Abstract
A new study shows that microRNA-9 regulates multiple processes near the organizing centers during early brain development in zebrafish, revealing previously unknown modes of action for microRNAs in the nervous system.
Some tiny RNAs, known as microRNAs (miRNAs) and barely noticed by scientists a few years ago, have recently emerged as important regulators of gene expression during animal development1, 2. Mature miRNAs are endogenous ~21-nucleotide noncoding RNAs that regulate mRNA translation or stability through sequence-specific base pairing with target 3′ untranslated regions (3′ UTRs)3. Vertebrate genomes contain hundreds of miRNAs. Some are specifically expressed in developing and mature nervous systems. Studies in several model systems have just begun to unravel specific roles for individual miRNAs in different aspects of neuronal development, including cell-fate determination in C. elegans, sensory organ precursor specification in Drosophila and morphological differentiation of mammalian neurons4. Although the nucleotide sequences of many miRNAs are highly conserved across different species, the extent of their functional conservation is unknown.
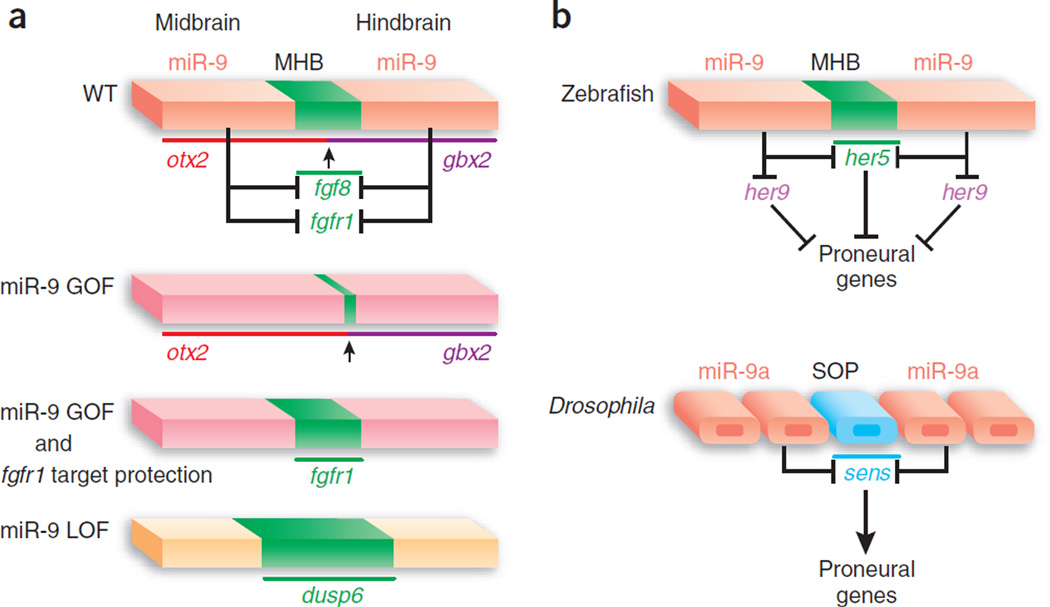
In this issue, Leucht et al.5 report that a specific miRNA, miR-9, is expressed in regions adjacent to the midbrain-hindbrain boundary (MHB), but not in cells in the MHB itself, and that it restricts the organizing activity of the MHB by targeting the fibroblast growth factor (Fgf ) signaling pathway (Fig. 1a). Moreover, miR-9 promotes neurogenesis in the midbrain-hindbrain (MH) domain near the MHB in the developing zebrafish brain5. These findings uncover multiple previously unknown functions for a single miRNA and provide further evidence for the crucial roles of miRNAs in controlling the precise gene expression that is essential for normal brain development.
Figure 1.
The functions of miR-9 near the MHB. (a) miR-9 delimits the MHB through inhibition of fgf8 and fgfr1. The Fgf signaling pathway is highly active in the MHB (highlighted in green) and miR-9 is expressed in adjacent domains (highlighted in pink and orange) but not in the MHB. The bars above each gene indicate their expression domains. GOF, gain of function; LOF, loss of function. (b) Differential actions of miR-9 in zebrafish and Drosophila. In fish, miR-9 suppresses the expression of its anti-target her5 and its target her9, which are suppressors of neurogenesis. In fly, miR-9a downregulates Senseless (sens), whose high level of expression in the presumptive sensory organ precursors promotes the formation of neuronal precursors. In both cases, miR-9 expression avoids the progenitor pool.
Several potent secreted signaling molecules are under tight spatial and temporal regulation during the regional specification of the neural plate6. For example, Fgf8 promotes midbrain development7 and is specifically expressed in cells at the MHB. The MHB functions as an organizing center to specify the tectum at its rostral side and the cerebellum at its caudal side. Fgf8 mediates some of the MHB’s patterning activities and maintains the MHB mainly via its receptor, Fgfr1. The strength of Fgf signaling is crucial for determining whether tectum or cerebellum tissues differentiate. Because Fgf8 is such a potent signaling molecule in the organizing center, its activity must be tightly controlled in the embryo. Both positive feedback loops involving Wnt1, Engrailed1/2 and Pax2/5 in the MHB and inhibitory feedback loops involving Sprouty around the MHB are employed to regulate the Fgf pathway8. The new findings of Leucht et al.5 indicate that miR-9 provides an additional, critical layer of regulation.
Leucht et al.5 first took a gain-of-function approach by injecting miR-9 duplex or miR-9-1 precursor (pre–miR-9-1) into zebrafish oocytes. A marked phenotype resulted: the size of the MHB was substantially reduced, as was the cerebellum5. The specificity of the phenotype was confirmed by a decrease in the expression of several MHB marker genes. Notably, this defect was similar to phenotypes observed in fgf8 (ace) mutants and fgfr1 knockdown embryos. Leucht et al.5 found that several key genes in the Fgf pathway, such as fgf8, fgfr1 and canopy1, contain putative targets for miR-9 in their 3′ UTRs, raising the possibility that overexpressed miR-9 downregulates the Fgf signaling pathway. Indeed, expression of a reporter gene that was injected into one-celled embryos was suppressed by miR-9 through the 3′ UTRs of fgf8, fgfr1 and canopy1.
Misregulation of the Fgf pathway underlies the observed defects in miR-9–overexpressing embryos (Fig. 1a). First, Fgf target genes such as dusp6 and pea3 were downregulated. Second, the midbrain marker otx2 was aberrantly expressed in the posterior part of the MH domain, similarly to fgf8 mutants. The homeobox genes otx2 and gbx2 regulate the establishment of the MHB at the boundaries of their expression; therefore, expression of otx2 in posterior MH progenitors may account for their conversion to alternative fates8. Finally, morpholino antisense oligonucleotides directed against the predicted miR-9 binding sites in the fgfr1 3′ UTR were injected into one-celled embryos to block the effect of overexpressed miR-9 duplex on fgfr1 3′ UTR. These ‘target protectors’ were sufficient to rescue the MHB defect and restore the expression of the MHB marker genes, identifying fgfr1 mRNA as a key in vivo target of miR-9.
An interesting twist in the story arises from the observation that endogenous miR-9 is expressed in the surrounding regions, but not in the MHB itself. This finding is consistent with the hypothesis that some genes, as so-called ‘anti-targets’, have evolved to be expressed in adjacent domains to avoid miRNA suppression9, 10. Therefore, we could conclude that miR-9 expressed in the surrounding regions functions to define the limits of the MHB. Indeed, when miR-9 was inactivated by a morpholino oligonucleotide, the levels of fgf8, fgfr1 and canopy1 mRNAs were increased. Moreover, the expression domain or level of some downstream Fgf target genes, such as dusp6 and pea3, was expanded or increased, indicating enhanced Fgf signaling.
Another important contribution by Leucht et al.5 is the finding that miR-9 simultaneously regulates multiple developmental processes; it not only delimits the MHB, but also promotes neurogenesis near this organizing center. The MHB contains a pool of progenitor cells that show delayed neuronal differentiation compared with other domains of the neural tube as a result of an active process of neurogenesis inhibition8. Her family genes, encoding homologs of the Drosophila basic helix-loop-helix transcription factors Hairy and Enhancer of split, are required to maintain the progenitor pool and inhibit neuronal differentiation within the MHB11. In addition to components of the Fgf pathway, Leucht et al.5 also identified her5 and her9 mRNAs as potential in vivo targets of miR-9, as shown by miR-9’s ability to repress the expression of a reporter gene through 3′ UTR elements of her5 and her9.
As they were analyzing the effects of the widespread overexpression of miR-9 on MHB function and maintenance during early embryogenesis, Leucht et al.5 observed that the phenotype could not be explained solely by the misregulation of the Fgf pathway. Because her5 and her9 are also potential targets of miR-9, the authors went on to examine defects in neurogenesis and found that either miR-9 overexpression or loss of her5 and her9 activity induced ectopic activation of the neurogenesis-promoting transcription factor neurogenin1 in the MHB. Moreover, the number of cells in the MH domain that are positive for HuC, a marker for postmitotic neurons, was markedly increased. These observations support the notion that miR-9 promotes MH neurogenesis.
Leucht et al.5 also tested whether the derepression of her5 in the miR-9–overexpressing embryos was sufficient to rescue the phenotype. Co-injection of a target protector specific to the putative miR-9 binding site in the her5 3′ UTR with miR-9 duplex was sufficient to restore the expression of MHB markers, confirming her5 as another key target of miR-9. In these experiments using target protectors, however, it is unclear whether fgf8 and fgfr1 were still downregulated by overexpressed miR-9 in the presence of the her5 target protector.
A stronger line of evidence to support a role for miR-9 in neurogenesis was provided by phenotype analysis of the effects of miR-9 loss of function; the size of the HuC-positive area at the anterior hindbrain was reduced on miR-9 blockade by a morpholino, presumably reflecting reduced production of HuC-positive postmitotic neurons, although an effect on cell survival could not be completely ruled out. The mRNA levels of her5 and her9 were not affected by the loss of miR-9 activity. However, their translation could be enhanced without a substantial change in mRNA steady-state level3. Consistent with this notion, reducing her9 activity could rescue the effect of miR-9 blockade on the size of HuC-positive area. On the basis of both the gain- and loss-of-function analyses, the authors concluded that a single miRNA, miR-9, coordinates events to delimit the organizing activity of the MHB and to promote neurogenesis in domains near this organizing center.
These findings by Leucht et al.5 provide the impetus to address several interesting questions about the evolution of miR-9 function in neurogenesis. In contrast to the authors’ conclusions, miR-9a functions to limit the production of sensory organ precursors (SOPs) in Drosophila12. The transcription factor Senseless, a positive regulator of proneural genes highly expressed in SOPs, was identified as a major in vivo target of miR-9a. Differential effects of miR-9 on neurogenesis in fish and flies can be partially explained by their distinct targets that function as either negative or positive regulators of neurogenesis in different species (Fig. 1b). Furthermore, miR-9a in flies is expressed in ectodermal cells, including the ones adjacent to the presumptive SOPs, whereas in fish it is expressed in the cells that undergo neuronal differentiation (Fig. 1b).
Although many miRNAs are highly conserved at the nucleotide-sequence level across species, accumulating evidence from a number of recent studies suggests that sequence conservation does not necessarily imply similar expression patterns or functional conservation with the same targets13, 14. miRNA binding sites seem to be gained and lost quickly over short evolutionary distances; indeed, about half of them in humans are not conserved in other organisms15. The regulation of the Fgf pathway by miR-9 is probably conserved, as predicted target sites are present in related genes in mammals. Some other targets may turn out to be species specific. These differences may be involved in the generation of the various brain morphologies during evolution. Future genetic analysis in model systems will probably uncover additional interesting modes of action for miR-9 and other miRNAs in early neurogenesis and other aspects of brain development.
References
- 1.Bushati N, Cohen SM. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Hobert O. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, et al. Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Gao F-B. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leucht C, et al. Nat. Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 6.Lumsden A, Krumlauf R. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 7.Crossley PH, et al. Nature. 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- 8.Raible F, Brand M. Trends Neurosci. 2004;27:727–734. doi: 10.1016/j.tins.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Farh KK, et al. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 10.Stark A, et al. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Geling A, et al. Development. 2004;131:1993–2006. doi: 10.1242/dev.01093. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, et al. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ason B, et al. Proc. Natl. Acad. Sci. USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V, Chen X. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, El Naqa IM. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]