Abstract
The formation of programmed DNA double-strand breaks (DSBs) at the beginning of meiotic prophase marks the initiation of meiotic recombination. Meiotic DSB formation is catalyzed by SPO11 and their repair takes place on meiotic chromosome axes. The evolutionarily conserved MEI4 protein is required for meiotic DSB formation and is localized on chromosome axes. Here, we show that HORMAD1, one of the meiotic chromosome axis components, is required for MEI4 localization. Importantly, the quantitative correlation between the level of axis-associated MEI4 and DSB formation suggests that axis-associated MEI4 could be a limiting factor for DSB formation. We also show that MEI1, REC8 and RAD21L are important for proper MEI4 localization. These findings on MEI4 dynamics during meiotic prophase suggest that the association of MEI4 to chromosome axes is required for DSB formation, and that the loss of this association upon DSB repair could contribute to turning off meiotic DSB formation.
KEY WORDS: Meiosis, Recombination, DNA double strand break, Synapsis
INTRODUCTION
Sexual reproduction relies on faithful chromosome transmission during meiosis to generate viable gametes. At the prophase of meiosis I, the programmed formation of DNA double-strand breaks (DSBs) is a key step that initiates homologous recombination events on each chromosome. Physical connections resulting from reciprocal exchanges (crossovers) generated by DSB repair between homologous chromosomes (homologs) play a mechanical role to ensure the accurate segregation of homologs at the first meiotic division (Hunter, 2007). As DSBs pose a potential threat to genome integrity, the timing and frequency of DSB formation, as well as their localization, are highly regulated. Meiotic DSBs are formed in a narrow time window at the leptotene stage of meiotic prophase after the completion of meiotic DNA replication (S phase). Moreover, DSBs occur preferentially in specific regions of the genome called hotspots. The number of DSBs per meiosis varies from tens in worms and flies to one hundred or more in yeast, plants and mammals (de Massy, 2013) and recent studies have identified several regulatory pathways for the control of timing and level of meiotic DSBs (Keeney et al., 2014). The evolutionarily conserved SPO11 protein, which carries the catalytic activity for DSB formation, is not sufficient on its own and other proteins that are essential for meiotic DSB formation have been identified in several organisms. Some of these DSB factors are proposed to act in sub-complexes and show limited sequence conservation during evolution (de Massy, 2013).
A more precise view of the factors involved in DSB formation has emerged from studies in S. cerevisiae, where at least nine proteins are required for Spo11-dependent DSB formation. Among these DSB factors Rec114, Mei4 and Mer2 form a complex (RMM complex), localized on chromosome axes before DSB formation and this complex is also conserved in S. pombe (Miyoshi et al., 2012). Recruitment of both Rec114 and Mei4 on meiotic chromosomes is dependent on Mer2, which associates with chromatin during meiotic S phase (Henderson et al., 2006; Li et al., 2006; Panizza et al., 2011). The phosphorylation of Mer2 by replication-associated kinases has been shown to allow the coordination between DNA replication and DSB formation (Murakami and Keeney, 2014). The DNA damage checkpoint turns off Rec114 activity, by a phosphorylation mechanism that is dependent on Tel1 and Mec1 (the budding yeast ATM and ATR orthologs), to limit DSB frequency (Carballo et al., 2013). The axis localization of the RMM suggests that DSB formation takes place on the axis. The chromosome axis is a proteinaceous structure that has been proposed to serve as a platform for different meiotic recombination steps. On this structure, called the axial element, chromatin is organized as an array of loops of both sister chromatids that are anchored to the axis at sites that correspond to cohesin-binding sites (Kleckner, 2006; Zickler and Kleckner, 1999). Axis formation is as yet poorly characterized but is known to involve chromosomal architectural proteins, such as condensins and type II DNA topoisomerases (Wood et al., 2010; Zickler and Kleckner, 1999), and other meiosis-specific factors. Among these, the S. cerevisiae Hop1 and Red1 proteins are spatially enriched in similar domains along chromosomes (Börner et al., 2008) and their localization is partly dependent on the meiosis-specific cohesin subunit Rec8 (Klein et al., 1999; Panizza et al., 2011). The S. cerevisiae rec8Δ strain shows region-specific variations in meiotic DSB levels (Kim et al., 2010; Kugou et al., 2009) that correlate with Hop1 localization in these regions (Panizza et al., 2011). In addition, Hop1 and Red1 are required for wild-type levels of meiotic DSBs (Hunter, 2007) and for the association of the RMM complex with the meiotic axis (Panizza et al., 2011). These data highlight a role for the axis structure in determining chromosomal domains for DSB formation. In addition, work in S. cerevisiae indicates that the DNA sequences where DSBs take place are located in chromatin loops. This finding led to the proposition of a mechanism to tether these sites to the axis (Blat et al., 2002; Panizza et al., 2011). Two recent reports in S. cerevisiae have shown that Spp1 tethers DSB sites to meiotic axes through its direct interaction with Mer2 (Acquaviva et al., 2013; Sommermeyer et al., 2013). In C. elegans, support for a functional link between DSB formation and higher-order chromosome structures is based on the parallel increase of the chromosome axis length and DSB activity in the absence of condensin I (Mets and Meyer, 2009).
In mammals, the knowledge on the regulation of meiotic DSB formation is more limited, partly because only three mouse proteins are known to be essential for DSB formation (SPO11, MEI1 and MEI4) (Baudat et al., 2013). MEI1 was discovered in a genetic screen and is required for DSB formation (Libby et al., 2003), but its molecular function is not understood yet. Our previous work has demonstrated that yeast Mei4 and Rec114 orthologs are evolutionarily conserved in most eukaryotes. We have shown that mouse MEI4 is required for meiotic DSB formation, and that MEI4 is located at discrete foci on meiotic chromosome axes (Kumar et al., 2010). The axis-associated protein HORMAD1, the yeast Hop1 ortholog and components of the cohesin complexes are required for wild-type DSB levels (Biswas et al., 2013; Daniel et al., 2011; Ishiguro et al., 2014; Llano et al., 2012). However, the nature and control of the interplay between DSB factors and chromosome axis components remain to be analyzed in the mouse.
In the present study, we investigated the assembly and disassembly dynamics of MEI4 foci on meiotic chromosomes by analyzing spermatocytes from mouse strains with defects in various steps linked to recombination, such as cohesin recruitment, meiotic progression, axis formation, DSB formation, DSB repair and synapsis. In addition, we also examined the timing of MEI4 association with meiotic chromosomes before entry in prophase I. Our results shed light on the interplay between meiotic axis components and DSB formation in the mouse and highlight key components involved in the generation and regulation of DSBs in mammalian meiosis. The correlation between the reduced levels of MEI4 axis association and DSBs in several mutants (Hormad1−/−, cohesin mutants and Mei1−/−) is consistent with a role for MEI4 in the context of the chromosome axis.
RESULTS
Timing of MEI4 recruitment
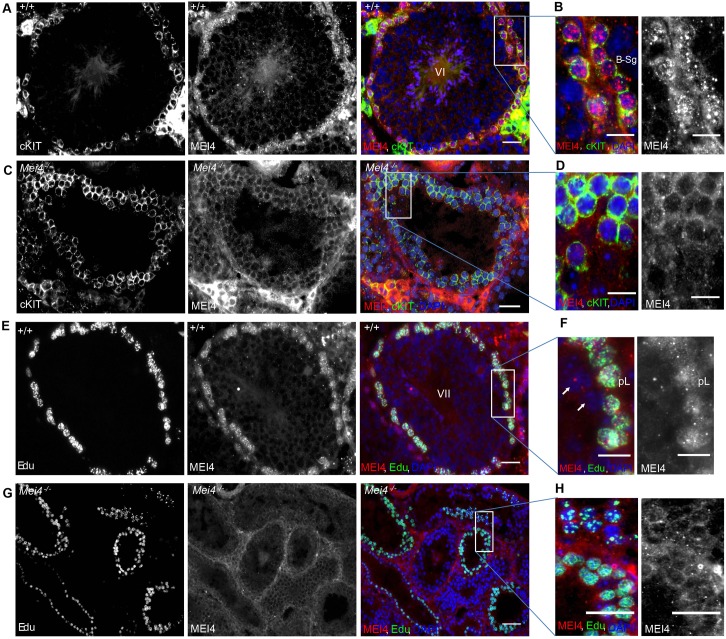
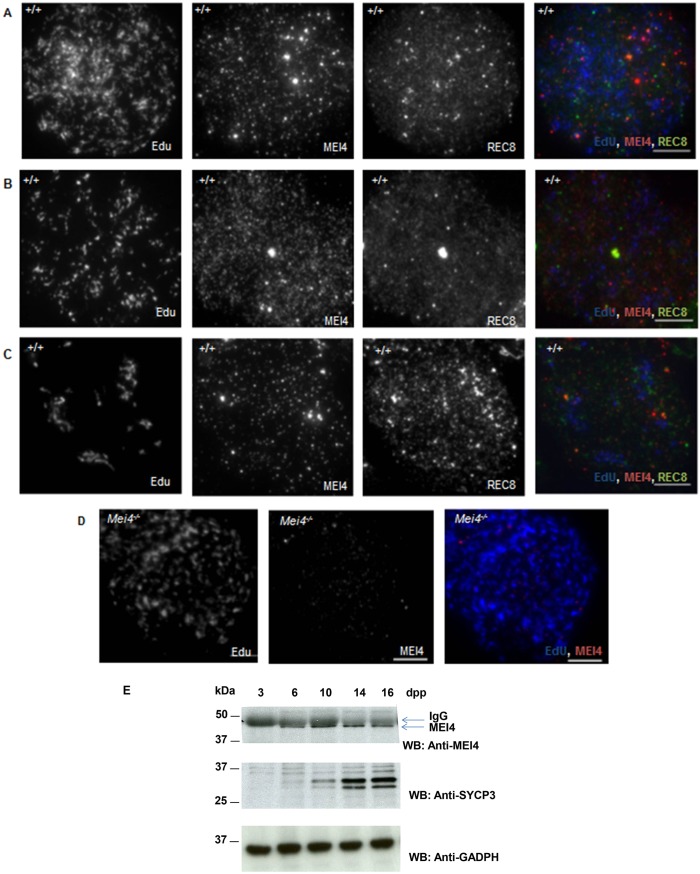
Mouse MEI4 is detected as multiple discrete foci on meiotic chromosome axes at the leptotene stage of meiotic prophase, when meiotic DSB formation takes place. These foci are also present in the absence of SPO11 (Kumar et al., 2010). Given its requirement for meiotic DSB formation, MEI4 should be cytologically detectable before DSB formation. Indeed, MEI4 nuclear foci can be observed before germ cells enter into meiosis, in B type spermatogonia that are identified based on their organization within the seminiferous tubules and on the expression of the tyrosine kinase receptor cKIT (Fig. 1A,B). This receptor is involved in cell proliferation, differentiation and is expressed in differentiated germ cell precursors of spermatocytes (spermatogonia A, intermediate and B types) and at the preleptotene stage (meiotic S phase), but not in leptotene spermatocytes. No MEI4 signal was detected in cells from Mei4−/− mice (Fig. 1C,D). We then tested whether MEI4 was detectable in testis sections during the meiotic S phase that precedes meiotic prophase by using 5-ethynyl-2′-deoxyuridine (EdU), a uridine analog that is incorporated during DNA replication and can be detected with a specific fluorescent dye. In wild-type preleptotene cells, identified as EdU-positive and SYCP3-negative cells, MEI4 was detected with a heterogeneous nuclear distribution with foci of variable intensities (Fig. 1E,F). These foci were absent in Mei4−/− meiocytes (Fig. 1G,H). The signal intensity variations between B type spermatogonia and preleptotene cells (compare Fig. 1A,B,E,F) might be due to differences in epitope accessibility, or could reflect distinct nuclear organizations of MEI4 in these cells. MEI4 foci were not detected in spermatocytes at late stages of meiotic prophase (i.e. pachytene spermatocytes in tubules at stage VII of development that are located towards the tubule lumen) (Fig. 1F), as previously shown by analysis of chromosome spreads (Kumar et al., 2010). MEI4 localization during meiotic S phase was more precisely analyzed in spreads of spermatocytes incubated with EdU by co-detecting the REC8 cohesin subunit, which establishes the cohesion between sister chromatids at this stage. In early, middle and late S phase spermatocytes, MEI4 was detected as multiple nuclear foci with no specific pattern (Fig. 2A–D). The intensity and number of MEI4 foci were variable, possibly owing to variations in the strength of interaction with chromatin, and when discrete MEI4 and REC8 foci were observed (Fig. 2C), no co-localization with REC8 was detected. These cytological findings are consistent with western blotting data during the first wave of spermatogenesis, when B type spermatogonia enter meiosis synchronously at day 8 after birth. Specifically, MEI4 expression in testis protein extracts started to be detected soon before meiosis entry (6 days postpartum, dpp) and gradually increased during progression into meiotic prophase (Fig. 2E). By contrast, it was undetectable at 3 dpp, when B type spermatogonia are absent.
Fig. 1.
MEI4 expression in pre-meiotic and preleptotene cells. Cryosections of testes from wild-type (A,B,E,F) and Mei4−/− (C,D,G,H) adult mice were immunostained with an anti-MEI4 (red) antibody in combination with an anti-cKIT antibody (green) or in vivo EdU labeling (green). In vivo EdU labeling for 1 h allows the visualization of cells undergoing DNA replication. DNA was stained with DAPI (blue). The stage of wild-type seminiferous epithelium (VI in panels A,B; VII in panels E,F) was determined based on DAPI staining. Some non-specific cytoplasmic signal is observed with the anti-MEI4 antibody. pL, preleptotene stage; B-sg, B type spermatogonia. White arrows in F indicate spermatocytes at the pachytene stage. Scale bars: 10 µm (A,C,E,G); 5 µm (B,D,F,H).
Fig. 2.
MEI4 and REC8 expression in preleptotene cells. MEI4 localization on meiotic chromosome spreads prepared after in vitro EdU labeling for 1 h of cells from wild-type (A–C) and Mei4−/− testes (D). In wild-type spreads, REC8 localization was concomitantly assessed. Preleptotene nuclei are in early (A), middle (B) and late (C) S phase, based on EdU localization relative to heterochromatin identified by intense DAPI staining (not shown). (E) MEI4 immunoprecipitation from protein extracts of juvenile testes at different times during the first wave of spermatogenesis with a non-crosslinked anti-MEI4 antibody. SYCP3 and GAPDH are used as controls. dpp, day postpartum. Scale bars: 10 µm.
Mei1 is required for the axis-localization of MEI4 foci
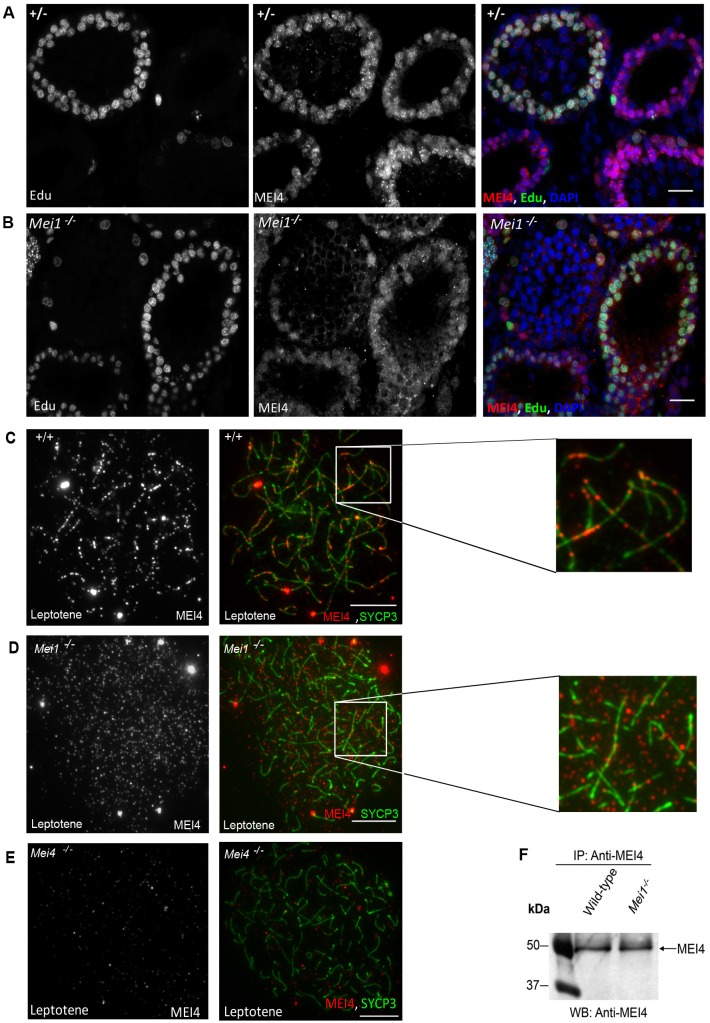
Mei1, like Spo11 and Mei4, is required for DSB formation in mice (Kumar and de Massy, 2010). The phenotypes of Mei1−/−, Spo11−/− and Mei4−/− mice are undistinguishable (Libby et al., 2003). In these mouse mutants, on the basis of monitoring DSB repair foci, programmed meiotic DSB formation does not occur. We have previously shown that MEI4 foci form in the absence of SPO11 (Kumar et al., 2010); by contrast, MEI1 involvement in the regulation of MEI4 activity is unknown. By monitoring meiotic cells in S phase as above, we found that MEI4 nuclear localization was similar in Mei1+/− and Mei1−/− mice (Fig. 3A,B). On spermatocyte spreads, at leptotene, Mei1−/− mice showed a slight reduction in the number of MEI4 foci throughout the nucleus (225±45 in Mei1−/− versus 326±39 in wild-type cells, mean±s.e.m., supplementary material Fig. S1A); however, the most striking effect was the drastic reduction in the axis localization of these foci (11±2% in Mei1−/− versus 58±5% in wild-type cells) (Fig. 3C–E; supplementary material Fig. S1B). The low level of MEI4 axis localization in Mei1−/− is compatible with a random overlap of the two signals as a similar level (9%) is obtained upon rotation of the MEI4 image by 180°. MEI4 protein levels were similar in wild-type and Mei1−/− testes as assessed by western blotting analysis (Fig. 3F). Taken together, these observations suggest that MEI1 plays a key role in the localization of MEI4 to chromosome axes after the meiotic S phase.
Fig. 3.
Recruitment of MEI4 on meiotic chromosome axes is MEI1-dependent. Immunostaining with an anti-MEI4 antibody (red) and DAPI (blue) after in vivo EdU labeling (green) of cryosections of testes from 21-day-old Mei1+/− (A) and Mei1−/− (B) mice. MEI4 and SYCP3 were monitored on chromosome spreads from wild-type (C), Mei1−/− (D) and Mei4−/− (E) mutant testes. (F) The absence of MEI1 did not affect MEI4 protein levels as determined by MEI4 immunoprecipitation in protein extracts of juvenile testes (18 dpp) from wild-type and Mei1−/− mice using a crosslinked anti-MEI4 antibody. Scale bars: 10 µm.
Stra8, a regulator of meiotic progression, is not required for MEI4 focus formation
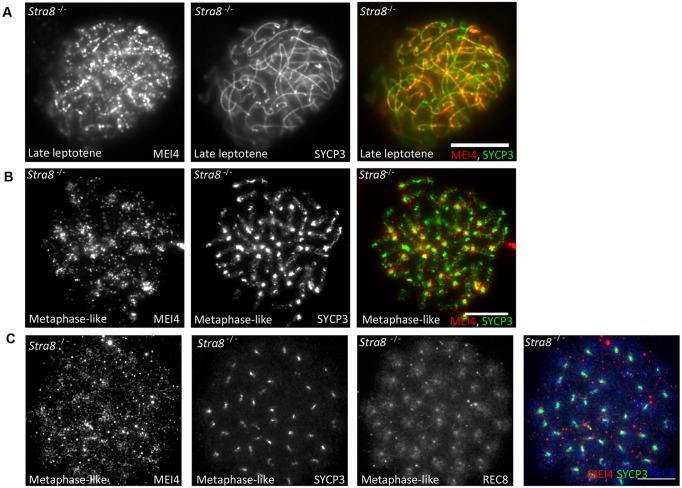
We then asked whether deregulation of meiosis initiation might affect the formation of MEI4 foci. One of the genes involved in the earliest steps of meiosis progression is Stra8. Two independent studies have revealed phenotypic variations between two Stra8 mutant strains generated in different genetic backgrounds. In one study Stra8−/− spermatocytes were defective in entry into meiotic prophase (Anderson et al., 2008), in another study some spermatocytes were shown to enter prophase but with recombination and synapsis defects (Mark et al., 2008). We reasoned that Stra8 mutant from this second study might have an alteration in the induction of meiotic DSBs and thus tested MEI4 localization. We however observed a wild-type localization of MEI4 in Stra8−/− leptotene spermatocytes with foci predominantly localized on unsynapsed chromosome axes (Fig. 4A). In this mutant and genetic background, some Stra8−/− spermatocytes reached a metaphase-like stage with 40 univalents, thought to derive from preleptone or leptotene cells that had skipped the normal events of meiotic prophase, including recombination (Mark et al., 2008). Interestingly, we did detect MEI4 foci on condensed chromatin of metaphase-like Stra8−/− cells (Fig. 4B,C) where SYCP3 was mainly localized to centromeres, like in wild-type metaphase I spermatocytes (Sun and Handel, 2008), and REC8 was localized on chromosome arms and concentrated in centromeric regions (Fig. 4C). Taken together, these data show that Stra8 is not required for MEI4 focus formation.
Fig. 4.
Formation of MEI4 foci does not require STRA8. MEI4 is associated with meiotic chromatin in Stra8−/− mouse spermatocytes. Immunodetection of MEI4 (red), SYCP3 (green) and REC8 (blue) on chromosome spreads of late leptotene stage (A) and metaphase-like (B,C) spermatocytes from Stra8−/− mice. Scale bars: 10 µm.
Meiotic-specific cohesins cooperate for MEI4 foci formation
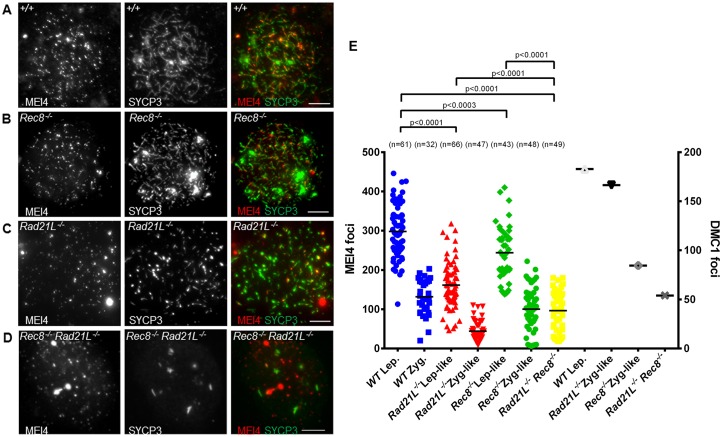
Studies in S. cerevisiae have highlighted that an important and complex interplay between cohesins, axial elements proteins and the RMM complex is required for meiotic DSB formation (Panizza et al., 2011). RAD21L and REC8, two meiotic-specific α-kleisin subunits of the mouse cohesin complexes, are expressed in meiotic cells in addition to the mitotic kleisin RAD21 (Haering and Jessberger, 2012). Genetic ablation of RAD21L or REC8 leads to a reduction of DSB activity based on monitoring DSB repair foci (RAD51 and/or DMC1). MEI4 foci were formed in Rec8−/− (we used here the mouse strain carrying a point mutation in the N-terminal region of REC8) (Bannister et al., 2004; Xu et al., 2005) and Rad21L−/− spermatocytes (Fig. 5A–C; supplementary material Fig. S1B), but at leptotene their number was reduced compared to wild-type cells (82±9% and 54±6%, mean±s.e.m., of wild-type levels, respectively) (Fig. 5E). The number of MEI4 foci further decreased from leptotene to zygotene in both mutant spermatocytes in which DSB repair and homologous synapsis are partially deficient (Bannister et al., 2004; Herrán et al., 2011). In the double Rad21L−/− Rec8−/− mutant, which has a greater reduction of the number of DSB repair foci and in which leptotene cells show a strong defect in axis formation (Ishiguro et al., 2014; Llano et al., 2012), the number of MEI4 foci was reduced to 32±5% of wild-type levels (Fig. 5D,E). These results suggest that both kleisins contribute to MEI4 focus formation or maintenance and have partially overlapping functions in the assembly of these foci. It is important to note that in the Rad21L−/− Rec8−/− mutant the assembly of the cohesin complex in prophase is compromised as SMC3, SMC1β and STAG3 cannot form axial structures (Llano et al., 2012). We also examined the role of SMC1β, a non-kleisin subunit, in MEI4 focus formation. SMC1β is a meiosis-specific SMC1-type cohesin subunit that is required for the maintenance, but not for the establishment of cohesion, possibly achieved by the SMC1α complexes. Smc1b−/− spermatocytes show a meiotic prophase phenotype similar to that of the single Rec8 or Rad21L mutants (Revenkova et al., 2004), with only a 1.5-fold reduction of DSB level compared to wild type (Biswas et al., 2013). The number of MEI4 foci was reduced to 58±9% of wild-type level but the proportion of foci with axis association was similar to wild type (55% in Smc1b−/− and 58% in wild type) (supplementary material Fig. S1A,B; Fig. S2A–C), consistent with the fact that cohesin complexes are needed for the formation or stabilization of MEI4 foci. One unique phenotype of Smc1b−/− meiocytes is the short length of chromosome axes (measured in oocytes) and the presence of longer chromatin loop extensions from the axis (Novak et al., 2008). It is striking that this is the only mutant, among those analyzed here, where MEI4 formed elongated stretches of higher intensity (supplementary material Fig. S2C). We indeed observed a substantial difference in the distribution of foci intensity in Smc1b−/− compared to wild type with Smc1b−/− spermatocytes having a greater proportion of foci with intensities greater than the one of the majority class (supplementary material Fig. S2D). This pattern is compatible with a coalescence of individual foci in Smc1b−/− spermatocytes.
Fig. 5.
The REC8 and RAD21L cohesin subunits are involved in MEI4 localization. The number of MEI4 foci at leptotene is lower in chromosome spreads from both Rec8−/− and Rad21L−/− than in wild-type mice (A–C); this effect was more pronounced in the double Rec8− Rad21L−/− mutant (D). Scale bars: 5 µm. (E) Quantification (the mean is indicated) of MEI4 foci in the different mouse strains at leptotene and zygotene stages. P-values are from two-sided Mann–Whitney tests. n, number of nuclei. The number of DMC1 foci detected and quantified in wild-type and mutant mice in a previous study (Ishiguro et al., 2014) is plotted on the right y-axis.
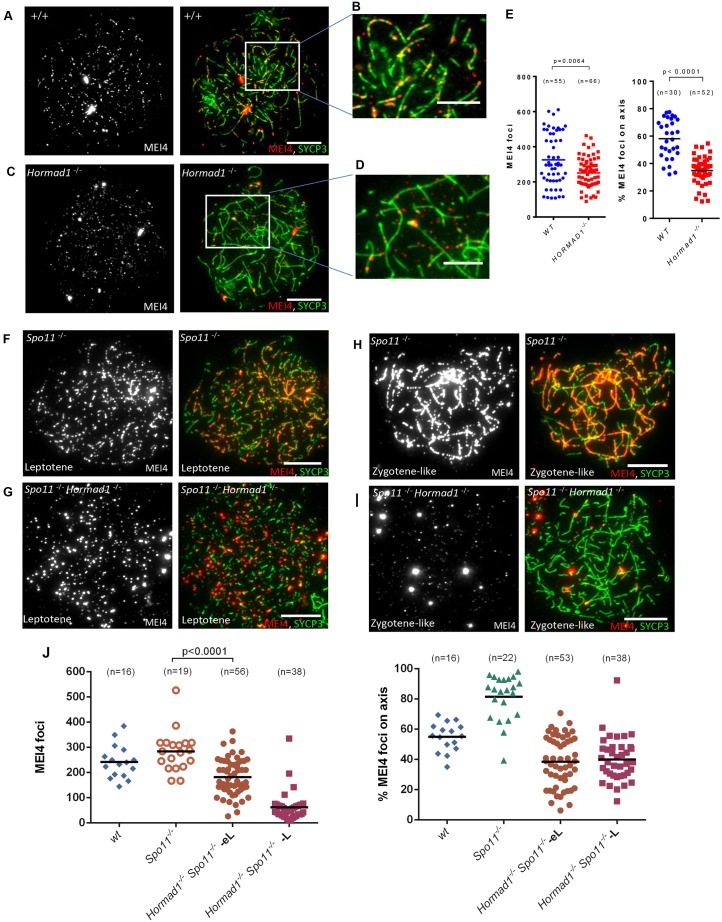
Hormad1 is required for the stabilization of MEI4 foci
We then tested the role of SYCP3 and HORMAD1, two meiotic chromosome structural proteins, in MEI4 foci formation. SYCP3 assembles on meiotic chromosome axes at the onset of meiotic prophase (Eijpe et al., 2003) and is not required for cohesin loading and for DSB formation, but is required for SYCP2 recruitment on axial elements and for full homologous synapsis (Kouznetsova et al., 2011; Pelttari et al., 2001; Yuan et al., 2000). In the absence of SYCP3, MEI4 foci were detected at wild-type level and the percentage of axis associated foci was only slightly reduced (50±4% in Sycp3−/− versus 58±5%, mean±s.e.m., in wild type cells) (supplementary material Figs S1A,B; Fig. S2E,F). HORMAD1 is not required for the timely assembly of meiotic chromosome axes, but is involved in DSB formation, as in Hormad1−/− mice, the DSB level shows a 2- to 4.8-fold reduction compared to wild-type controls, based on SPO11-oligonucleotides quantification, and this is correlated with a reduction in DSB repair foci (Daniel et al., 2011). Analysis of MEI4 foci in Hormad1−/− spermatocytes showed a reduction of their number at leptotene (77±12% of wild type level) (Fig. 6A,C,E) and only 35% of them were associated with axes (Fig. 6B,D,E). Thus, the absolute number of axis-associated foci in Hormad1−/− cells was 4-fold lower than in wild-type spermatocytes, in good correlation with the level of SPO11-oligonucleotides. This reduction of MEI4 foci was not due to a lower amount of MEI4 protein in Hormad1−/− spermatocytes, as shown by western blot analysis after immunoprecipitation of MEI4 protein from wild-type and Hormad1−/− testicular cells (supplementary material Fig. S3A). As HORMAD1 is first detected at leptotene (Wojtasz et al., 2009), whereas MEI4 foci are already observed in preleptotene cells (Fig. 1E,F; Fig. 2A–C; Fig. 3A), the formation of MEI4 foci at the preleptotene stage could be independent of HORMAD1. In fact, labeling with EdU and an anti-MEI4 antibody highlighted the presence of comparable number of MEI4 foci in wild-type and Hormad1−/− preleptotene cells (supplementary material Fig. S3B,C). As DSB formation occurs at the onset of leptotene, during axis development, HORMAD1 might be specifically required for MEI4 stabilization upon DSB formation, but not for establishment of MEI4 axis association at the onset of DSB formation. In this case, the ablation of HORMAD1 should not have any effect on the formation of MEI4 foci in the absence of DSBs. However, the number of MEI4 foci and the fraction of axis-associated foci were higher in Spo11−/− spermatocytes compared to in Hormad1−/− Spo11−/− cells at early leptotene (Fig. 6F,G,J), indicating that HORMAD1 plays a role in the axis association and stabilization of MEI4 foci even in the absence of meiotic DSBs. The instability of MEI4 foci in the absence of meiotic DSBs and HORMAD1 is exacerbated when prophase progresses as observed by comparing Hormad1−/− Spo11−/− and Spo11−/− spermatocytes at middle/late leptotene and zygotene-like stages (Fig. 6H–J).
Fig. 6.
HORMAD1 stabilizes MEI4 foci on meiotic axes. Immunodetection of MEI4 and SYCP3 on surface-spread meiotic chromosomes from wild-type (A,B), Hormad1−/− (C,D), Spo11−/− (F,H) and Spo11−/−/Hormad1−/− (G,I) spermatocytes. Scale bars: 10 µm. Quantification of the total number of MEI4 foci and of the percentage of axis-associated foci in leptotene spermatocytes from wild-type and Hormad1−/− (E) and from wild-type, Spo11−/− and Spo11−/− Hormad1−/− mice (J). In J, quantification was performed using spreads prepared from frozen testes. As the quality of foci detection is lower compared to spreads prepared from fresh testes, a smaller number of foci was detected compared with the data in E. MEI4 foci were counted separately in early leptotene (eL) and middle or late leptotene (L) Spo11−/−/Hormad1−/− spermatocytes (J). P-values are from two-sided Mann–Whitney tests); n, number of nuclei; the mean is indicated by the black bar.
Strand exchange activity and synapsis elongation are not required for MEI4 foci disassembly
In wild-type cells, MEI4 foci disassemble concomitantly with DSB repair and initiation of synapsis, as indicated by the strong reduction in their number from leptotene to zygotene. In fact, the extremely low level of colocalization between MEI4 and DMC1 is compatible with the successive activity of these proteins at DSB sites (Kumar et al., 2010). We therefore tested whether the loading of the strand exchange protein DMC1 or of the synaptonemal complex central element component SYCE2 was necessary for the dissociation of MEI4 foci. The number of MEI4 foci and their association with the axis at leptotene were not significantly different between wild-type and Dmc1−/− or Syce2−/− spermatocytes (supplementary material Fig. S1A,B). Moreover, the number of MEI4 axis-associated foci was reduced from leptotene to the zygotene-like stage in both mutants (from 164±15 to 17±3 for Dmc1−/− and from 183±24 to 37±12, mean±s.e.m., for Syce2−/− spermatocytes) (supplementary material Fig. S3D,E). This suggests that DMC1 loading and synaptonemal complex elongation are not required for the process leading to the reduction of MEI4 axis association. The simplest interpretation of these observations is that MEI4 is displaced from the axes at the time or soon after DSB formation, before DMC1 loading.
DISCUSSION
Association of MEI4 with chromosome axes
In meiotic cells, chromosomes are organized as an array of loops along a proteinaceous axis that can be visualized by microscopic analysis during meiotic prophase (Kleckner, 2006). This organization is developed before and during leptotene when components of meiotic chromosome axes, such as cohesins and then SYCP2, SYCP3, HORMAD1 and HORMAD2, are present. Several cohesin complex components (SMC3, SMC1α, RAD21, RAD21L and REC8) are recruited to chromosomes before meiotic prophase during meiotic S phase. Other subunits of the cohesin complexes (SMC1β and STAG3) are detected later, at leptotene, along with SYCP2, SYCP3, HORMAD1 and HORMAD2, as multiple foci that elongate to form the axis (Eijpe et al., 2003). Meiotic DSBs occur as axes develop during leptotene. Other components involved in axis structure are type II DNA topoisomerases and condensins, specifically the condesin II complex. The role of condensins in meiotic chromosome organization and the impact on meiotic DSB formation and crossover control has been demonstrated in C. elegans (Mets and Meyer, 2009).
In this context, the dynamics of MEI4 foci suggest that, in differentiated spermatogonia, MEI4 associates with chromatin before meiosis entry and this association is still detected in meiotic S phase, the preleptotene stage. Whether these foci are organized through interactions with other proteins and whether there is a maturation process remains to be determined. At the preleptotene stage, several components of cohesin complexes assemble onto chromosomes and mediate sister chromatin cohesion but we have not detected a colocalization between MEI4 and the REC8 kleisin. Analysis of colocalization of MEI4 with other proteins and/or higher resolution studies might help to further understand the dynamics of these foci at these early stages. Mouse MEI4 might be recruited to chromatin by specific protein interactions, as shown in S. cerevisiae for the role of Mer2 (Murakami and Keeney, 2008; Murakami and Keeney, 2014; Panizza et al., 2011), although no mouse Mer2 ortholog has been identified yet. At the onset of the leptotene stage, chromosome axes develop and the majority of MEI4 foci are localized on axes. We report here a major role for MEI1 in mediating the association of MEI4 to chromosome axes. MEI1 is a protein which contains putative armadillo repeats and has similarity to importins but is of unknown activity. Mei1−/− mice show a strong reduction or absence of meiotic DSB activity (a strong reduction of γH2AX, undetectable axis associated RAD51 and RPA foci) (Libby et al., 2002; Reinholdt and Schimenti, 2005) similar to what is observed in Spo11−/− mice and in an Arabidopsis thaliana mutant for the Mei1 ortholog Prd1 (De Muyt et al., 2007). Mei1−/− mutant spermatocytes show the most dramatic reduction of MEI4 axis localization among those tested in our study (supplementary material Figs S1, S4), and the residual colocalization detected is likely occurring by chance. We propose that this loss of MEI4 axis association could be responsible for the reduction or loss of DSB activity, but more complex scenarios with additional consequences due to the absence of MEI1 cannot be excluded. This role of MEI1 is independent from that of HORMAD1 described below, as HORMAD1 localization is not affected in Mei1−/− spermatocytes (data not shown).
The strong reduction of MEI4 foci at leptotene in the absence of the two kleisins REC8 and RAD21L, correlated with DMC1 foci reduction (Fig. 5), indicates that there is a requirement for these proteins in the establishment or maintenance of MEI4 foci, and could be a direct or indirect effect of their ablation. Observations in S. cerevisiae, where cohesins affect Hop1 organization (Panizza et al., 2011), are compatible with an indirect effect. One other key component highlighted in this study is HORMAD1, which is required for the establishment and maintenance (see below) of MEI4 axis association independently from DSB activity as demonstrated by the comparative analysis of Spo11−/− and Hormad1−/− Spo11−/− mutant spermatocytes (Fig. 6F–J).
MEI4 maintenance and turnover on chromosome axes
MEI4 dynamics during meiotic prophase progression are particularly interesting and suggest a negative regulation of meiotic DSB formation as DSB repair and synapsis progress. Our earlier observations highlighted several interesting features of MEI4 foci (Kumar et al., 2010). MEI4 axis-associated foci are SPO11-independent and, in wild-type spermatocytes, they disappear as prophase progresses, in parallel with DSB repair and synapsis, with a dramatic reduction of the number of axis-associated foci in leptotene compared with zygotene stages. Colocalization studies have indicated that MEI4 foci are excluded from DSB repair foci (which are marked by DMC1) (Kumar et al., 2010). Consistent with this early displacement of MEI4 upon DSB repair, we show here that MEI4 foci are displaced in Dmc1−/− spermatocytes, suggesting that MEI4 displacement occurs before the step of DMC1 loading on the single-stranded DNA at DSB ends. Some events linked to the early stages of DSB recognition and/or processing might contribute to delocalization of MEI4 through a locally regulated process. In S. cerevisiae, several components have been shown to be involved in meiotic DSB regulation (Keeney et al., 2014). Among components involved are the ATM and ATR kinases which are involved in DSB signaling and for which orthologs in S. cerevisiae (Tel1 and Mec1, respectively) have been shown to regulate DSB formation (Garcia et al., 2015; Zhang et al., 2011) and to downregulate a Mei4 partner, Rec114, through phosphorylation (Carballo et al., 2013). However, in S. cerevisiae, Rec114 phosphorylation decreases DSB activity but does not lead to Rec114 depletion from axes as assayed by chromatin immunoprecipitation. The dynamics of Mei4 localization in S. cerevisiae (Li et al., 2006; Maleki et al., 2007), S. pombe (Lorenz et al., 2006) and the DSB-1 and DSB-2 proteins in C. elegans, which might play similar functions to Mei4 and Rec114 (Rosu et al., 2013; Stamper et al., 2013), share similar properties to those we have described in mice, although differences in the mechanism of regulatory pathways might operate in these species.
Importantly, in Spo11−/− spermatocytes, MEI4 foci are stable and persist on unsynapsed axes, but are undetectable on non-homologously synapsed regions (Kumar et al., 2010) (supplementary material Fig. S4). This observation suggests that another process that is independent of DSB formation and processing, but potentially shares common properties with these events, leads to the displacement of MEI4 directly or indirectly. The interesting common feature between these two contexts (DSB repair in wild-type and synapsis in Spo11−/−) is the depletion of HORMAD1 (Wojtasz et al., 2009) (supplementary material Fig. S4). HORMAD1 has been proposed to be a component of a negative-feedback loop that monitors homologous synapsis (Daniel et al., 2011; Wojtasz et al., 2009), and our results suggest that this regulation might involve MEI4. MEI4 could thus be displaced locally at DSB repair sites but also at distance, as a consequence of synapsis elongation and HORMAD1 displacement. The negative regulation of synapsis on DSB activity has also been directly detected in S. cerevisiae (Thacker et al., 2014). HORMAD1 is phosphorylated (Kogo et al., 2012; Shin et al., 2010; Wojtasz et al., 2009) and could be a target of the ATM kinase, which has been proposed to play a role in the downregulation of DSB formation in Drosophila and mice, based on the detection of increased DSB levels in Atm mutants (Joyce et al., 2011; Lange et al., 2011).
It would thus not be surprising if there were several levels and pathways of regulation of DSB formation involving various components of the chromosome axis. The precise mechanism(s) of signal transmission along chromosomes will be exciting to unravel, and it might involve properties of chromosome axis proteins and/or kinases and different substrates, including structural proteins and/or recombination proteins. These interplays between chromosome structure and recombination are important for many steps of meiotic recombination from DSB formation to crossover control and thus for the proper segregation of chromosomes at the first meiotic division.
MATERIALS AND METHODS
Mice
For MEI4 protein expression analysis and immunofluorescence assays, testes were isolated from wild-type and Mei4−/− littermates or from other progeny derived from the same type of mating. Homozygous mutant mice strains (Rec8−/−, Rad21L−/−, Rec8−/−Rad21L−/−, Smc1b−/−, Sycp3−/−, Hormad1−/−, Spo11−/−Hormad1−/−, Syce2−/−, Dmc1−/−, Spo11−/−, Mei1−/− and Stra8−/−) are as described previously (Bannister et al., 2004; Baudat et al., 2000; Bolcun-Filas et al., 2007; Daniel et al., 2011; Ishiguro et al., 2014; Li and Schimenti, 2007; Mark et al., 2008; Pittman et al., 1998; Revenkova et al., 2004; Roig et al., 2010; Yuan et al., 2000). All experiments were carried out according to the CNRS guidelines and approved by the Ethics Committee of Languedoc-Roussillon (CEEA-LR-11028).
Antibodies
Antibodies used in the study are: rabbit polyclonal anti-MEI4 (Kumar et al., 2010), rat monoclonal anti-cKIT (abcam, ab112177), guinea pig anti-SYCP3 (Kumar et al., 2010), rabbit anti-SYCP3 (Abcam, ab15093), rat polyclonal anti-SYCP3 (Morimoto et al., 2012) (for experiments reported in Fig. 5), guinea pig anti-HORMAD1 (Wojtasz et al., 2009), mouse polyclonal anti-REC8 (Ishiguro et al., 2011) and mouse monoclonal anti-GAPDH (Abcam, ab8245) antibodies.
Preparation of testis protein extracts
Protein extracts were prepared from testis tissue isolated from juvenile mice (3, 6, 10, 14 and 16 dpp) or 8-week-old adult mice. After removal of the tunica albuginea, testes were homogenized in lysis buffer (25 mM HEPES, pH 7.9, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 2 mM DTT) supplemented with Complete protease inhibitor (Roche) and phosphatase inhibitor (Roche). Cell extracts were sonicated at 4°C for five cycles of 30 s on and 30 s off and centrifuged at 20,000 g, 4°C, for 15 min. The resulting supernatants were used for immunoprecipitation and western blotting experiments.
Antibody crosslinking, immunoprecipitation and western blotting
Rabbit anti-MEI4 serum was first purified on G-protein–sepharose beads (GE healthcare). 30–50 µg of purified IgGs were then crosslinked to Dynabeads Protein A (Invitrogen) using 20 mM dimethyl pimelidate dihydrochloride (Sigma, D-8388). For immunoprecipitation, wild-type and Mei4−/− testis protein extracts were incubated with either crosslinked anti-MEI4 antibodies or without crosslinking at 4°C with gentle rotation for 3 h. After washing the beads with the lysis buffer for four times, elution was performed by boiling beads in Laemmli sample buffer.
Proteins from testis extracts or after immunoprecipitation were separated on 10% SDS polyacrylamide gels and transferred onto PVDF membranes (Thermo scientific). Membranes were probed with anti-MEI4 (1∶500), anti-SYCP3 (1∶500) or anti-GAPDH (1∶5000) antibodies. Primary antibodies were detected by incubation with TrueBlot anti-rabbit (1∶10,000; Rockland Immunochemicals Inc.) or anti-guinea pig or mouse IgG HPR-conjugated antibodies and chemiluminescent substrate (Thermo scientific).
EdU incorporation
In vivo EdU incorporation was performed by injecting 300 µg EdU in PBS per mouse intraperitoneally in wild-type and Mei4−/− mice. Mice were killed 1 h later and testes were processed to prepare cryosections. Alternatively, testis cell suspensions were incubated with 10 µM Edu in DMEM with 10% FCS at 37°C for 1 h for in vitro labeling. EdU incorporation was detected using Alexa Fluor 488 and the Click-iT EdU imaging kit according to the manufacturer's instructions (Invitrogen).
Preparation of testis cryo-sections and meiotic spreads
Immunolocalization of MEI4 was performed on cryosections and meiotic chromosome spreads. For cryosections, adult testes were fixed at room temperature in 4% formaldehyde in PBS pH 7.4, 0.1% Triton X-100 for 20 min. Then testes were incubated in 30% sucrose at 4°C overnight and were frozen on dry ice in Tissue-Tek O.C.T. (Sakura). Sections of 8 µm thickness were cut, dried onto slides and stored at −80°C. Meiotic nuclear spreads from mouse testes were prepared according to the dry-down technique, as described previously (Peters et al., 1997).
Immunofluorescence assays
Immunostaining of cryosections was performed by fixing tissues in methanol: acetone (1∶1) at −20°C prior to incubation in blocking buffer (10% donkey serum and 0.5% Triton X-100 in PBS). Then, sections were incubated (at room temperature, overnight) in a humid chamber with affinity-purified rabbit anti-MEI4 (1∶50) and rat anti-cKIT (1∶200). Slides were washed in PBS three times and incubated with secondary antibodies (goat anti-rabbit IgG conjugated to Alexa Fluor 594 and anti-rat IgG conjugated Alexa Fluor 488) at 37°C for 1 h. After DAPI (4-9-6-diamidino-2-phenylindole, 2 µg/ml) staining, slides were mounted with Prolong Gold mounting medium (Life Technologies).
Immunofluorescence assays on meiotic spreads were performed as described (Kumar et al., 2010). Spreads were incubated at room temperature overnight with the following antibodies: affinity-purified rabbit anti-MEI4 (1∶50) (Kumar et al., 2010), guinea pig anti-SYCP3 (1∶500), guinea pig anti-HORMAD1 (1∶400) and mouse monoclonal anti-phosphorylated H2AX (Upstate) (1∶20,000). Secondary antibodies [goat anti-guinea pig IgG conjugated to Alexa Fluor 488 (Molecular Probes), Cy3-conjugated donkey anti-rabbit IgG and Cy5-conjugated donkey anti-mouse IgG antibodies (Jackson Immunoresearch Laboratories)] were added at 37°C for 1 h. Nuclei were visualized by staining with DAPI. Digital images were captured using a cooled charge-coupled device (CCD) camera (Coolsnap HQ; Photometrics) attached to a Leica DM 6000B microscope and the Metamorph imaging software. After data acquisition, images were processed with ImageJ. Upon setting fixed thresholds on images of identical exposures, total MEI4 foci were counted using the ‘analyze particle’ tool with size (0.025–5.0) and circularity (0.0–1.0) parameters. MEI4 axis association was estimated as the number of MEI4 foci that coincided with SYCP3 or HORMAD1 signals. In Mei1−/− spermatocytes, to evaluate the stochastic distribution of MEI4 foci with respect to chromosome axes, MEI4 foci on axes were counted with or without flipping the MEI4 images by 180°. For foci intensity in Smc1b−/− compared to wild type, we measured the intensity of the axis-associated MEI4 signal by calculating the integrated pixel density of each MEI4 focus in Smc1b−/− and wild type (1091 and 1083 foci were measured for each genotype).
Statistical analysis
Comparisons of the number of foci in the different conditions were done using the two-tailed non-parametric Mann–Whitney test.
Supplementary Material
Acknowledgements
We thank the RAM network (animal facility network) of Montpellier and the IGH/IGF mouse facility, Florence Arnal, Fred Gallardo, Dominique Haddou, Francis Guido, the MRI (Montpellier Imaging Facility) with Julien Cau and Julio Mateos Langerak. We thank all members of the B.d.M. laboratory for stimulating discussions. We thank Manuel Mark for expertise in histological analysis.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
R.K. and B.d.M. designed the research. R.K. and K.I. performed experiments. N.G., K.I., Y.W., A.K., C.H., E.S., J.S. K.D. and A.T. contributed to reagents and material. R.K. and B.d.M. analyzed the results and wrote the manuscript.
Funding
C.H. and A.K. were funded by grants from the Swedish Cancer Society and the Swedish Research Council. K.I. is supported by a Grant-in-Aid for Scientific Research on Innovative Areas, Y.W. is supported by a Grant-in-Aid for Specially Promoted Research from MEXT, Japan. R.K. received financial support from ANR, EDF and a post-doctoral fellowship from the Axa Research Fund. K.D. and A.T. were funded by the Deutsche Forschungsgemeinschaft [grant numbers SPP1384: TO 421/4-1, TO 421/4-2, and Heisenberg Fellowship: TO 421/5-1]. B.d.M. was funded by grants from the Fondation pour le Recherche Médicale, Association pour la Recherche contre le Cancer, Agence Nationale pour la Recherche [grant number ANR-09-BLAN-0269-01], Centre National pour la Recherche Scientifique (CNRS) and the European Research Council Executive Agency under the European Community's Seventh Framework Programme (FP7/2007-2013) [grant number 322788]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.165464/-/DC1
References
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, Géli V. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–218. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/S1097--2765(00)00098--8. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Biswas U, Wetzker C, Lange J, Christodoulou EG, Seifert M, Beyer A, Jessberger R. Meiotic cohesin SMC1β provides prophase I centromeric cohesion and is required for multiple synapsis-associated functions. PLoS Genet. 2013;9:e1003985. doi: 10.1371/journal.pgen.1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/S0092--8674(02)01167--4. [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, De Rooij DG, Cooke HJ. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 2007;176:741–747. doi: 10.1083/jcb.200610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Barot A, Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA. 2008;105:3327–3332. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, Borde V, Klein F, Cha RS. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet. 2013;9:e1003545. doi: 10.1371/journal.pgen.1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M. et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat. Cell Biol. 2011;13:599–610. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 2013;47:563–599. doi: 10.1146/annurev--genet--110711--155423. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Vezon D, Gendrot G, Gallois JL, Stevens R, Grelon M. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 2007;26:4126–4137. doi: 10.1038/sj.emboj.7601815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J. Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ. Tel1-mediated interference suppresses clustered meiotic double-strand-break formation. Nature. 2015 doi: 10.1038/nature13993. [Epub ahead of print] doi: 10.1038/nature13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Jessberger R. Cohesin in determining chromosome architecture. Exp. Cell Res. 2012;318:1386–1393. doi: 10.1016/j.yexcr.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrán Y, Gutiérrez-Caballero C, Sánchez-Martín M, Hernández T, Viera A, Barbero JL, de Álava E, de Rooij DG, Suja JA, Llano E. et al. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 2011;30:3091–3105. doi: 10.1038/emboj.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Berlin; Heidelberg: Springer-Verlag; 2007. pp. 381–442. [Google Scholar]
- Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011;12:267–275. doi: 10.1038/embor.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Shibuya H, Hernández-Hernández A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J. et al. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, Pedersen M, Tiong S, White-Brown SK, Paul A, Campbell SD, McKim KS. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J. Cell Biol. 2011;195:359–367. doi: 10.1083/jcb.201104121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 2014;48:187–214. doi: 10.1146/annurev--genet--120213--092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412--006--0055--7. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092--8674(00)80609--1. [DOI] [PubMed] [Google Scholar]
- Kogo H, Tsutsumi M, Ohye T, Inagaki H, Abe T, Kurahashi H. HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes Cells. 2012;17:439–454. doi: 10.1111/j.1365--2443.2012.01600.x. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Benavente R, Pastink A, Höög C. Meiosis in mice without a synaptonemal complex. PLoS ONE. 2011;6:e28255. doi: 10.1371/journal.pone.0028255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T. et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol. Biol. Cell. 2009;20:3064–3076. doi: 10.1091/mbc.E08--12--1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, De Massy B. Initiation of meiotic recombination in mammals. Genes. 2010;1:521–549. doi: 10.3390/genes1030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Bourbon HM, de Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 2010;24:1266–1280. doi: 10.1101/gad.571710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3:e130. doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hooker GW, Roeder GS. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics. 2006;173:1969–1981. doi: 10.1534/genetics.106.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O'Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev. Biol. 2002;242:174–187. doi: 10.1006/dbio.2001.0535. [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc. Natl. Acad. Sci. USA. 2003;100:15706–15711. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano E, Herrán Y, García-Tuñón I, Gutiérrez-Caballero C, de Álava E, Barbero JL, Schimenti J, de Rooij DG, Sánchez-Martín M, Pendás AM. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J. Cell Biol. 2012;197:877–885. doi: 10.1083/jcb.201201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412--006--0053--9. [DOI] [PubMed] [Google Scholar]
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–486. doi: 10.1007/s00412--007--0111--y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Féret B, Vernet N, Codreanu CA, Chambon P, Ghyselinck NB. STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J. Cell Sci. 2008;121:3233–3242. doi: 10.1242/jcs.035071. [DOI] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell. 2012;47:722–733. doi: 10.1016/j.molcel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 2012;198:165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S. Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev. 2008;22:286–292. doi: 10.1101/gad.1642308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014;158:861–873. doi: 10.1016/j.cell.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Höög C. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J. Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Pelttari J, Hoja MR, Yuan L, Liu JG, Brundell E, Moens P, Santucci-Darmanin S, Jessberger R, Barbero JL, Heyting C. et al. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 2001;21:5667–5677. doi: 10.1128/MCB.21.16.5667--5677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/A:1018445520117. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/S1097--2765(00)80069--6. [DOI] [PubMed] [Google Scholar]
- Reinholdt LG, Schimenti JC. Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma. 2005;114:127–134. doi: 10.1007/s00412--005--0346--4. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, Keeney S. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 2010;6:e1001062. doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 2013;9:e1003674. doi: 10.1371/journal.pgen.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YH, Choi Y, Erdin SU, Yatsenko SA, Kloc M, Yang F, Wang PJ, Meistrich ML, Rajkovic A. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 2010;6:e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 2013;9:e1003679. doi: 10.1371/journal.pgen.1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Handel MA. Regulation of the meiotic prophase I to metaphase I transition in mouse spermatocytes. Chromosoma. 2008;117:471–485. doi: 10.1007/s00412--008--0167--3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–246. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S. et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Höög C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/S1097--2765(00)80404--9. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kim KP, Kleckner NE, Storlazzi A. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. USA. 2011;108:20036–20041. doi: 10.1073/pnas.1117937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.