google Scholar]
- 201. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20(6):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Voigt RM, Forsyth CB, Green SJ, et al. Circadian disorganization alters intestinal microbiota. PLoS One. 2014;9(5):e97500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Cani PD, Delzenne NM. The gut microbiome as therapeutic target. Pharmacol Ther. 2011;130(2):202–212. [DOI] [PubMed] [Google Scholar]
- 204. Chen Z, Guo L, Zhang Y, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest. 2014;124(8):3391–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Solt LA, Wang Y, Banerjee S, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Woldt E, Sebti Y, Solt LA, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19(8):1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Hirota T, Lee JW, Lewis WG, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 2010;8(12):e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Maier B, Wendt S, Vanselow JT, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23(6):708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Zhang EE, Liu AC, Hirota T, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139(1):199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Chen Z, Yoo SH, Park YS, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA. 2012;109(1):101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111(45):16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]