Abstract
The mechanical stability of force-bearing proteins is crucial for their functions. However, slow transition rates of complex protein domains have made it challenging to investigate their equilibrium force-dependent structural transitions. Using ultra stable magnetic tweezers, we report the first equilibrium single-molecule force manipulation study of the classic titin I27 immunoglobulin domain. We found that individual I27 in a tandem repeat unfold/fold independently. We obtained the force-dependent free energy difference between unfolded and folded I27 and determined the critical force (∼5.4 pN) at which unfolding and folding have equal probability. We also determined the force-dependent free energy landscape of unfolding/folding transitions based on measurement of the free energy cost of unfolding. In addition to providing insights into the force-dependent structural transitions of titin I27, our results suggest that the conformations of titin immunoglobulin domains can be significantly altered during low force, long duration muscle stretching.
Introduction
The mechanical stability of force-bearing proteins has drawn increasing attention due to their crucial roles in tissue development and maintenance.(1) Such proteins typically consist of modular building-block domains. However, how these domains respond to force is not fully understood. Although the recent development of atomic force microscopy (AFM) technology has made it possible to unfold a single protein by applying high forces and study its refolding at decreased forces, AFM stretching experiments have so far been restricted to a short experimental time scale of seconds to minutes due to rapid mechanical drift of the instrument, despite numerous successes.(2, 3) Yet, probing equilibrium unfolding/folding transitions of single proteins under constant forces has still been challenging for complex proteins that have slow transition rates.
Titin's 27th immunoglobulin (Ig) domain in the I-band (I27) has been studied extensively by AFM, which revealed that I27 has an excellent mechanical stability to resist unfolding at >200 pN forces in direct pulling mode depending on the pulling speed.(3-5) In this mode, unfolding is indicated by sudden decrease in tension at controlled cantilever-surface distance. Bell's model describing the force-dependent unfolding rate(6).
| (1) |
has been used to fit the loading rate or pulling speed dependent unfolding force distributions. Here, f is the tensile force, ku0 is the unfolding rate at zero force, xu is the unfolding transition distance from the native state to the transition state, kB is the Boltzmann constant, and T is the absolute temperature. Bell's model assumes the molecule has a folded state and an unfolded peptide chain state, which are separated by a transition state. ku0 ∼ 3.3 × 10−4 s−1 has been determined in previous AFM experiments for I27. Further, the folding rate of I27 at zero force was measured to be .(4) From these results, the free energy cost of I27 unfolding at zero force was estimated by . ΔG0 of I27 was also measured to be ∼11kBT in previous chemical denaturation experiments.(4, 7) Taken together, ΔG0 of I27 is around 8–11 kBT. However, these previous measurements rely on the validity of unfolding rate extrapolation to zero force or denaturant-free condition, which do not provide the force-dependent free energy cost of I27 unfolding, ΔG(f).
Unfolding and folding processes of tandem repeats of I27 have also been studied using AFM in the force-clamping mode, which clamps the force at a set value through feedback control. Using this technique unfolding/folding is indicated by extension changes at the clamped force.(8, 9)Using this technique, individual unfolding steps were observed when force was clamped at large values of ∼100 pN. When force was subsequently jumped to lower values (<30 pN), a previously unknown folding behavior was observed. After an initial drop upon force jumping, the extension remained at a plateau for some time with large fluctuation, followed by a single large extension drop with all domains folded. The initial extension drop indicates an entropic collapse of the peptide chain due to force decrease. The second phase could be understood as a search for folding pathways at the lower force. All domains are folded after the last step. Theoretical model and Langevin dynamics simulation were used to understand the experimental results.(10, 11) Because of the limited instrumental stability of AFM and the use of large force probe (typical dimension ∼100 μm), the extension drop in the folding process may include small folding steps that were not distinguishable in these experiments. The large force probe of AFM may also affect the collapse rates.(12-15)
The titin I27 domain unfolds under large forces, and folds under zero or much smaller forces. Only nonequilibrium unfolding and folding processes were studied in previous AFM experiments. To reconstruct the free energy landscape from nonequilibrium process, complex theoretical analysis is needed(16-18) and the reliability of the analysis needs more verification. Magnetic tweezers can apply constant forces or programmable changing forces to single-molecule tethers and measure the extension change with high stability (therefore long experimental time scale),(19-21) which have been successfully used to study the force-dependent protein unfolding(22, 23) and interactions between proteins at low forces.(24-26) The superior force stability of magnetic tweezers makes them powerful tools to study the slow folding and unfolding processes of proteins at low forces.
Here, we report the first study of equilibrium folding and unfolding dynamics of I27 under constant force over an unprecedented experimental time scale using in-house made ultrastable magnetic tweezers. From such equilibrium measurements and simple theoretical analysis based on elasticity of polypeptide and folded protein, the force-dependent free energy of protein folding and free energy landscape were obtained.
Methods
Theoretical Framework
Force-dependent protein folding free energy ΔG(f) of I27 can be determined by the force-dependent probability of an I27 domain to be in the unfolded state p(f), or by force-dependent unfolding rate ku(f) and folding rate kf(f):
| (2) |
Measurement of p(f) can only be done over a narrow force range where folding and unfolding transitions both occur in the experimental time scale. The force range can be extended if ku(f) andkf(f) are known over a wider force range.
ΔG(f) includes a force-independent term ΔG0 and a force-dependent term ΔΦ(f):
| (3) |
where , (27) and Δx(f) = xchain(f) – xI27(f) is the extension difference between the unfolded polypeptide chain, xchain(f), and the folded I27, xI27(f), at the same force f (Figure 1).xchain(f) has been shown to follow the worm-like chain (WLC) polymer model and can be obtained by numerical inversion of equation(28)
Figure 1.
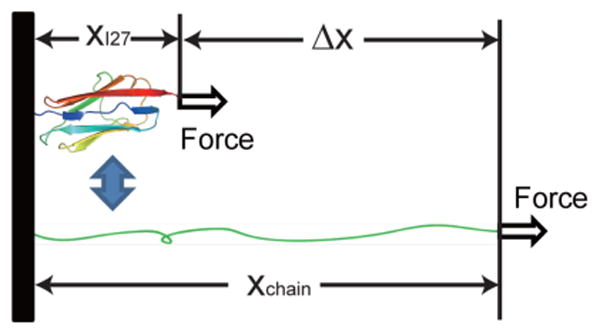
Sketch of an I27 in its folded and unfolded state under force. Extensions of folded I27 (xI27) and unfolded polypeptide (xchain) and their difference (Δx) are dependent on force.
| (4) |
where persistence length of A ∼ 0.8 nm at low forces(29) and contour length of L ∼ 33 nm(4) (SI: Force–extension curves). A folded protein domain can be approximated as a rigid body. Under force, a tethered rigid body can only rotate to align with the force direction in competition with thermal fluctuation. On the basis of the resolved structure of I27 (pdb: 1tit),(30) its rigid body size, the distance between its N- and C-termini, is l0∼ 4 nm. Its extension, which is the average projected length along the force direction, has an analytical solution identical to the monomer force–extension curve in the freely joint-chain (FJC) model: xI27(f = l0 cloth(fl0/kB T) – kB T/f (SI: Force–extension curves).
Materials and Experimental Method
In wild type I27 protein, there are two cysteines at residues 47 and 63. For long time equilibrium measurement, they may form disulfide bond when I27 is unfolded. To avoid the formation of disulfide bonds between cysteines, cysteines were mutated to alanines to get construct I27(C47A,C63A). Hereafter I27 was used to indicate I27(C47A,C63A) throughout the paper. Protein construct of HaloTag-(I27)8-Avitag-Histag was expressed in Escherichia coli bacteria together with BirA plasmid. Therefore, the Avitag in protein construct was already ligated with biotin in bacteria. Purified protein construct was used in the single-molecule magnetic tweezers experiments described in the paper.
Functionalized coverslips were used to make flow chambers and anchor the protein construct. Coverslips were first roughly cleaned in detergent by sonication for 5 min, rinsed by DI water, and cleaned in DI water by sonication for 5 min. Then the coverslips were treated by oxygen plasma cleaner for 15 min. Cleaned coverslips were incubated in solution of 1% 3-aminopropyltriethoxysilane (APTES, cat. A3648, Sigma) in methanol for 1 h. The coverslips were then washed by methanol and DI water, and dried at 100 °C for 30 min. Flow chambers were made by sandwiching the APTES functionalized coverslip and another coverslip with parafilm in between. Then 1% glutaraldehyde (cat. G-7526, Sigma) in DI water was flowed into the chamber and incubated for 1 h. After rinsing by 200 μL DI water, Polybead Amino Microspheres (cat. 17145, Polysciences) with diameter of 3.0 μm were flowed into chamber and incubated for 20 min to get stuck on the glutaraldehyde-coated coverslip. These beads were used as reference to eliminate spatial drift during experiments. HaloTag Amine (O4) Ligand (cat. P6741, Promega) in DI water was flowed into chamber to coat the coverslip surface for 1 h. 1% BSA in Tris Buffer pH 7.4 and 150 mM NaCl was flowed into chamber to block nonspecific interactions and incubated overnight at room temperature. Around 1 nM protein (I27)8 in HEPES buffer was flowed into chamber and incubated for 10 min. Streptavidin-coated paramagnetic beads Dynabead M270 (cat. 65305, Invitrogen) were flowed into the chamber to form protein tethers. The specificity of the surface functionalization and tethering chemistry have been tested by many experiments on various protein and DNA tethers. One example in Supporting Information (SI) shows unfolding of a chimeric tether of HaloTag-(I27)8-DNA-biotin, where the fingerprint DNA overstretching transition followed by sequential unfolding of all eight I27 domains were observed (SI: Specificity of the surface functionalization and tethering chemistry, Figure S4).
Because of the slow transition rates of I27 at low forces, it has been challenging to study the equilibrium mechanical unfolding/folding transitions in previous AFM experiments directly. In our study, we used ultrastable magnetic tweezers that were developed in-house. Magnetic tweezers were built on an inverted microscope (IX71, Olympus). 100× oil-immersion objective (UPLFLN100XO2, Olympus) was used to take images of the protein-tethered bead. Before measurement, a piezo objective actuator (F100, Madcitylab) was used to adjust the focal plane of objective to build an image library at a series of different focal planes separated by 50 nm. Two permanent magnetic rods were placed above the sample, and a motorized stage (VT-40, Micos) was used to move the magnets to control the force.(21) During realtime measurements, the piezo objective actuator was used to actively correct the drift of focal plane in real time by monitoring a fixed reference bead, which is critical to long term stability of magnetic tweezers. These tweezers have a spatial resolution of ∼2 nm and a sampling rate of 200 Hz,(21) which can record multiple hours extension time courses of single molecules under constant forces. The force stability of the magnetic tweezers is demonstrated in SI: Force stability of magnetic tweezers, Figure S1.
The accuracy of our magnetic tweezers' capability to determine extension change is demonstrated by using the well-known DNA overstretching transition at ∼65 pN as ruler (SI: Figure S4). The rotation of the paramagnetic bead at different forces contributes to the height change of the bead,(31) making it difficult to directly measure the force–extension curve of short tethers (SI, Figure S2); while the extension change at a constant force can be accurately measured. The force was calibrated by the bead fluctuation(19, 20) at forces <15 pN and then extrapolated to higher forces through a precalibrated force–distance curve describing how force applied to a M270 bead changes with the distance between the magnets and the bead. This method has ∼10% uncertainty mainly due to bead size heterogeinity as detailed in(21) (also see SI: Force stability of magnetic tweezers and Figure S1A,B). All experiments were carried out in buffer solution of 10 mM HEPES (pH 7.3), 150 mM NaCl at room temperature of 21 ± 1°C.
Results
Force-Dependent Folding Rate and Folding Free Energy of I27
We tethered the construct of eight tandem I27 repeats between a chloroalkane-coated coverslip through a covalent HaloTag-chloroalkane bond(32-34) at the N-terminus and a streptavidin-coated paramagnetic bead M270 through a strong streptavidin–biotin bond at the C-terminus (Figure 2A inset). The HaloTag-ligand and streptavidin–biotin interactions provide strong bonds allowing force-manipulation of single molecules for long duration.(35) HaloTag can occasionally unfold under large force with large step of >40 nm, which is easily distinguished from unfolding of I27. Streptavidin–biotin bond has been extensively used to study DNA mechanics such as DNA overstretching transitions at high force(36) and protein unfolding(23, 25) without any nonspecific unfolding steps. Therefore, this tethering chemistry not only is highly specific, but also does not induce unfolding steps that may interfere with the unfolding signal of I27.
Figure 2.

Force-dependent unfolding and folding of (I27)8. (A) Unfolding occurs during force-increase scan at a loading rate of 3.5 pN/sec and constant force of ∼80 pN. Folding occurs after jumping to lower forces of 4.1 ± 0.4 pN, 3.3 ± 0.3 pN, and 2.6 ± 0.3 pN. The number of folded domains is counted in a following force-increase scan. The red arrow indicates a spontaneous I27 unfolding at 4.1 pN. Raw data recorded at a sampling rate of 200 Hz (black) are smoothed using one-second time window (red). Inset shows schematic of the unfolding and folding transitions of a (I27)8molecule tethered between a coverslip and a bead under force. (B) From another protein tether, the extension drops during folding at constant forces of 4.2 ± 0.4 pN and 3.4 ± 0.3 pN were calculated by the difference between the average extensions in the first 0.3-s time window right after force drop from 80 pN and in the last 0.3-s time window at the low forces (see Figure S5 for further details). Error bar is the standard deviation of the extension fluctuation within the time-window. The result shows that the extension drop at each force is proportional to the number of folded I27 (red lines are linear fitting result with R2 = 0.98). (C) Eight folding time traces at 4.1 pN, six traces at 3.3 pN, and six traces at 2.6 pN were combined to get the averaged folding time trace (black data) at each force, respectively. The force-dependent folding rates kf(f) are determined by fitting with exponential decay function (red line) as 0.012 s–1 at 4.1 pN, 0.081 s–1 at 3.3 pN, and 0.29 s–1 at 2.6 pN, with <3% fitting error. (D) Four independent unfolding time courses of (I27)8 at 6.0 ± 0.6 pN. Curves are shifted vertically for clarity. (E) Force-dependent folding rate kf(f) and unfolding rate ku(f) and the unfolding free energy cost. Open circles indicate kf(f) obtained from fitting to time traces in (C), while open squares indicate kf(f) obtained by dwell time analysis for extension fluctuations in (D) and Figure 3A. The data of kf(f) are fitted using theoretical prediction by eq 5 (black line), with a best fitting parameter kf0 = 2.1 ± 0.4 s–1 determined. The solid squares indicate ku(f) obtained by dwell time analysis for extension fluctuations in (D) and Figure 3A. Black dashed line shows ku(f) predicted by Bell's model using parameters reported in previous AFM experiments. The red solid line shows ΔG(f) obtained by eq 3.
To determine the extension trajectory of (I27)8 during the force-dependent unfolding and folding processes, we increased force from ∼1.7 pN to ∼80 pN at a loading rate of r = 3.5 pN/sec and then kept constant force of ∼80 pN for 2 min (Figure 2A). During this process, sequential unfolding of all eight I27 repeats with a step size of 23.3 ± 2.3 nm were observed, which agrees with previous AFM experiments.(4) The I27 domains refolded after force was subsequently reduced to 4.1 pN or below. At 4.1 pN, all eight unfolded I27 refolded in ∼6 min, indicated by eight extension decrease steps. Interestingly, one of the folded domains spontaneously unfolded at 4.1 pN (red arrow in Figure 2A). Successful refolding was confirmed in a subsequent constant loading rate (r = 3.5 pN/sec) force-increase scan, during which seven unfolding steps similar to those observed in the first force-increase scan were observed. Seven, rather than eight unfolded steps, were observed because one of the domains already unfolded at 4.1 pN, preceding the force-increase scan. Repeating such folding and unfolding procedure at lower forces of 3.3 and 2.6 pN, we found that all eight unfolded I27 domains refolded within 1 min.
For an initially completely unfolded (I27)8, folding after jumping to lower forces involved two distinct stages of extension decrease: a subsecond rapid extension drop followed by a second stage slower progressive extension decrease. The rapid extension drop can be explained by entropic collapse of the unfolded polypeptide chain upon switching to a lower force, although it cannot be accurately measured by magnetic tweezers due to the bead rotation caused by force change (see Materials and Experimental Method). The nature of the second stage of the slower extension decrease requires more analysis. At 4.1 pN, clear folding steps in the second stage were seen. However, at 3.3 pN and 2.6 pN, due to faster folding rates and reduced extension difference between unfolded and folded I27 at lower forces, observation of folding steps became difficult. From another protein tether, by measuring the extension drop of the second stage folding process during different time window from 5 s to 10 min (SI: Figure S5), and counting the number of successfully folded I27 domains from the number of unfolding steps in the following force-increase scan, we found that the second stage extension drop is proportional to the number of folded I27 during the folding process at constant forces (Figure 2B). The slopes of fitting lines give folding step sizes of 10.6 ± 0.5 nm at 4.2 pN and 9.1 ± 0.5 nm at 3.3 pN, respectively. Two additional independent measurements are given in SI: Figure S6 to demonstrate the reproducibility. Overall, these results indicate that the second stage of progressive extension decrease is entirely caused by folding of I27 domains.
To obtain kf(f), the second stage of extension decrease time traces were measured at three low forces of 4.1, 3.3, and 2.6 pN. At each force, at least 6 folding time traces (SI: Figure S7) extracted from multiple unfolding/refolding cycles were averaged to obtain an averaged folding time course (Figure 2D), which could be fitted by an exponential decay function with a single rate constantkf(f). This result indicates that the folding kinetics of (I27)8 can be described by statistically independent folding of individual I27 domains. Our approach shows that kf(f) drastically decreases as force increases: over a small force range from 2.6 to 4.1 pN, kf(f) decreases from ∼0.29 to ∼0.01 s–1 (Figure 2E).
The sensitive force dependence of kf(f) implies a force-dependent barrier that slows down the folding transition.(11) With a reasonable assumption that the folding transition state is a collapsed state with a similar dimension to the folded I27, the force-dependent barrier is just where is the extension difference between an I27 in the unfolded state (xchain(f)) and transition state was measured by the folding/unfolding stepsize of I27 at constant forces. It could be converted into force–extension curves of polypeptide chain by adding back the theoretical force–extension curve of folded I27. The resulting force–extension curve is consistent with WLC model with previously reported bending persistence length of ∼0.8 nm at low forces(29) (SI Figure S3). This barrier results in a prediction for the force-dependent folding rate as
| (5) |
Using the WLC polymer model with a persistence length of 0.8 nm for the peptide chain and l0 = 4 nm for the rigid body size of the transition state, the predicted kf(f) reasonably reproduces the experimental data with a single fitting parameter kf0 = 2.1 ± 0.4 s–1 (black solid curve in Figure 2E). Here we note that Bell's model (eq 1) is not a good approximation to describe the folding kinetics. This is because Δx(f) cannot be approximated as a constant due to the distinct force responses between the unfolded and the nearly folded transition states, as detailed in SI: Discussion of free energy landscape.
Unfolding of I27 involves a small transition distance xu ∼ 0.25 nm,(4) which is expected due to the rigid body effect of folded I27 (SI: Discussion of the free energy landscape). It implies an insensitive force dependence of the unfolding rate ku(f) at low forces smaller than kBT/xu ∼ 16 pN according to the Bell's model (eq 1). On the basis of the previously reported values of xu and the zero force unfolding rate k0∼ 3.3 × 10–4 s–1, the Bell's model predicts a slow unfolding rate of I27 of <10–3 s–1 at forces smaller than 10 pN. Figure 2D shows four typical unfolding time traces until tether breakage obtained from independent experiments at 6.0 ± 0.6 pN from initial conformations with all eight I27 domains in folded states. In two of the time traces, occasional refolding steps were observed, indicated by the stepwise extension drop. By using the pseudo dwell time analysis(37) (SI: Figure S8), the unfolding rate and folding rate were estimated to be (3.2 ± 0.7) × 10–4 s–1 and (3.3 ± 1.6) × 10–5 s–1 at 6 pN, respectively. The measured unfolding rate is consistent with the prediction by the Bell's model.
As reasoned above, ku(f) can be approximated as a constant at small force range <16 pN. Using the value of 3.2 × 10–4 s–1 determined at ∼6 pN and kf(f) discussed above, ΔG(f) obtained using eq 2 is shown in Figure 2E. The critical force fc where folding and unfolding rates are balanced is determined to be fc ∼ 5.4 ± 0.1 pN located at the intersection of ku(f) and kf(f), corresponding to ΔG(fc) = 0.
Equilibrium Folding and Unfolding Dynamics of I27
ΔG(f) can also be measured by analyzing equilibrium folding and unfolding transitions under constant forces. Figure 3A shows equilibrium transitions among multiple states of (I27)8 in an 8 h time trace recorded at 4.5 pN. The histogram of the smoothed extension (right panel of Figure 3A) reveals multiple extension peaks corresponding to varying number of unfolded I27 repeats among eight I27 repeats. Figure 3B shows histogram of transition stepsizes which can be fitted by Gaussian function. A transition stepsize of 10.9 ± 1.0 nm (standard deviation) is consistent with the WLC model with a persistence length of ∼0.8 nm for the unfolded peptide chain, which predicts an extension difference of ∼10.5 nm between folded I27 and unfolded polypeptides at ∼4.5 pN.
Figure 3.

Equilibrium folding and unfolding transitions of (I27)8 at 4.5 pN. (A) Eight hours of measurement of (I27)8 extension under a constant force of 4.5 pN. Raw data recorded at 200 Hz and smoothed data using five-second time window are indicated by black and gray, respectively. Right panel shows the histogram of the smoothed extension with distinct peaks corresponding to different numbers of unfolded I27. (B) The transition stepsize is determined by Gaussian fitting to the histogram of the transition stepsizes as 10.9 ± 1.0 nm. (C,D) Exponential fitting of the normalized histograms of pseudo folding and unfolding dwell times obtained from 66 folding events and 66 unfolding events gives kf(f) = (2.2 ± 0.2) × 10–3 s–1 and ku(f) = (3.7 ± 0.4) × 10–4 s–1 at 4.5 pN, respectively. (E) Binomial fitting of the probability of the number of unfolded I27 in (I27)8 gives the unfolding probability of an I27 as p = 0.131 ± 0.005 at 4.5 pN.
The transition rates of one I27 can be obtained by pseudo dwell time analysis.(37) Briefly, in a tether containing multiple independent repeats, the unfolding (folding) rate for one I27 is calculated by dividing the apparent unfolding (folding) rate of the tether with the number of the current folded (unfolded) domains (SI: Figure S8). The histograms of the pseudo dwell times were fitted with single exponential decay functions (Figure 3C,D), which gives kf(f) = (2.2 ± 0.2) × 10–3 s–1 and ku(f) = (3.7 ± 0.4) × 10–4 s–1 as well as ΔG(f) = ln(kf(f)/ku(f)) = 1.8 ± 0.2 kBT at f = 4.5 pN. The determined folding rate at 4.5 pN perfectly agrees with ku(f) predicted by eq 5 in Figure 2E.
The probability of having n unfolded I27 domains in eight independent I27 repeats, P8(n), can be directly read out from the extension distribution peaks (Figure 3A, right panel), which should follow the binomial distribution: . Here is the binomial coefficient, and the single free parameter p denotes the probability of an I27 domain in unfolded state at 4.5 pN. Fitting of the binomial distribution to the normalized probability of the number of unfolded I27 repeats gives p = 0.131 ± 0.005 (Figure 3E). Therefore, ΔG(f = 4.5 pN) =1.89 ± 0.05 kBT by eq 2, which agrees well with the value obtained from the pseudo dwell time analysis. The zero force free energy difference ΔG0 is calculated to be 8.3 ± 1.0 kBT using eq 3, which is within the range reported in previous bulk chemical denaturant(4, 7) and AFM single-molecule measurements for wild type I27 based on extrapolation.(4)
Free Energy Landscape of a Two-State Model of I27
It is useful to understand the mechanically induced protein structural transitions in a free energy landscape.(10, 11) In the simplest model involving a folded and a completely unfolded states, the free energy landscape GF(x) at a given external force constraint of F is simply
| (6) |
where
are the constrained free energies of an folded and unfolded I27, respectively. Here fI27(x) is the force–extension curve of folded I27 which is the inverse function of eq S3 that is numerically solved by MATLAB, and fchain(x) is the force–extension curve of unfolded I27 given by eq 4. Note that here we use capital F to distinguish the external force constraint from the tension f in the molecule. At equilibrium, the average tension equals the external force.
GF(x) calculated at several external forces show two energy minimums corresponding to the two structural states, separated by an energy barrier (Figure 4). It should be noted that the energy barrier in a two-state model is different from a more realistic picture that the transition state corresponds to an ensemble of intermediate states connecting the native and completely unfolded states. We also note that the locations of the energy minimums change with force as predicted theoretically.(10, 11) This change is due to the force-responses of folded and unfolded I27. The details of the derivation and physical meanings of the free energy landscape, as well as its advantages and limitations are detailed in SI: Discussion of the free energy landscape.
Figure 4.
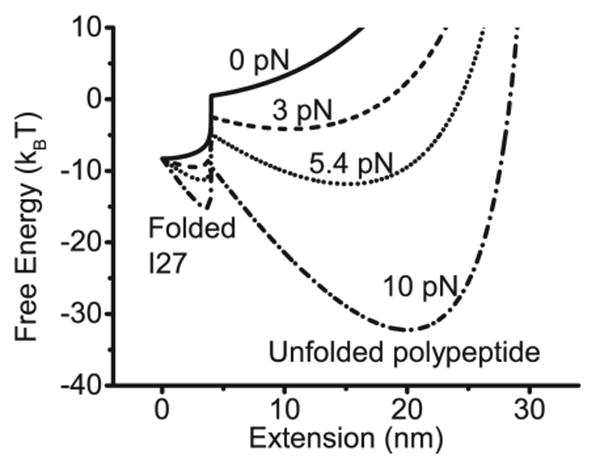
Free energy landscape of I27 GF(x) calculated at several constant forces using ΔG0 = 8.3 kBT determined in our experiment. A folded I27 is modeled as a rigid rod with contour length of 4 nm, and an unfolded I27 polypeptide chain is modeled as a WLC polymer with contour length of 33 nm and persistence length of 0.8 nm.
Summary and Discussion
In summary, we have investigated the equilibrium unfolding and folding transitions of I27. Several important results emerged, including the determinations of force-dependent folding free energy ΔG(f), force-dependent folding rate kf(f), force-dependent free energy landscape GF(x), and the independent folding of individual I27 units at low forces. A critical force of only ∼5.4 pN, where unfolding and folding have equal probability, was determined. This force is 1–2 orders of magnitude smaller than the unfolding forces reported in previous nonequilibrium AFM stretching experiments. As I27 is a typical all-β protein domain consisting of seven β-strands organized in two β-sheets, our results provide important insights into the mechanical stability of the broad class of all-β protein domains such as the actin binding protein Filamin A.(23, 38) The experimental and theoretical approaches developed in this work can also be applied to studies of proteins in other major classes such as all-α, α+β, and α/β domains.(39)
Although the existence of a less stable folding intermediate state(40) cannot be excluded due to our limited spatial and time resolutions, our results reveal that the folding process of I27 can be described by a two-state model. Under an assumption of independence of the I27 repeats in the (I27)8 construct, which is supported by the pseudodwell time analysis, we obtained the force-dependent free energy difference between a folded I27 and an unfolded I27 peptide chain. The folding process of tandem unfolded I27 repeats reveals independent folding steps of individual I27, which were not detected in previous force-clamping AFM experiments.(8, 9) We reason that this is because of the better stability and the smaller force probe of magnetic tweezers that make magnetic tweezers more suitable to distinguish small refolding step-sizes at low forces.
Our results of the force-dependent folding and unfolding transitions of I27 have important physiological implications on muscle elasticity. The ultra large muscle protein titin consists of more than 300 Ig and fibronectin domains.(41) This protein plays an important role in muscle contraction, acting as a molecular spring to provide the passive elasticity of muscle and to help maintain myosin position in the middle of sarcomere.(42) It has been suggested that titin's function relies on the mechanical unfolding and refolding of many of its Ig domains during mechanical stretching.(2) The small critical force of ∼5.4 pN determined for I27 in our experiments suggests that the conformations of the titin Ig domains may be significantly altered during low force, long duration muscle stretching in activities such as yoga. In addition, mechanically exposed cryptic sites buried in these Ig domains may cause subsequent interactions with other proteins, which may be important for the elastic regulations of muscle. This possibility is consistent with several recent studies reporting that the force-dependent conformational changes of mechanosensing proteins can drastically alter their interactions with other proteins.(25, 26, 43)
Supplementary Material
Acknowledgments
The authors thank Matthew J. Lang for critical reading of the manuscript. This research was supported by grants from the National Research Foundation through the Mechanobiology Institute Singapore to J.Y., the Fundamental Research Funds for the Central Universities (Grant 2013121005) and the National Nature Science Foundation of China (Grants 11474237) to H.C., and NSF (Grant 1252857) and NIH (Grants HL66030 and HL61228) to J.M.F.
Footnotes
Author Contributions: H.C. and G. Y. contributed equally to this work.
The authors declare no competing financial interest.
Supporting Information: Figure S1–S9; Force stability of magnetic tweezers; Extension and stepsize measurement; Force–extension curves; Specificity of the surface functionalization and tethering chemistry; Derivation of the free energy landscape; Discussion of the free energy landscape. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Moore SW, Roca-Cusachs P, Sheetz MP. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rief M, Gautel M, Oesterhelt F, Fernandez J, Gaub H. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 3.Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, Schulten K, Fernandez JM. Nature. 1999;402:100–103. doi: 10.1038/47083. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 4.Carrion-Vazquez M, Oberhauser A, Fowler S, Marszalek P, Broedel S, Clarke J, Fernandez J. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [Free @ PNAS], [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams PM, Fowler SB, Best RB, Toca-Herrera JL, Scott KA, Steward A, Clarke J. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 6.Bell G. Science. 1978;200:618–627. doi: 10.1126/science.347575. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 7.Clarke J, Cota E, Fowler SB, Hamill SJ. Structure. 1999;7:1145–1153. doi: 10.1016/s0969-2126(99)80181-6. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 8.Fernandez J, Li H. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Manyes S, Brujic J, Badilla CL, Fernandez JM. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkovich R, Garcia-Manyes S, Urbakh M, Klafter J, Fernandez JM. Biophys J. 2010;98:2692–2701. doi: 10.1016/j.bpj.2010.02.053. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkovich R, Garcia-Manyes S, Klafter J, Urbakh M, Fernandez JM. Biochem Biophys Res Commun. 2010;403:133–137. doi: 10.1016/j.bbrc.2010.10.133. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkovich R, Hermans RI, Popa I, Stirnemann G, Garcia-Manyes S, Berne BJ, Fernandez JM. Proc Natl Acad Sci U S A. 2012;109:14416–14421. doi: 10.1073/pnas.1212167109. [Free @ PNAS], [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai H, Kath JE, Zorgiebel FM, Sun M, Ghosh P, Hatfull GF, Grindley ND, Marko JF. Proc Natl Acad Sci U S A. 2012;109:16546–16551. doi: 10.1073/pnas.1203118109. [Free @ PNAS], [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elms PJ, Chodera JD, Bustamante CJ, Marqusee S. Biophys J. 2012;103:1490–1499. doi: 10.1016/j.bpj.2012.06.051. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popa I, Kosuri P, Alegre-Cebollada J, Garcia-Manyes S, Fernandez JM. Nat Protoc. 2013;8:1261–1276. doi: 10.1038/nprot.2013.056. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris NC, Song Y, Kiang CH. Phys Rev Lett. 2007;99:068101. doi: 10.1103/PhysRevLett.99.068101. [CrossRef], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberhofer H, Dellago C. J Comput Chem. 2009;30:1726–1736. doi: 10.1002/jcc.21290. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 18.Lannon H, Haghpanah JS, Montclare JK, Vanden-Eijnden E, Brujic J. Phys Rev Lett. 2013;110:128301. doi: 10.1103/PhysRevLett.110.128301. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 19.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 20.Gosse C, Croquette V. Biophys J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J. Biophys J. 2011;100:517–523. doi: 10.1016/j.bpj.2010.12.3700. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Garcia-Manyes S, Sarkar A, Badilla CL, Fernandez JM. Biophys J. 2009;96:3810–3821. doi: 10.1016/j.bpj.2009.01.043. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Zhu X, Cong P, Sheetz MP, Nakamura F, Yan J. Biophys J. 2011;101:1231–1237. doi: 10.1016/j.bpj.2011.07.028. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang H, Lee GU. J Am Chem Soc. 2007;129:6640–6646. doi: 10.1021/ja071215c. ACS Full Text [PubMed], [CAS. [DOI] [PubMed] [Google Scholar]
- 25.Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. CAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mege RM, Yan J. Nat Commun. 2014;5:4525. doi: 10.1038/ncomms5525. CAS. [DOI] [PubMed] [Google Scholar]
- 27.Rouzina I, Bloomfield VA. Biophys J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marko J, Siggia E. Macromolecules. 1995;28:8759–8770. [ACS Full Text ], [CAS] [Google Scholar]
- 29.Rief M, Pascual J, Saraste M, Gaub HJ. Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [CrossRef], [CAS] [DOI] [PubMed] [Google Scholar]
- 30.Improta S, Politou AS, Pastore A. Structure. 1996;4:323–337. doi: 10.1016/s0969-2126(96)00036-6. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 31.Lipfert J, Kerssemakers JW, Jager T, Dekker NH. Nat Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 32.Los GV, Wood K. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi Y, Kawakami M. Langmuir. 2010;26:10433–10436. doi: 10.1021/la101658a. [ACS Full Text], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 34.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popa I, Berkovich R, Alegre-Cebollada J, Badilla CL, Rivas-Pardo JA, Taniguchi Y, Kawakami M, Fernandez JM. J Am Chem Soc. 2013;135:12762–12771. doi: 10.1021/ja4056382. ACS Full Text, [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu H, Chen H, Zhang X, Qu Y, Marko JF, Yan J. Nucleic Acids Res. 2011;39:3473–3481. doi: 10.1093/nar/gkq1278. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Y, Kuske R, Li H. Biophys J. 2008;95:782–788. doi: 10.1529/biophysj.107.128298. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Chandrasekar S, Sheetz MP, Stossel TP, Nakamura F, Yan J. Sci Rep. 2013;3:1642. doi: 10.1038/srep01642. CAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cecconi C, Shank EA, Bustamante C, Marqusee S. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 41.Labeit S, Kolmerer B. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 42.Trinick J. Curr Biol. 1996;6:258–260. doi: 10.1016/s0960-9822(02)00472-4. [CrossRef], [PubMed], [CAS] [DOI] [PubMed] [Google Scholar]
- 43.Alegre-Cebollada J, Kosuri P, Giganti D, Eckels E, Rivas-Pardo JA, Hamdani N, Warren CM, Solaro RJ, Linke WA, Fernandez JM. Cell. 2014;156:1235–1246. doi: 10.1016/j.cell.2014.01.056. [CrossRef], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


