Abstract
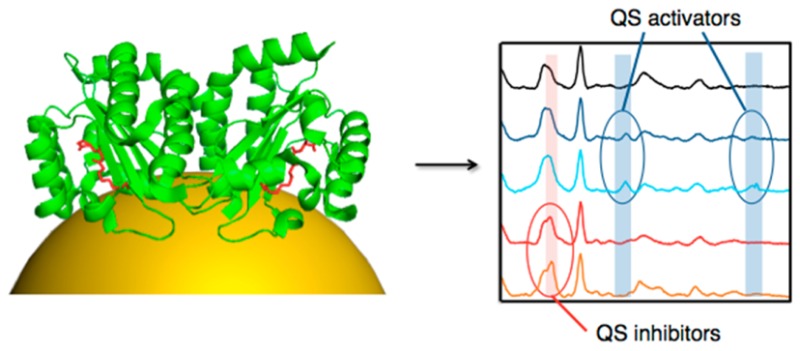
Many members of the LuxR family of quorum sensing (QS) transcriptional activators, including LasR of Pseudomonas aeruginosa, are believed to require appropriate acyl-homoserine lactone (acyl-HSL) ligands to fold into an active conformation. The failure to purify ligand-free LuxR homologues in nonaggregated form at the high concentrations required for their structural characterization has limited the understanding of the mechanisms by which QS receptors are activated. Surface-enhanced Raman scattering (SERS) is a vibrational spectroscopy technique that can be applied to study proteins at extremely low concentrations in their active state. The high sensitivity of SERS has allowed us to detect molecular interactions between the ligand-binding domain of LasR (LasRLBD) as a soluble apoprotein and modulators of P. aeruginosa QS. We found that QS activators and inhibitors produce differential SERS fingerprints in LasRLBD, and in combination with molecular docking analysis provide insight into the relevant interaction mechanism. This study reveals signal-specific structural changes in LasR upon ligand binding, thereby confirming the applicability of SERS to analyze ligand-induced conformational changes in proteins.
Keywords: nanoplasmonics, SERS, quorum sensing, protein receptors, Pseudomonas aeruginosa
Surface-enhanced Raman scattering (SERS) spectroscopy is an ultra-sensitive and ultra-rapid analytical technique that can achieve detection limits down to the single molecule level thanks to nanoplasmonic effects.1,2 As a vibrational spectroscopy technique, SERS can provide detailed information about the structure and conformation of a molecular target, which has been used to identify molecules, proteins and their mutual interactions.3 A major advantage of SERS is that it can be performed under physiological conditions, thereby preserving the active state of proteins and revealing protein kinetics and dynamics. SERS does not require high protein concentrations as is the case for NMR or X-ray crystallography and it can be applied without prior knowledge of the structure of the protein.4 Hence, SERS has emerged as an attractive analytical tool to investigate protein–protein and protein–ligand interactions.5,6
In Gram-negative bacteria, quorum sensing (QS) is mostly achieved through the biosynthesis and sensing of N-acyl l-homoserine lactones (acyl-HSLs) by LuxI/R-type QS protein receptor systems.7,8 In such QS circuits, LuxI-type synthases catalyze the production of acyl-HSLs that freely diffuse into and out of cells, where they bind LuxR-type proteins that serve as signal receptors. When acyl-HSLs reach a threshold concentration, the acyl-HSL-LuxR complex activates the expression of QS-dependent genes. Biochemical and structural studies have shown that LuxR homologues are homodimers, and each monomer folds into two functional domains: an amino-terminal ligand-binding domain (LBD) and a carboxyl-terminal DNA-binding domain (DBD), joined by a short linker region. The cognate ligand binds to a narrow cavity in the LBD, formed by a cluster of hydrophobic and aromatic residues, and it is believed that this triggers a conformational change in the polypeptide, presumably enabling the binding of the protein to specific promoters, ultimately leading to transcriptional activation of QS genes.9,10 Despite of the availability of a wealth of information, the precise molecular mechanisms of LuxR receptor activation still remain mostly unknown. This is largely due to the failure to purify the relevant regulators in nonaggregated form outside the cellular environment, in the absence of an appropriate acyl-HSL.
Much of the information regarding acyl-HSL-regulated QS systems has been obtained through the study of Pseudomonas aeruginosa bacteria. P. aeruginosa contains two pairs of LuxI/LuxR-type systems, termed LasI/LasR and RhlI/RhlR. LasI and RhlI are synthases that catalyze production of N-(3-oxododecanoyl)-homoserine lactone (C12-HSL) and N-(butyryl)-homoserine lactone (C4-HSL), which are sensed by the LasR and RhlR transcriptional receptors, respectively.11,12 LasR is known to initiate the QS regulatory system in this bacterial species,13 and therefore, it has attracted significant interest as a target for QS modulation.14−16 The X-ray structure of the LBD of LasR forming a complex with its natural ligand C12-HSL17 (Figure 1A) revealed a symmetric homodimer with the molecule buried in a pocket enclosed by α-helices (α3, α4 and α5) and the concave side of the central β-sheet of each LBD.17,18
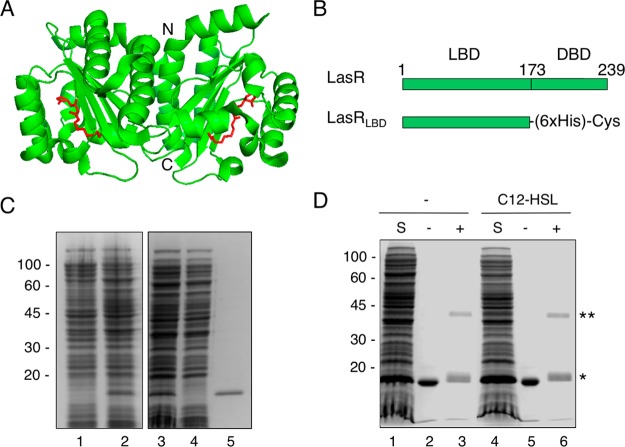
Figure 1.
(A) Co-crystal structure of LasRLBD (green) bound to C12-HSL (red) (PDB identity code 2UV0). The amino (N-) and carboxy (C)-terminal regions of the polypeptide are indicated. (B) Schematic representation of full-length LasR and the corresponding LBD bearing a hexahistidine-tag (6×His) and a C-terminal cysteine (Cys). (C) SDS-PAGE and Coomassie blue staining of LasRLBD expression and purification. Lane 1, uninduced total fraction; lane 2, IPTG-induced total fraction; lane 3, IPTG-induced soluble fraction; lane 4, affinity chromatography flow-through; lane 5, eluted protein. (D) Cross-linking assay of LasRLBD. The polypeptide expressed in bacteria in the absence (lanes 1–3) or in the presence (lanes 4–6) of C12-HSL was solubilized (S), affinity purified and subjected to crosslinking (+) with 0.5 mM DSS (lanes 3,6) or not (lanes 2,5). Bands corresponding to dimeric (**) and monomeric (*) forms are indicated on the right.
We demonstrate in this paper the application of SERS spectroscopy, combined with structural analysis, molecular docking studies and assays of biological activity, to detect molecular interactions between LasR and diverse activators and inhibitors of P. aeruginosa QS. To this end the LBD of LasR (LasRLBD) bearing a hexahistidine (6×His) tag and a cysteine in its carboxyl-terminus was expressed and purified in a soluble, ligand-free (i.e., apoprotein) active form. Oriented attachment of the polypeptide onto plasmonic gold nanoparticle thin films and label-free SERS allowed us to detect structural changes of LasRLBD as a result of its interaction with cognate C12-HSL ligands, C4-HSL agonists and Furanone C30 or acetylsalicylic acid (SA) antagonists. Remarkably, highly sensitive and reproducible SERS spectra allowed us to discriminate between activators and inhibitors of QS, through distinctive vibrational features. Our study thus provides insight into the molecular interactions between LasR and modulators of P. aeruginosa QS, while featuring SERS as a powerful tool for the dynamic study and screening of protein–ligand interactions.
Results and Discussion
Expression of Apo LasRLBD and Analysis of Functional Activity
We schematically show in Figure 1 the lactone-binding domain (LBD) and the DNA-binding domain (DBD) of LasR. To carry out the SERS investigation of conformational changes in LasR upon ligand interaction, we focused on the LBD region of the protein as the DBD is not required for ligand binding activity.17,19 Therefore, the LBD of lasR gene, coding from amino-acids encompassing Met-1 to Lys-173,17 was PCR amplified from the pMHLAS plasmid20 and cloned into the pET21a(+) vector in frame with a 6×His tag for affinity purification and a C-terminal cysteine that is known to strongly bind onto gold surfaces through the formation of Au–S bonds (Figure 1B). The resulting construct, termed pET21-LasRLBD, was transformed in Escherichia coli BL21(DE3)pLysS cells for protein expression. E. coli bacteria have been widely used as suitable hosts to study the influence of different acyl-HSLs in the expression of LuxR homologues as they do not produce acyl-HSLs endogenously.9
The SERS study of the interactions of apo LasRLBD with QS modulators required the expression and purification of the polypeptide in the absence of its cognate C12-HSL ligand. However, it has been shown that overexpression of recombinant LuxR homologues in E. coli without the appropriate signaling molecules in the bacterial growth medium renders these proteins insoluble and prone to degradation by cellular proteases.9,17 Therefore, we first assessed the stability and solubility of the LasRLBD polypeptide produced in E. coli BL21(DE3)pLysS bacteria grown without C12-HSL ligand (see Materials and Methods). Briefly, bacterial cultures in log phase (OD600–0.6) were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 28 °C for 4 h and then total proteins and soluble protein fractions were separated on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and stained with Coomassie Blue. As shown in Figure 1C, a protein band with the expected molecular weight corresponding to LasRLBD (∼19 kDa) can be seen in the total protein extract from IPTG-induced cells (Figure 1C, lane 2) but not in the total protein extract from uninduced cells (Figure 1C, lane 1). The expression of LasRLBD was also confirmed by Western blot and immunostaining with anti-His tag antibodies (data not shown). Importantly, apo LasRLBD was found in the soluble fraction (Figure 1C, lane 3), which allowed us to purify it upon elution from a nickel resin (Figure 1C, lane 5). Similar levels of apo LasRLBD relative to the rest of proteins in the soluble fraction (Figure 1C, lane 3) and in the total protein extract (Figure 1C, lane 2), suggested that the polypeptide was highly soluble. The amount of soluble apo LasRLBD (Figure 1D, lane 1) was similar to that obtained from bacteria grown in the presence of the C12-HSL ligand (Figure 1D, lane 4), again confirming that the polypeptide is stable and soluble in its apoprotein form. Importantly, increasing either the expression of apo LasRLBD or its concentration resulted in decreased solubility (data not shown).
Dimerization of LasR, as well as other LuxR-type receptors, is a requisite for transcriptional activation of target QS genes.21,22 The oligomeric state of apo LasRLBD was assessed by chemical cross-linking with disuccinimidyl suberate (DSS), which is a commonly used strategy for the characterization of dimers of LuxR homologues.23−25 To this end, affinity purified LasRLBD obtained from IPTG-induced BL21(DE3)pLysS bacteria grown in the presence or in the absence of C12-HSL was cross-linked with DSS and analyzed by SDS-PAGE. As shown in Figure 1D, cross-linked dimeric complexes of LasRLBD were detected at similar levels regardless of the presence (lane 6) or absence (lane 3) of the ligand, indicating that LasRLBD does exist in the form of homodimers even when no C12-HSL is present. We then investigated the activity of purified apo LasRLBD as a quorum-quencher of C12-HSL molecules, by employing an E. coli MT102 biosensor strain harboring the pMHLAS reporter system that expresses green fluorescent protein (GFP) upon activation of LasR by exogenous addition of C12-HSL20 (Supporting Information Figure S1). We hypothesized that functional LasRLBD would sequester extracellular C12-HSLs, reducing the levels of free ligand that would be available for binding and activating intracellular LasR, resulting in decreased GFP expression (Supporting Information Figure S1). To test this hypothesis, C12-HSL was preincubated with either affinity purified LasRLBD or BSA for 1 h, and the mixtures were subsequently added to liquid cultures of E. coli MT102 bacteria bearing pMHLAS and further incubated for 90 min to allow GFP expression. The expression of GFP was then evaluated by epifluorescence microscopy and quantified by fluorescence spectroscopy. As shown in Figure 2, the preincubation of C12-HSL (0.5 nmol) with apo LasRLBD significantly reduced the levels of GFP whereas preincubation with BSA (0.5 nmol) as a control did not affect GFP expression. This indicated that apo LasRLBD did indeed bind C12-HSLs, preventing full induction of the acyl-HSL reporter system.
Figure 2.
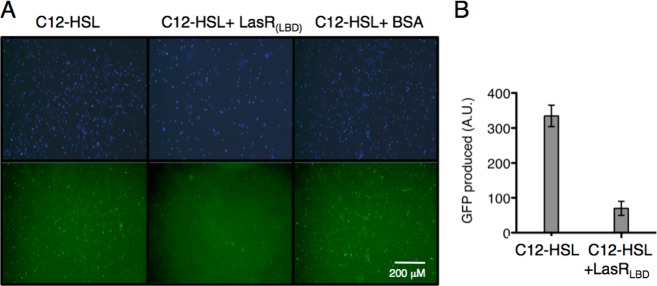
(A) Fluorescence microscopy images of MT102/pMHLAS bacteria treated with C12-HSL previously incubated with either LasRLBD or BSA, as labeled. Upper and lower panels show DAPI staining of bacteria and GFP expression, respectively. (B) Induction of GFP expression from pMHLAS in E. coli MT102 after incubation of bacteria with C12-HSL or C12-HSL and LasRLBD polypeptide. Error bars represent standard deviation calculated for three independent experiments.
A first conclusion of this study is thus related to the general belief that LasR and other LuxR family members require the presence of appropriate acyl-HSL molecules to properly fold into their active conformations, which is mostly based on the production of large amounts of soluble and stable protein upon supplementing the bacterial growth medium with cognate acyl-HSLs. This strategy has been widely applied toward the biochemical and structural characterization of several LuxR homologues including LasR,22 TraR21,26 or QscR.27 This idea has been however challenged by several groups,28,29 who provided experimental evidence demonstrating that LasR folds into an active conformation in the absence of C12-HSLs and can remain in a properly folded signal-free state. Our results indeed confirm these findings as we demonstrate that LasRLBD can be produced as a soluble, dimeric and functional protein in extracts of bacteria that were grown in the absence of cognate C12-HSL ligands.
LasR Modulation by QS Activators and Inhibitors
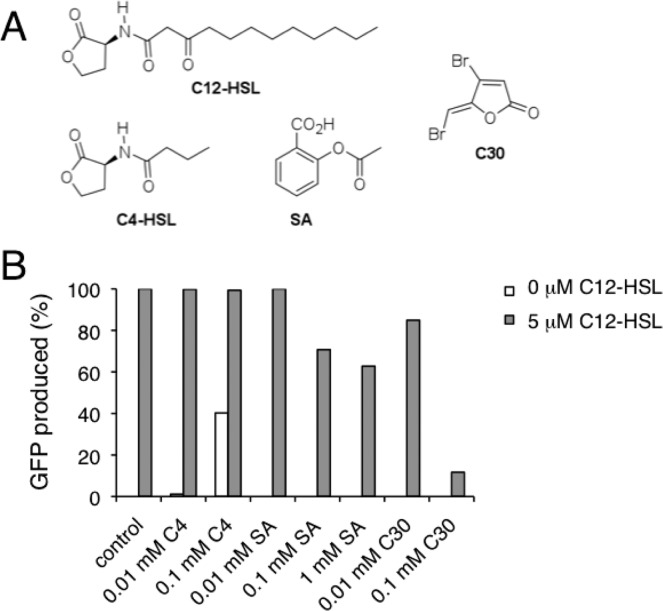
C4-HSL is the natural signaling ligand of P. aeruginosa RhlR QS receptor, and it has been shown to slightly activate the expression of a LasR-dependent lasB-lacZ translational fusion in E. coli.30 On the other hand, Furanone C30 and SA are synthetic compounds that inhibit LasR signaling,31,32 presumably by interacting with the acyl-HSL binding pocket of LasR as shown by molecular modeling studies.11,32,33 We analyzed the activity of these LasR modulating compounds by means of the E. coli MT102 biosensor bearing pMHLAS. A liquid culture of E. coli containing pMHLAS plasmid was divided into several subcultures, and incubated with C4-HSL, Furanone C30 or SA at different concentrations for 20 min. Subsequently, they were either treated or untreated with C12-HSL for 90 min, as explained in the Materials and Methods section, and GFP expression was determined by fluorescence spectroscopy. As shown in Figure 3B, C4-HSL was capable to induce GFP expression, though to a lesser extent than C12-HSL, demonstrating an agonistic activity over LasR. On the other hand, SA and Furanone C30 displayed strong inhibitory effects on GFP expression, while not affecting bacterial growth (not shown), which confirms their role as LasR antagonists.
Figure 3.
(A) Compounds used in LasR modulation studies: N-3-oxododecanoyl-homoserine lactone (C12-HSL), N-butyryl-homoserine lactone (C4-HSL, C4), acetylsalicylic acid (SA) and 4-bromo-5-(bromomethylene)-2(5H)-furanone (C30). (B) Evaluation of GFP expression from pMHLAS in E. coli MT102 bacteria upon incubation with the indicated amounts of C4, SA or C30 and subsequently treated without (white bars) or with (gray bars) C12-HSL. The mean from at least two independent experiments was normalized to control inductions (control) lacking C4, SA and C30.
SERS Analysis of Apo LasRLBD
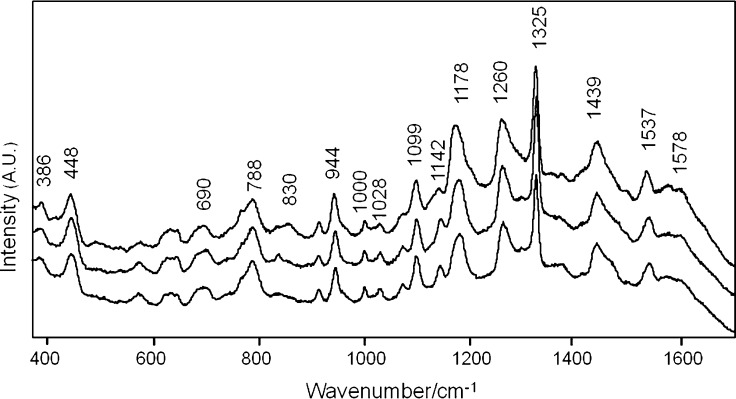
The SERS substrate was a gold nanoparticle thin film fabricated using polyelectrolyte layer-by-layer (LbL) assembly, by means of alternate deposition of a positively charged polyelectrolyte, poly(diallyldimethylammonium chloride) (PDDA), and negatively charged, citrate-stabilized Au NPs (60 nm diameter), on a glass slide. The resulting LbL film comprising three bilayers (PDDA-Au NPs) displayed a long-range uniform Au deposition (Supporting Information Figure S2), displaying a broad extinction band centered at 850 nm due to significant plasmon coupling and a less intense band at around 550 nm from single particles.34 Since nonspecific binding of LasRLBD to the Au nanoparticle surface would lead to nonhomogenous SERS signals, a cysteine amino acid was genetically introduced at the C-terminus of the polypeptide, immediately after the hexa-histidine tag. The high affinity toward Au-thiol binding has been widely employed for site-directed attachment of functional antibody fragments, enzymes or proteins on Au surfaces.35,36 In addition, the hexa-histidine tag has been reported to bind the polypeptide to gold surfaces through metal-histidine coordination.37 Thus, the attachment of LasRLBD onto the Au nanoparticle-based substrate should take place through its C-terminus, placing the polypeptide in the conformation illustrated in Figures 1A and 5. As shown in Figure 4, highly reproducible SERS spectra were obtained, most likely due to the targeted binding of LasRLBD to the gold surface. The main features of the SERS spectra were assigned according to the existing literature (Table 1). The SERS spectra (Figure 4) display the characteristic feature of Au–S vibrational mode at 386 cm–1 confirming the specific interaction between the C-terminal cysteine and the Au surface.38 In addition, the spectra show vibrational bands that could also be assigned to this amino acid (448, 690, and 788 cm–1) and to the histidine amino acid (1099, 1439, and 1578 cm–1) abundantly present in the hexa-histidine tag.39 The targeted attachment of LasRLBD orients the L3 loop toward the Au surface (Figure 5). Therefore, the following vibrational bands can be ascribed to amino acids present in the L3 loop and in its vicinity, such as Tyr-47 (peaks at 830, 1178, 1260, and 1537 cm–1), Phe-167 (peaks at 1000, 1028, and 1178 cm–1), Asp-43, Asp-46 and Glu-48, 124, and 168 (peaks at 944 and 1439 cm–1).39−43 Furthermore, the enhancement of modes corresponding to the asymmetric stretching of CCαN (1142 cm–1) suggests interactions of nitrogen atoms in histidine, arginine, lysine or glutamine amino-acids with the gold surface.39 The intense peak at 1325 cm–1 can be assigned to an amide III (in phase combination of the N–H bending and C–H stretching vibrations).39 We confirmed that the SERS spectral peaks shown in Figure 4 were characteristic of LasRLBD, as neither the buffer nor the gold nanoparticle substrate produced significant Raman signals (data not shown). Importantly, SERS spectra recorded at 1, 3, and 6 h after binding the polypeptide to the gold substrate did not show any significant changes, thus proving that the protein is stable under the assay conditions (Supporting Information Figure S3).
Figure 5.
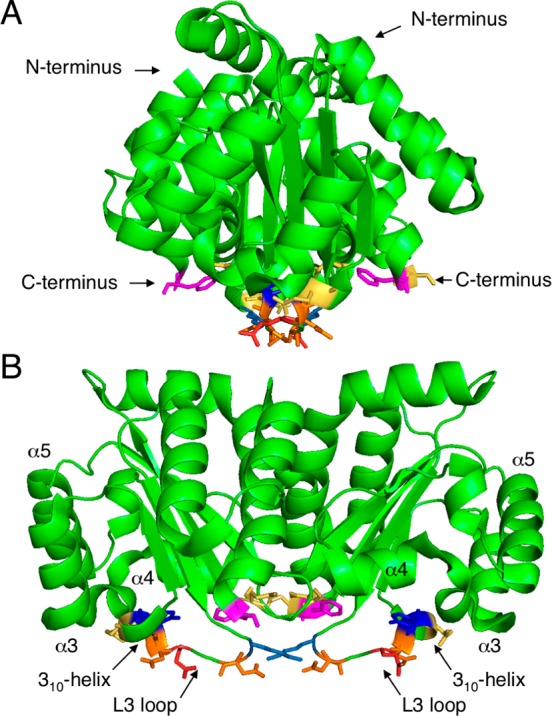
Schematic representation of possible contact points between LasRLBD and a gold surface. Side (A) and front (B) view of LasRLBD (PDB identification code 2UV0). Highlighted are relevant structural features (α3, α4, α5, 310-helix, L3 loop) and interacting amino acids: Lys-42 (light blue), Asp-43 (orange), Gln 45 (red), Asp-46 (orange), Tyr-47 (blue), Glu-48 (yellow), Phe-167 (magenta), and Glu-168 (yellow).
Figure 4.
SERS spectra of apo LasRLBD measured in three different regions of a gold nanoparticle-based substrate. Excitation with a 785 nm laser line was used to avoid protein damage.
Table 1. Experimental Vibrational Frequencies (cm–1) for LasRLBD with Tentative Assignments.
| LasR/cm–1 | band assignment |
|---|---|
| 386 | Au–S |
| 448 | Cys |
| 690 | Cys |
| 788 | Cys |
| 830 | Tyr |
| 944 | Glu, Asp |
| 1000 | Phe |
| 1028 | Phe |
| 1099 | His, CαN |
| 1142 | CCαN |
| 1178 | Tyr, Phe |
| 1260 | Tyr |
| 1325 | Amide III |
| 1439 | His, Glu, Lys |
| 1537 | Tyr |
| 1578 | NH and NH3+ def. |
Detection of Interactions between LasRLBD and QS Modulators by SERS
SERS was then used to analyze whether incubation of apo LasRLBD with either its cognate C12-HSL or with agonist C4-HSL ligands can induce conformational changes in the polypeptide. We attached apo LasRLBD (ca. 0.5 nmol in 10 mM sodium phosphate buffer at pH 8.0, PB buffer) to the Au nanoparticle-based substrate, followed by incubation with C12-HSL or C4-HSL (100 μM). As shown in Figure 6, the SERS spectra of LasRLBD forming a complex with either C12-HSL or C4-HSL, resemble that of ligand-free LasRLBD but display some remarkable differences: reduction in SERS intensity at 448, 690, and 788 cm–1 corresponding to the C-terminal cysteine and appearance of additional peaks at 408, 1405 and 1630 cm–1. The peak at 408 cm–1 was assigned to the deformation of the C–N vibrational mode,40 whereas the 1630 cm–1 band was assigned to an Amide I peak characteristic of a relatively uncommon secondary structure called the 310-helix.44,45 Interestingly, LasRLBD possesses a 310-helix conformed by the amino acids from Gly-68 to Arg-71, which follows the L3 loop (Figure 5) and has been shown to participate in the lactone-binding pocket.17 Finally, the band at 1405 cm–1 was assigned to the COO– symmetric vibration of aspartic and glutamic amino acids, both present in the L3 loop and 310-helix (Figure 5).43 As neither C12-HSL nor C4-HSL or the PB buffer produced any significant SERS signals at the concentration used in the binding assay when tested separately (data not shown), we can state that, the observed peaks are protein-specific. Altogether, these results show that incubation of apo LasRLBD with both QS activators gives rise to changes in the vibrational SERS spectra of the polypeptide, thereby indicating conformational changes linked to ligand interaction.
Figure 6.
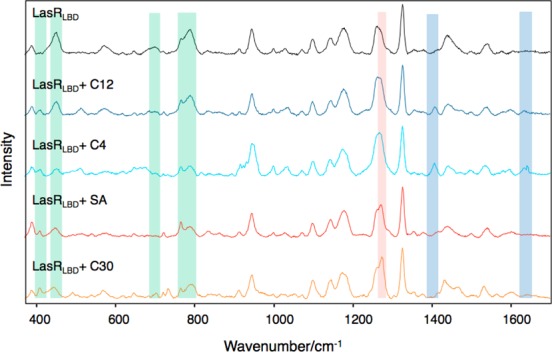
SERS spectra (after baseline correction) of LasRLBD polypeptide incubated with C12-HSL, C4-HSL, SA or Furanone C30. The green, red, and blue areas indicate common, inhibitor-specific, and activator-specific SERS fingerprints, respectively. Illumination with a 785 nm laser line was used to avoid protein damage.
Analysis of the interactions of LasRLBD with SA (100 μM) and Furanone C30 (10 μM) antagonists (Figure 6) showed changes in the SERS spectra, which were similar to those obtained with the acyl-HSLs, such as the appearance of the C–N stretching at 408 cm–1 and the decrease of the cysteine peaks at 448, 690, and 788 cm–1. Additionally, the LasR antagonists induced a new peak at 1267 cm–1, assigned to C=O stretching vibrations of an α-helix,46 which was not observed for C12-HSL or C4-HSL. This specific SERS signal could be attributed to movements of the α-helices α3, α4 and/or α5, which constitute the ligand-binding pocket (Figure 5). Importantly, the antagonists did not induce the 1405 and 1630 cm–1 peaks associated with structural movements involving the L3 loop and the 310-helix, thus confirming these SERS signals as signatures of the QS activators. SA was not detected by SERS on bare gold nanoparticle substrates at the concentration used in the binding assay (data not shown). On the other hand, Furanone C30 did yield strong SERS signals when analyzed separately (Supporting Information Figure S6). Interestingly, the SERS peaks corresponding to Furanone C30 itself were not detected upon incubation with LasRLBD, indicating that it did not reach the gold surface in the presence of the polypeptide, likely due to binding to LasRLBD. Finally, SERS measurements performed at 1, 3, and 6 h yielded similar spectra for each group of QS modulators (Supporting Information Figures S4 and S5) demonstrating the reliability of the data. These experiments also revealed that conformational changes in LasRLBD were evident within the first hour of incubation. The results, summarized in Table 2, clearly show SERS detection of structural changes in LasRLBD, associated with interactions of both activators and inhibitors of QS, and that each group of QS modulators produced specific SERS fingerprints.
Table 2. SERS Fingerprints of LasRLBD upon Binding to C12-HSL or C4-HSL (QS Activators) and SA or Furanone C30 (QS Inhibitors).
| QS activators | QS inhibitors | assignments |
|---|---|---|
| 408 | 408 | δ(CN) |
| 448 | 448 | Cys |
| 690 | 690 | Cys |
| 788 | 788 | Cys |
| 1267 | v(C=O) (α-helix) | |
| 1405 | COO–sym. stretch. | |
| 1630 | Amide I (310-helix) |
Molecular Docking analysis of LasRLBD–Ligand Interactions
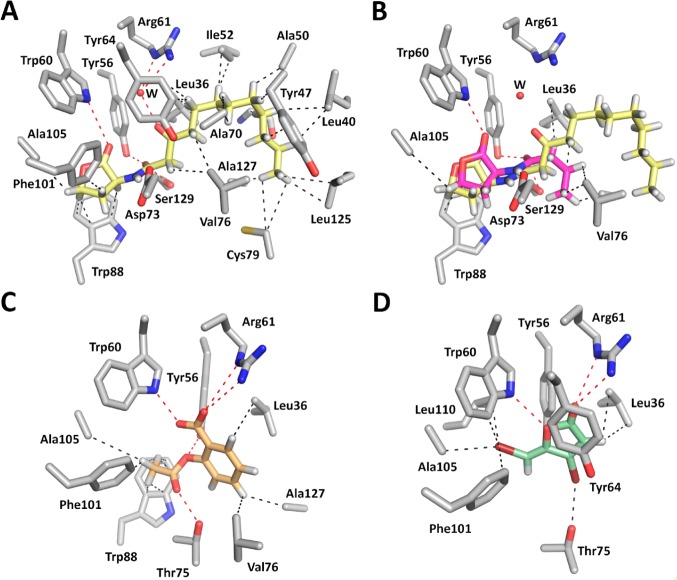
Molecular docking analysis was used to study the binding mode of C4-HSL, SA and Furanone C30 to LasRLBD, so as to obtain an insight into their functional roles as QS modulators. These studies were carried out using the program GOLD (version 5.2) and the protein geometries found in the crystal structure of the LasRLBD/C12-HSL binary complex (PDB identification code 2UV0). C12-HSL was initially docked as a control and the result compared to the PDB entry 2UV0. The efficacy of a compound to activate LasR was directly correlated with its ability to maintain the binding interactions observed in the LasRLBD/C12-HSL binary complex (Figure 7A). As illustrated in Figure 7B, the binding mode of the agonist C4-HSL is similar to that of the natural ligand as it retains most of the binding interactions of the homoserine lactone and amide moieties of C12-HSL, including four intermolecular hydrogen bonds involving Tyr-56, Trp-60, Asp-73 and Ser-129, CH−Π interactions with Trp-88 and a lipophilic contact with Ala-105. Interestingly, despite of the different lengths of their acyl chains, the two QS activators did not show any significant distinct SERS signals. This suggests that the molecular interactions of C12-HSL and C4-HSL with the recognition center of LasRLBD detected by SERS mainly depend on the homoserine lactone moieties. On the other hand, it has been proposed that exposure of buried hydrophobic residues in the lactone-binding pocket to the bulk solvent could be responsible for protein instability and degradation within the cell.18 Therefore, as C4-HSL lacks a full-length acyl chain it cannot meet most of the hydrophobic interactions established by C12-HSL (cf.Figures 7A and 7B), thus explaining why the noncognate C4-HSL molecule is not able to sustain full activation of LasR (Figure 3B).
Figure 7.
Modeling of QS ligand interactions with LasRLBD. (A) Binding mode of C12-HSL (yellow) in the recognition center of LasR receptor (gray). Hydrogen bonding (red) and lipophilic interactions (black) are shown. (B) Comparison between the predicted binding mode of C4-HSL (purple) and the natural autoinducer C12-HSL (yellow). (C) Plausible binding mode of SA (light orange). (D) Predicted binding mode of Furanone C30 (green).
Regarding LasR antagonists, molecular docking analysis revealed that both SA (Figure 7C) and Furanone C30 (Figure 7D) would establish similar polar contacts with the lactone-binding pocket of LasRLBD, involving Tyr-56, Trp-60, Thr-75 and Arg-61 residues. Small differences were however observed in the lipophilic contacts of both ligands, in particular involving Leu-110, Val-76 and Ala-127 residues. The shared topology may be responsible for inducing similar SERS spectra as experimentally observed (Figure 6). Although each compound (Figure 7C,D) seemed to establish some of the strongest contacts predicted for the homoserine lactone moieties (Figure 7B), the type of chemical interactions and their respective strengths were different (Supporting Information Table S1), which may explain their inhibitory effect on LasR signaling (Figure 3B). We speculate that SA and Furanone C30 do not establish contacts with key amino-acid residues in LasRLBD, thus failing to properly stabilize the polypeptide, which would result in an accelerated proteolytic degradation. Indeed, it has been reported that the half-life of LuxR protein of Vibrio fischeri is reduced up to 100-fold in the presence of Furanone C30, suggesting that furanones control expression of acyl-HSL-dependent QS by destabilizing LuxR.31 Muh and co-workers used ultrahigh-throughput screening to search for inhibitors in a library of 200 000 compounds. The screening identified triphenyl (TP) compounds that are structurally different from acyl-HSLs, bearing either agonist or antagonist activity of the LasR-dependent fluorescent reporter. In silico docking studies suggested that hydrogen bond interactions of these molecules with Trp-60 and Asp-73 influenced whether the TP compounds act as an agonist or an antagonist.9 The authors indicated that only those molecules establishing contacts with both Trp-60 and Asp-73 function as agonists. Interestingly, our modeling study showed that whereas C12-HSL and C4-HSL were predicted to maintain interactions with Trp-60 and Asp-73, SA and Furanone C30 were predicted to interact with Trp-60 only (Figure 7). In a follow-up study, Zou and Nair obtained crystal structures of LasRLBD complexing TP compounds of agonistic effect in LasR, demonstrating that these molecules do establish hydrogen bond interactions with Trp-60 and Asp-73 amino acids. Unfortunately, the authors could not demonstrate the importance of these residues in the molecular mechanism for an agonist to antagonist switching as they could not determine the X-ray crystallographic structure of LasRLBD bound to the antagonistic triphenyl compound TP-5, due to instability of this complex.19
Conclusions
Upon demonstration by biochemical and biological activity assays that the LBD of LasR receptor proteins can fold into a functional conformation in the absence of its cognate acyl-HSL ligand, SERS spectroscopy allowed us to obtain direct experimental evidence of conformational changes in LasRLBD upon interaction with activators and inhibitors of P. aeruginosa quorum sensing. SERS spectra indicated structural rearrangements in the polypeptide that were specific to either activators (310-helix and L3 loop) or inhibitors (α-helices α3, α4 and/or α5). Moreover, the binding of QS modulators to LasRLBD influenced the interaction of the C-terminal cysteine of the polypeptide with an Au nanoparticle film, which thus acted as an efficient SERS substrate to sense protein–ligand interactions. Docking studies allowed us to provide insight into the molecular interactions between QS modulators and LasRLBD that may be involved in the differential SERS fingerprints obtained. As a prospect, SERS can be used in combination with structure-based approaches to understand protein–ligand systems, thereby providing a fast, cost-effective, and robust technique for screening interactions between proteins and drug molecules.
Materials and Methods
Materials
Tetrachloroauric(III) acid trihydrate (HAuCl4·3H2O), trisodium citrate dihydrate, poly(diallyldimethylammonium chloride) (PDDA, average Mw 100 000–200 000), hydrogen peroxide (H2O2, 28%) and sulfuric acid (H2SO4, 98%) were supplied by Aldrich. N-(3-Oxododecanoyl)-l-homoserine lactone (C12-HSL), acetylsalicylic acid (SA), (Z)-4-bromo-5-(bromomethylene)-2(5H)-furanone (C30), HIS-Select HF Nickel affinity gel and modifying enzymes were purchased from Sigma-Aldrich. N-Butyryl-l-homoserine lactone (C4-HSL) was from Cayman Chemical. Restriction enzymes were supplied by New England Biolabs. Disuccinimidyl suberate (DSS) and Zeba Spin desalting columns, 7k MWCO, were from Pierce. Other reagents as salts, buffers and enzymes were acquired from Sigma-Aldrich or Merck. All chemicals were used as received. Pure grade ethanol and Milli-Q water were used as solvents. DNA treatment with restriction enzymes, ligation and transformation of E. coli were performed using standard methods. For DNA isolation, the Spin Miniprep Kit (Qiagen) was used and DNA fragments were excised and purified from agarose gels using the PCR purification kit (Qiagen).
Methods
Plasmid Constructs and Protein Expression
DNA encoding from Met-1 to Lys-173 of lasR gene was amplified from plasmid pMHLAS using the oligonucleotides LasR-NdeI F (CACTGCCATATGGCCTTGGTTGACGGTTTTCTTG) and LasR-HindIII-6×HC R (CCTAGTAAGCTTTCAGCAGTGGTGGTGGTGGTGGTGTTTGCTGACCGGATGTTCGAAGGCCAGTC) (Sigma Genosys) and cloned on the NdeI and HindIII sites of vector pET21a(+) (Novagen). The reverse primer (LasR-HindIII-6×HC R) was designed to add a hexa-histidine tag and a cysteine amino acid residue at the C-terminus of the polypeptide. The resulting plasmid was sequenced at the CACTI sequencing facilities (Universidade de Vigo, Spain).
Protein Expression and Purification
The fusion protein LasR(1–173)-His6-Cys (LasRLBD) was expressed in E. coli BL21(DE3)pLysS cells grown in Luria–Bertani (LB) broth plus 100 μg/mL ampicillin, 100 μg/mL chloramphenicol and 2% glucose until an OD600 of 0.4. The media was then replaced with fresh LB plus antibiotics without glucose, and protein expression was induced with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) at 28 °C during 4 h. The bacterial pellet was lysed in CelLytic B buffer (Sigma) supplemented with 5 mM MgCl2, 1 mM PMSF, protease inhibitor cocktail (Sigma), and deoxyribonuclease I (Sigma-Aldrich). LasRLBD was purified by Ni2+ affinity chromatography using the HIS-Select HF nickel affinity gel (Sigma-Aldrich) following the commercial protocol. Protein elution was performed using 300 mM imidazole in PBS. The protein was desalted with Zeba Spin columns equilibrated in 10 mM sodium phosphate buffer at pH 8.0, and the sample was kept at 4 °C for immediate use. Proteins were not stored at 4 °C for more than 1 day.
Protein Cross-Linking
Purified LasRLBD protein was preincubated with 20 μM C12-HSL or the equivalent volume of ethanol during 45 min at room temperature. When indicated, 0.5 mM disuccinimidyl suberate (DSS) was added to the samples and the mixtures were incubated at room temperature for 40 min. The reaction was stopped by the addition of 20 mM Tris-HCl pH 8.0. Subsequently, samples were analyzed on a 12% SDS-PAGE gel, and proteins were visualized by Coomassie Brilliant Blue staining.
Quorum Sensing Reporter System and Quorum Quenching Assay
Affinity purified apo LasRLBD (approximately 0.5 nmol) and BSA (0.5 mmol) in PBS or PBS were incubated with 5 μM C12-HSL for 1 h at room temperature. The mixture was subsequently added to a cellular suspension of MT102 E. coli bacteria carrying pMHLAS in LB (OD600 of 0.4) and further incubated for 90 min at 30 °C to allow GFP expression. Fluorescence was monitored using a Leica fluorescence microscope, and GFP expression was measured at 488 nm using the EnVision ELISA plate reader (PerkinElmer). Analysis of the functional activity of QS ligands was performed as follows: 300 mL of bacteria grown to an OD600 of 0.4 was incubated with 0.01 mM or 0.1 mM C4-HSL, 0.01 mM, 0.1 mM or 1 mM SA, and 0.01 mM or 0.1 mM C30 for 20 min at 30 °C under shaking. Then, 5 μM C12-HSL was added and bacteria were subsequently incubated for 90 min at 30 °C. GFP expression was measured as previously indicated.
Synthesis and Characterization of Gold Nanoparticle Thin Films
Citrate-stabilized AuNPs (∼60 nm in diameter) were prepared by the kinetically controlled seeded growth method.47 Briefly, 150 mL of 2.2 mM trisodium citrate in Milli-Q water was heated to boiling under vigorously stirring. After 15 min, 1 mL of 25 mM HAuCl4 was injected into the boiling reaction mixture, incubated for 10 min and the reaction mixture was cooled down to 90 °C. Subsequently, 1 mL of a 25 mM HAuCl4 aqueous solution was injected into the reaction mixture. After additional 30 min incubation, 55 mL of 60 mM sodium citrate was added. The final solution was used as seed, and the process was repeated again (but with just two injections of the HAuCl4 aqueous solution) six times to yield 60 nm Au NPs. Glass slides were washed in piranha solution for 30 min and then copiously rinsed with pure water and stored in water until use. Au NPs films were formed by immersing the glass slides in an aqueous PDDA solution (1 mg/mL, 0.05 M NaCl) for 15 min, rinsed with water, dried and then immersed in the Au NPs solution for 1 day, followed by rinsing with water. The process was repeated two more times to obtain the final material. For substrate characterization, UV–vis-NIR spectra were recorded using an Agilent 8453 spectrophotometer. SEM images were obtained using a JEOL JSM-6700F FEG scanning electron microscope operating at an acceleration voltage of 10.0 kV.
Surface-Enhanced Raman Scattering
SERS measurements were conducted with a Renishaw InVia Reflex system. The spectrograph used a high-resolution grating (1800 grooves cm–1) with additional band-pass filter optics, a confocal microscope and a 2D-CCD camera. Laser excitation was carried out at 785 nm (diode) with a 20× objective (N.A. 0.4), 4.7 mW power and 10 s acquisition time. The substrates were immersed in a solution of 10 mM sodium phosphate buffer at pH 8.0 containing ca. 0.5 nmol of purified LasRLBD and incubated for 16 h at 4 °C. Then, the different ligand molecules were added at the indicated concentrations and incubation was carried out at room temperature until SERS measurements.
Docking Studies
The program GOLD 5.248 was used and protein geometries were found in the crystal structure of the binary complex LasRLBD/C12-HSL (PDB identification code 2UV0). The receptor was used in the dimer form. The ligand and all water molecules were removed from the crystal structure with the exception of water molecule W2134 for docking with ligands C12-HSL and C4-HSL. This water molecule is close to the carbonyl group of C12-HSL side chain and seems to be responsible for the appropriate arrangement of ligand side chains. Ligand geometries were minimized using the AM1 Hamiltonian as implemented in Gaussian 0949 and used as MOL2 files. Each ligand was docked in 25 independent genetic algorithm (GA) runs, and for each of these a maximum number of 100 000 GA operations were performed on a single population of 50 individuals. Operator weights for crossover, mutation and migration in the entry box were used as default parameters (95, 95, and 10, respectively), as well as the hydrogen bonding (4.0 Å) and van der Waals (2.5 Å) parameters. The position of C12-HSL in the crystal structure was used to define the active-site and the radius was set to 10 Å. The “flip ring corners” flag was switched on, while all the other flags were off. The GOLD scoring function was used to rank the ligands in order of fitness.
Acknowledgments
This work has been funded by the European Research Council (ERC Advanced Grant #267867 Plasmaquo). We are grateful to Michael Givskov for kindly providing the pMHLAS vector. V.L.-P. acknowledges an FPI scholarship from the Spanish MINECO. C.G.-B. acknowledges funding from the Spanish MINECO (SAF2013-42899-R) and Xunta de Galicia (GRC2013-041).
Supporting Information Available
Various figures, schemes and tables including additional SERS spectra showing stability and reproducibility of substrates. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.5b01800.
Author Contributions
# These authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Moskovits M. Surface-Enhanced Raman Spectroscopy: A Brief Retrospective. J. Raman Spectrosc. 2005, 36, 485–496. [Google Scholar]
- Willets K. A.; Van Duyne R. P. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [DOI] [PubMed] [Google Scholar]
- Schlucker S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chem., Int. Ed. 2014, 53, 4756–4795. [DOI] [PubMed] [Google Scholar]
- Siddhanta S.; Narayana C. Surface Enhanced Raman Spectroscopy of Proteins: Implications for Drug Designing Invited Review Article. Nanomater. Nanotechnol. 2012, 2, 1–10. [Google Scholar]
- Ahijado-Guzman R.; Gomez-Puertas P.; Alvarez-Puebla R. A.; Rivas G.; Liz-Marzan L. M. Surface-Enhanced Raman Scattering-Based Detection of the Interactions between the Essential Cell Division Ftsz Protein and Bacterial Membrane Elements. ACS Nano 2012, 6, 7514–7520. [DOI] [PubMed] [Google Scholar]
- Karthigeyan D.; Siddhanta S.; Kishore A. H.; Perumal S. S. R. R.; Agren H.; Sudevan S.; Bhat A. V.; Balasubramanyam K.; Subbegowda R. K.; Kundu T. K.; et al. SERS and MD Simulation Studies of a Kinase Inhibitor Demonstrate the Emergence of a Potential Drug Discovery Tool. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 10416–10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case R. J.; Labbate M.; Kjelleberg S. Ahl-Driven Quorum-Sensing Circuits: Their Frequency and Function among the Proteobacteria. ISME J. 2008, 2, 345–349. [DOI] [PubMed] [Google Scholar]
- Galloway W. R. J. D.; Hodgkinson J. T.; Bowden S. D.; Welch M.; Spring D. R. Quorum Sensing in Gram-Negative Bacteria: Small-Molecule Modulation of Ahl and Al-2 Quorum Sensing Pathways. Chem. Rev. 2011, 111, 28–67. [DOI] [PubMed] [Google Scholar]
- Churchill M. E. A.; Chen L. L. Structural Basis of Acyl-Homoserine Lactone-Dependent Signaling. Chem. Rev. 2011, 111, 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Nair S. K. Quorum Sensing: How Bacteria Can Coordinate Activity and Synchronize Their Response to External Signals?. Protein Sci. 2012, 21, 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C.; Greenberg E. P. Listening in on Bacteria: Acyl-Homoserine Lactone Signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 685–695. [DOI] [PubMed] [Google Scholar]
- Schuster M.; Sexton D. J.; Diggle S. P.; Greenberg E. P. Acyl-Homoserine Lactone Quorum Sensing: From Evolution to Application. Annu. Rev. Microbiol. 2013, 67, 43–63. [DOI] [PubMed] [Google Scholar]
- Pesci E. C.; Pearson J. P.; Seed P. C.; Iglewski B. H. Regulation of Las and Rhl Quorum Sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P.; Gray K. M.; Passador L.; Tucker K. D.; Eberhard A.; Iglewski B. H.; Greenberg E. P. Structure of the Autoinducer Required for Expression of Pseudomonas-aeruginosa Virulence Genes. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh U.; Hare B. J.; Duerkop B. A.; Schuster M.; Hanzelka B. L.; Heim R.; Olson E. R.; Greenberg E. P. A Structurally Unrelated Mimic of a Pseudomonas aeruginosa Acyl-Homoserine Lactone Quorum-Sensing Signal. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 16948–16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlee B. R.; Geske G. D.; Blackwell H. E.; Handelsman J. Identification of Synthetic Inducers and Inhibitors of the Quorum-Sensing Regulator Lasr in Pseudomonas aeruginosa by High-Throughput Screening. Appl. Environ. Microbiol. 2010, 76, 8255–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley M. J.; Muraglia E.; Bazzo R.; Carfi A. Molecular Insights into Quorum Sensing in the Human Pathogen Pseudomonas aeruginosa from the Structure of the Virulence Regulator Lasr Bound to Its Autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. [DOI] [PubMed] [Google Scholar]
- Zou Y. Z.; Nair S. K. Molecular Basis for the Recognition of Structurally Distinct Autoinducer Mimics by the Pseudomonas aeruginosa Lasr Quorum-Sensing Signaling Receptor. Chem. Biol. 2009, 16, 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiratisin P.; Tucker K. D.; Passador L. LasR, a Transcriptional Activator of Pseudomonas aeruginosa Virulence Genes, Functions as a Multimer. J. Bacteriol. 2002, 184, 4912–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M.; Riedel K.; Rasmussen T. B.; Heydorn A.; Andersen J. B.; Parsek M. R.; Rice S. A.; Eberl L.; Molin S.; Hoiby N.; et al. Inhibition of Quorum Sensing in Pseudomonas aeruginosa Biofilm Bacteria by a Halogenated Furanone Compound. Microbiology 2002, 148, 87–102. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Winans S. C. The Quorum-Sensing Transcriptional Regulator Trar Requires Its Cognate Signaling Ligand for Protein Folding, Protease Resistance, and Dimerization. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M.; Urbanowski M. L.; Greenberg E. P. Promoter Specificity in Pseudomonas aeruginosa Quorum Sensing Revealed by DNA Binding of Purified LasR. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 15833–15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. P.; Luo Z. Q.; Smyth A. J.; Gao P.; von Bodman S. B.; Farrand S. K. Quorum-Sensing Signal Binding Results in Dimerization of Trar and Its Release from Membranes into the Cytoplasm. EMBO J. 2000, 19, 5212–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.; Todd D. E.; Whitehead N. A.; McGowan S. J.; Bycroft B. W.; Salmond G. P. C. N-Acyl Homoserine Lactone Binding to the Carr Receptor Determines Quorum-Sensing Specificity in Erwinia. EMBO J. 2000, 19, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu D. J.; Ramachandran R.; Geissinger J. S.; Stevens A. M. Probing the Impact of Ligand Binding on the Acyl-Homoserine Lactone-Hindered Transcription Factor Esar of Pantoea stewartii Subsp Stewartii. J. Bacteriol. 2011, 193, 6315–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. G.; Pappas T.; Brace J. L.; Miller P. C.; Oulmassov T.; Molyneauz J. M.; Anderson J. C.; Bashkin J. K.; Winans S. C.; Joachimiak A. Structure of a Bacterial Quorum-Sensing Transcription Factor Complexed with Pheromone and DNA. Nature 2002, 417, 971–974. [DOI] [PubMed] [Google Scholar]
- Lintz M. J.; Oinuma K. I.; Wysoczynski C. L.; Greenberg E. P.; Churchill M. E. A. Crystal Structure of Qscr, a Pseudomonas aeruginosa Quorum Sensing Signal Receptor. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 15763–15768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappington K. J.; Dandekar A. A.; Oinuma K. I.; Greenberg E. P. Reversible Signal Binding by the Pseudomonas aeruginosa Quorum-Sensing Signal Receptor LasR. MBio 2011, 2, e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen A.; Jakobsen T. H.; Bjarnsholt T.; Givskov M.; Welch M.; Ferkinghoff-Borg J.; Sams T. Kinetic Model for Signal Binding to the Quorum Sensing Regulator LasR. Int. J. Mol. Sci. 2013, 14, 13360–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P.; Pesci E. C.; Iglewski B. H. Roles of Pseudomonas aeruginosa Las and Rhl Quorum-Sensing Systems in Control of Elastase and Rhamnolipid Biosynthesis Genes. J. Bacteriol. 1997, 179, 5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M.; Rasmussen T. B.; Henzter M.; Andersen J. B.; Steinberg P.; Kjelleberg S.; Givskov M. Halogenated Furanones Inhibit Quorum Sensing through Accelerated Luxr Turnover. Microbiology 2002, 148, 1119–1127. [DOI] [PubMed] [Google Scholar]
- El-Mowafy S. A.; Abd El Galil K. H.; El-Messery S. M.; Shaaban M. I. Aspirin Is an Efficient Inhibitor of Quorum Sensing, Virulence and Toxins in Pseudomonas aeruginosa. Microb. Pathog. 2014, 74, 25–32. [DOI] [PubMed] [Google Scholar]
- Yang L.; Rybtke M. T.; Jakobsen T. H.; Hentzer M.; Bjarnsholt T.; Givskov M.; Tolker-Nielsen T. Computer-Aided Identification of Recognized Drugs as Pseudomonas aeruginosa Quorum-Sensing Inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial S.; Pastoriza-Santos I.; Perez-Juste J.; Liz-Marzan L. M. Plasmon Coupling in Layer-by-Layer Assembled Gold Nanorod Films. Langmuir 2007, 23, 4606–4611. [DOI] [PubMed] [Google Scholar]
- Backmann N.; Zahnd C.; Huber F.; Bietsch A.; Pluckthun A.; Lang H. P.; Guntherodt H. J.; Hegner M.; Gerber C. A Label-Free Immunosensor Array Using Single-Chain Antibody Fragments. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.; Lee J. M.; Jung Y.; Habtemariam T.; Salah A. W.; Fermin C. D.; Kim M. Combination of Cysteine- and Oligomerization Domain-Mediated Protein Immobilization on a Surface Plasmon Resonance (Spr) Gold Chip Surface. Analyst 2011, 136, 2506–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogot J. M.; England H. J.; Strouse G. F.; Logan T. M. Single Peptide Assembly onto a 1.5 nm Au Surface via a Histidine Tag. J. Am. Chem. Soc. 2008, 130, 16156–16157. [DOI] [PubMed] [Google Scholar]
- Wang H.; Levin C. S.; Halas N. J. Nanosphere Arrays with Controlled Sub-10-Nm Gaps as Surface-Enhanced Raman Spectroscopy Substrates. J. Am. Chem. Soc. 2005, 127, 14992–14993. [DOI] [PubMed] [Google Scholar]
- Arif M.; Karthigeyan D.; Siddhanta S.; Kumar G. V.; Narayana C.; Kundu T. K. Analysis of Protein Acetyltransferase Structure-Function Relation by Surface-Enhanced Raman Scattering (Sers): A Tool to Screen and Characterize Small Molecule Modulators. Methods Mol. Biol. 2013, 981, 239–261. [DOI] [PubMed] [Google Scholar]
- Aliaga A. E.; Aguayo T.; Garrido C.; Clavijo E.; Hevia E.; Gomez-Jeria J. S.; Leyton P.; Campos-Vallette M. M.; Sanchez-Cortes S. Surface-Enhanced Raman Scattering and Theoretical Studies of the C-Terminal Peptide of the Beta-Subunit Human Chorionic Gonadotropin without Linked Carbohydrates. Biopolymers 2011, 95, 135–143. [DOI] [PubMed] [Google Scholar]
- Wei F.; Zhang D. M.; Halas N. J.; Hartgerink J. D. Aromatic Amino Acids Providing Characteristic Motifs in the Raman and Sers Spectroscopy of Peptides. J. Phys. Chem. B 2008, 112, 9158–9164. [DOI] [PubMed] [Google Scholar]
- Podstawka E.; Ozaki Y.; Proniewicz L. M. Part I: Surface-Enhanced Raman Spectroscopy Investigation of Amino Acids and Their Homodipeptides Adsorbed on Colloidal Silver. Appl. Spectrosc. 2004, 58, 570–580. [DOI] [PubMed] [Google Scholar]
- Barth A.; Zscherp C. What Vibrations Tell Us About Proteins. Q. Rev. Biophys. 2002, 35, 369–430. [DOI] [PubMed] [Google Scholar]
- Pande J.; Pande C.; Gilg D.; Vasak M.; Callender R.; Kagi J. H. R. Raman, Infrared, and Circular-Dichroism Spectroscopic Studies on Metallothionein—A Predominantly Turn-Containing Protein. Biochemistry 1986, 25, 5526–5532. [DOI] [PubMed] [Google Scholar]
- Gullekson C.; Lucas L.; Hewitt K.; Kreplak L. Surface-Sensitive Raman Spectroscopy of Collagen I Fibrils. Biophys. J. 2011, 100, 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G. V. P.; Reddy B. A. A.; Arif M.; Kundu T. K.; Narayana C. Surface-Enhanced Raman Scattering Studies of Human Transcriptional Coactivator P300. J. Phys. Chem. B 2006, 110, 16787–16792. [DOI] [PubMed] [Google Scholar]
- Bastus N. G.; Comenge J.; Puntes V. Kinetically Controlled Seeded Growth Synthesis of Citrate-Stabilized Gold Nanoparticles of up to 200 nm: Size Focusing versus Ostwald Ripening. Langmuir 2011, 27, 11098–11105. [DOI] [PubMed] [Google Scholar]
- CCDC Web page. http://www.ccdc.cam.ac.uk/products/life_sciencies/gold/.
- Frisch M. J. T., G W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.. et al. Gaussian 09, Revision A.2 ed.; Gaussian, Inc.: Wallingford, CT, 2009.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.