Abstract
Steroid-resistant nephrotic syndrome (SRNS) is the second most frequent cause of ESRD in the first two decades of life. Effective treatment is lacking. First insights into disease mechanisms came from identification of single-gene causes of SRNS. However, the frequency of single-gene causation and its age distribution in large cohorts are unknown. We performed exon sequencing of NPHS2 and WT1 for 1783 unrelated, international families with SRNS. We then examined all patients by microfluidic multiplex PCR and next-generation sequencing for all 27 genes known to cause SRNS if mutated. We detected a single-gene cause in 29.5% (526 of 1783) of families with SRNS that manifested before 25 years of age. The fraction of families in whom a single-gene cause was identified inversely correlated with age of onset. Within clinically relevant age groups, the fraction of families with detection of the single-gene cause was as follows: onset in the first 3 months of life (69.4%), between 4 and 12 months old (49.7%), between 1 and 6 years old (25.3%), between 7 and 12 years old (17.8%), and between 13 and 18 years old (10.8%). For PLCE1, specific mutations correlated with age of onset. Notably, 1% of individuals carried mutations in genes that function within the coenzyme Q10 biosynthesis pathway, suggesting that SRNS may be treatable in these individuals. Our study results should facilitate molecular genetic diagnostics of SRNS, etiologic classification for therapeutic studies, generation of genotype-phenotype correlations, and the identification of individuals in whom a targeted treatment for SRNS may be available.
Keywords: SRNS, steroid-resistant nephrotic syndrome, nephrosis, kidney failure, genetic disease, FSGS
CKDs take one of the highest tolls on human health. They insidiously lead to ESRD, necessitating dialysis or kidney transplantation for survival. More than 20 million individuals in the United States have CKD, and the treatment cost exceeds $29 billion Medicare expenses annually. The prevalence of CKD has been rising continuously during the last 20 years.1 Nephrotic syndrome (NS) is a CKD defined by significant proteinuria (>40 mg/m2 per hour) with resulting hypoalbuminemia, which in turn causes edema. In young adults and children, NS is classified by its response to a standardized steroid therapy as steroid-sensitive NS (SSNS) versus steroid-resistant NS (SRNS).
SRNS constitutes the second most frequent cause of ESRD in the first two decades of life (North American Pediatric Renal Trials and Collaborative Studies, 2008). For most patients, no curative treatment is available. The most frequent renal histologic feature of SRNS is FSGS. Moreover, in patients with FSGS the risk of recurrence after kidney transplantation is estimated to be 11%–50%, an event that again leads to terminal renal failure.2–4 The risk for recurrence of FSGS in patients with inherited forms of SRNS is lower than that in patients with nonhereditary FSGS.5,6
SRNS is one of the most intractable diseases in nephrology. Its etiology and pathogenesis have been a conundrum for decades. However, discovery of >27 recessive or dominant genes that, if mutated, cause SRNS has recently provided fundamental insights into mechanisms of this disease.7 Identification of these single-gene causes of SRNS has revealed that the glomerular podocyte and the glomerular slit membrane that it maintains are the primary sites at which the pathogenesis of SRNS unfolds.8 The power of identification of such single-gene causes of SRNS lies in the fact that recessive mutations almost always convey full penetrance and thereby represent the cause of SRNS (i.e., the etiology) rather than conveying only an increased risk for the disease. Thus, if strict genetic criteria are being followed for the decision in which mutations are considered causative, identification of the causative mutation allows for (1) unequivocal molecular genetic diagnostics, (2) establishment of genotype-phenotype correlations, (3) transfer of mutations into genetic animal models with detailed study of their detrimental effects, (4) etiologic stratification of participants for therapeutic studies by specific causative gene and mutation, and (5) discovery of specific mutations that may be amenable to treatment.
We have previously shown that 85% of SRNS cases with onset by 3 months of age and 66% with onset by 1 year of age can be explained by recessive mutations in one of four genes only (NPHS1, NPHS2, LAMB2, or WT1).9 However, few data are available on the fraction of monogenic causes of SRNS manifesting later in life, no data are available on large international cohorts,5,10–13 and the generation of data on single-gene causes of SRNS in large cohorts has been impractical until recently.
Because no large cohorts have been studied for the known 27 SRNS-causing genes regarding the frequency of mutations that are considered causative (as defined by strict genetic criteria), we developed a strategy of high-throughput, barcoded exon sequencing using the Fluidigm platform with consecutive next-generation sequencing.14–16 Using this approach we have established a high-throughput, low-cost strategy for exon sequencing of all 27 known SRNS-causing genes for proof of principle in a small cohort of 96 individuals.16 To generate data on the percentage of single-gene causes of SRNS in more individuals, we applied this technique to a large cohort of 2016 individuals (1783 families) with SRNS. We found that a high fraction of SRNS manifesting before 25 years of age is caused by single-gene mutations and is inversely correlated to age of onset. We observed that specific genotype-phenotype correlations exist for PLCE1 mutations and that certain founder mutations prevail in specific geographic regions. Our data demonstrate the heuristic power of mutation analysis in SRNS and will guide the expectations regarding the frequency and nature of causative mutations throughout selected regions of the world. Our high-throughput sequencing strategy will accelerate the generation of causation-based genotype data for large cohorts of therapeutic studies.
Results
Identification of Causative Mutations in an International SRNS Cohort of 1783 Families
We performed exon sequencing in an international cohort of 1783 different families (2016 individuals) with SRNS for NPHS2 and WT1 (exons 8 and 9) (Figure 1, Supplemental Table 1). Of the 1783 families, 253 presented with congenital NS and were additionally screened for mutations in NPHS1. In the context of discovering and studying novel genes, about 400 of the 1783 families underwent Sanger sequencing for the rare genes PLCE1, LAMB2, SMARCAL1, INF2, COQ6, TRPC6, ITGA3, CUBN, ADCK4, and ARHGDIA.17–25 Disease-causing mutations were detected in 170 families for NPHS2, 93 families for NPHS1, and 78 families for WT1. In an additional 51 families, causative mutations were detected in 1 of the other rare genes, thereby unveiling the causative mutation in a total of 392 families (Table 1).
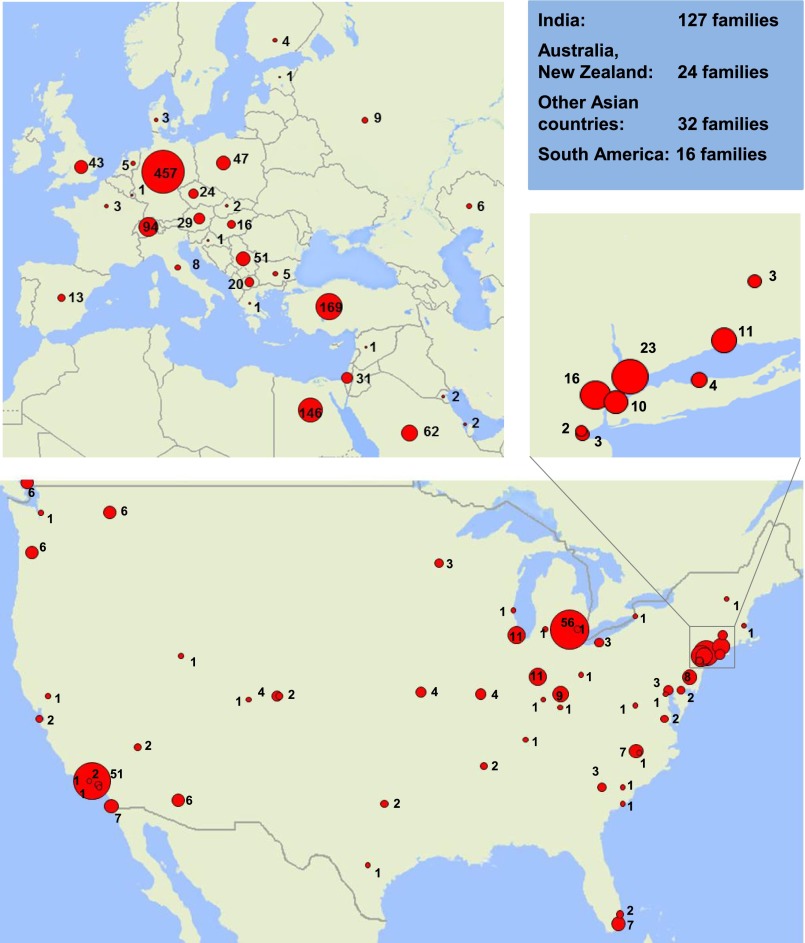
Figure 1.
Geographic distribution of the study cohort of 1783 families with SRNS. The number of affected families in the respective countries or cities is represented by the surface area (red) of a circle. Data from countries not represented on the map are shown in the blue box. Insert shows higher resolution of the New York Metropolitan area.
Table 1.
International cohort of 526 of the 1783 families, in whom a single-gene cause of SRNS was detected in 1 of 21 monogenic causes of SRNS (27 genes examined)
| Gene Causing SRNS | Mode of Inheritance | SRNS Families Molecularly Diagnosed by Sanger Sequencing (Published Previously), na | SRNS Families Molecularly Diagnosed by Multiplex PCR (n) | Total SRNS Families with Molecular Diagnosis (% of Families) |
|---|---|---|---|---|
| NPHS2 | AR | 170 (42) | 7 | 177 (9.93) |
| NPHS1 | AR | 93 (61) | 38 | 131 (7.34) |
| WT1 | AD | 78 (50) | 7 | 85 (4.77) |
| PLCE1 | AR | 23 (16) | 14 | 37 (2.17) |
| LAMB2 | AR | 10 (6) | 10 | 20 (1.12) |
| SMARCAL1 | AR | 1 (0) | 15 | 16 (0.89) |
| INF2 | AD | 2 (0) | 7 | 9 (0.5) |
| TRPC6 | AD | 1 (1) | 8 | 9 (0.53) |
| COQ6 | AR | 6 (5) | 2 | 8 (0.45) |
| ITGA3 | AR | 3 (3) | 2 | 5 (0.28) |
| MYO1E | AR | 0 (0) | 5 | 5 (0.28) |
| CUBN | AR | 1 (1) | 4 | 5 (0.28) |
| COQ2 | AR | 0 (0) | 4 | 4 (0.22) |
| LMX1B | AD | 0 (0) | 4 | 4 (0.22) |
| ADCK4 | AR | 3 (3) | 0 | 3 (0.17) |
| DGKE1 | AR | 0 (0) | 2 | 2 (0.11) |
| PDSS2 | AR | 0 (0) | 2 | 2 (0.11) |
| ARHGAP24 | AD | 0 (0) | 1 | 1 (0.06) |
| ARHGDIA | AR | 1 (1) | 0 | 1 (0.06) |
| CFH | AR | 0 (0) | 1 | 1 (0.06) |
| ITGB4 | AR | 0 (0) | 1 | 1 (0.06) |
| Total | 392 (189) | 134 | 526 (29.5) |
AD, autosomal dominant; AR, autosomal recessive.
Number in parenthesis show “molecularly solved” families with causative mutation detected that were published before from our cohort (see literature).
The same entire cohort of 1783 families with SRNS (2016 affected individuals) was screened for all known 27 monogenic causes of SRNS (Supplemental Table 2), including the genes screened individually before by microfluidic multiplex PCR and next-generation sequencing (see Concise Methods). We thereby identified the molecular diagnosis in 134 families. We detected the disease-causing mutations in 526 of 1783 families (29.5%) (Table 1, Figure 2). Through segregation analysis in multiplex families, we identified causative mutations in an additional 89 affected family members. We detected mutations in 21 of the 27 known SRNS genes and discovered 129 novel mutations (48 truncating alleles), thereby adding an additional 11.6% to the previously 1115 reported mutations for these genes in the Human Gene Mutation Database (http://www.hgmd.org). All detected disease-causing mutations are listed in Supplemental Tables 3–5, together with clinical phenotypes of the affected individuals. No disease-causing mutations were found in MEFV, CD2AP, NEIL1, PTPRO, SCARB2, or ACTN4. There was no sex difference for the likelihood of detecting the disease-causing mutation. The median age of onset in dominant SRNS genes was significantly higher (36 months; range, 0–444 months [0–37 years]) compared with the age of onset in recessive genes (median age of onset, 12 months; range, 0–720 months [0–60 years]) (Supplemental Table 1). We conclude that more than one quarter of all cases with SRNS worldwide that manifest before age 25 years is caused by a single-gene mutation in 1 of 21 SRNS genes in an international cohort. In a control group of 185 children with SSNS, no disease-causing mutation could be detected in the 27 SRNS genes by microfluidic multiplex PCR, confirming that the presence of a causative monogenic mutation in 1 of the 27 genes is likely to exclude steroid sensitivity of NS.5
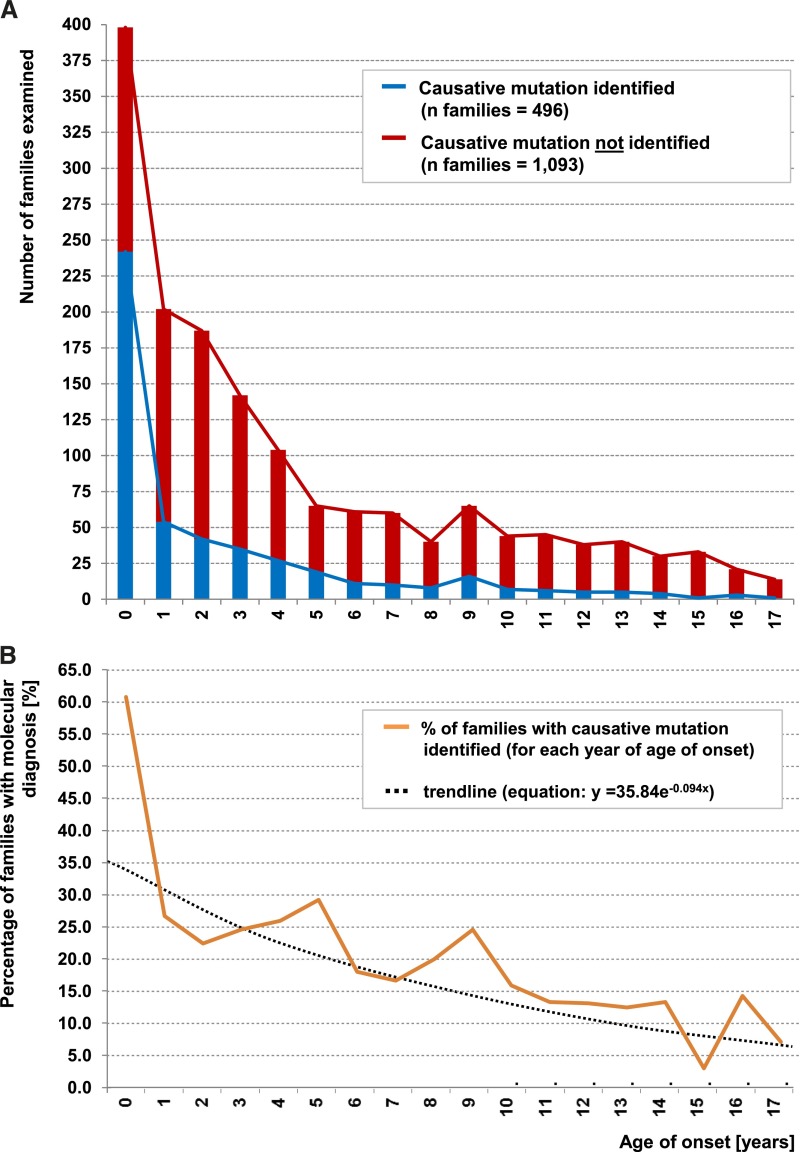
Figure 2.
Age of onset distribution (in years) for 1589 of 1783 examined SRNS families. The displayed 1589 families represent the number of families with available data for age of onset of proteinuria <18 years. (A) Red curve and histograms represent the number of families in each age of onset (years) of SRNS for 1093 families without molecular genetic diagnosis. Blue curve and histograms show number of 496 families with causative mutations identified for each age of onset. (B) Graph indicates percentage of solved families per year of age of onset (from A). Black dotted line represents a binomial fit of age-related percent of families with causative mutation. (Data are not displayed for 72 individuals who were older than 17 years at onset of SRNS and for 122 families with no available information for age of onset. In families with >1 affected family member, the mean age of onset from all affected individuals was used.)
Age Distribution of Single-Gene Causes of SRNS
In the entire international cohort of 1783 families with SRNS, the fraction of families in whom we detected the single-gene cause was 29.5% (Table 1). We found a negative correlation between the likelihood of identifying the causative gene and the age of onset of proteinuria (Figure 2A). We detected the disease-causing mutation in 61.3% of children in the first year of life. This fraction decreased to approximately 25% at age 2–5 years, to 17% at age 7–12 years, and to about 10% after an age of onset >12 years (Figure 2B). The same trend was observed in clinically relevant age groups. The fraction of families with detection of the single-gene cause was as follows (percentage of families with causative mutation in parentheses) (Figure 3A): onset in the first 3 months of life (69.4%); 4–12 months (49.7%); first year combined (61.3%); young children, age 1–6 years (25.3%); older children, age 7–12 years (17.8%); adolescents, age 13–18 years (10.8%); and young adults, age 19–25 years (21.4%). We conclude that the earlier the age of onset, the more likely it is that the causative mutations can be detected. However, even in young adults the detection rate was still >10%.
Figure 3.
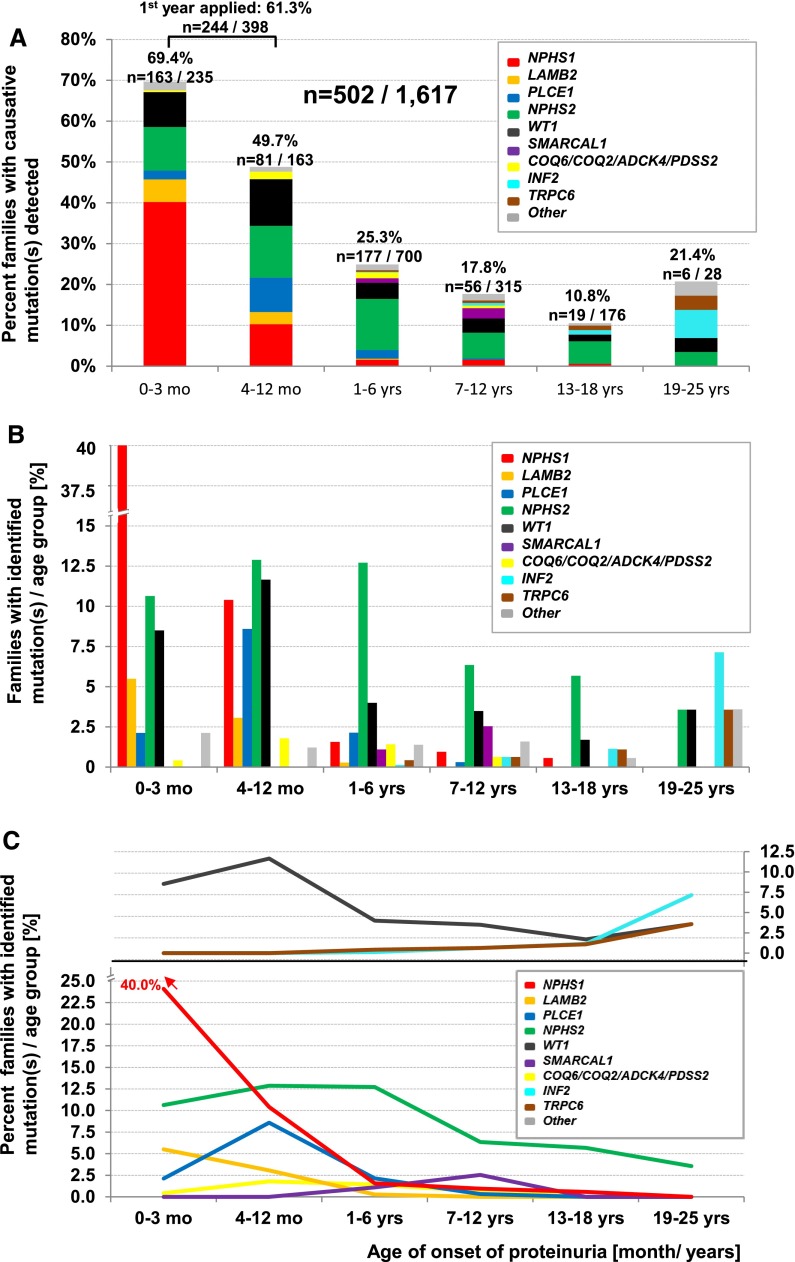
Detection of the causative mutation in 502 families in an international cohort of 1617 families with SRNS in 21 SRNS-causing genes in relation to age of onset of proteinuria in clinically relevant age groups. The displayed 502 families represent the number of families with detected causative mutation and available data for age of onset of proteinuria <26 years. (A) Percentage of families with causative mutation detected per gene per age group. Histograms indicate fraction (in percentage) of families with causative gene detected in n families per families examined (on top of histograms) per age group. (B) Percentages shown in A are represented by separate bars to highlight distribution across age groups as further delineated in C. Note that mutations in NPHS1 or LAMB2 cause early-onset SRNS, whereas mutations in INF2 or TRPC6 cause late-onset SRNS. (C) Percentages of families with causative mutation detected (from B) are interconnected by lines between age groups and shown in different colors for each causative gene (lower panel for recessive genes, upper panel for dominant genes). NPHS1 mutations (red) have an early age of onset and are rarely found in patients older than 6 years. The dominant genes INF2 and TRPC6 manifest in early adulthood and WT1 (black) shows a biphasic distribution (upper panel). (For families with detected disease-causing mutation, data are not shown for 11 families where no age of onset was available, and for 10 families with an age of onset older than 25 years. For families with no detection of the disease-causing mutation, data are not displayed for 111 families for whom no data for age of onset were available and for 34 families with age of onset older than 25 years. In families with >1 affected family member the mean age of onset from all affected individuals was calculated.).
Distribution of Causative Gene by the Age of Onset
We then evaluated the distribution of the detected causative genes by age of onset of SRNS (Figure 3B). The distribution of causative genes within the first 3 months of life was as follows: 40% for NPHS1, 10.6% for NPHS2, 8.5% for WT1, 5.5% for LAMB2, and 4.7% for all other genes together (Figure 3A). NPHS2 was the most frequent gene mutated in individuals with onset of SRNS between 1 and 18 years (range in different age groups, 5.7%–12.7%). Because the relative distribution of the causative genes was age dependent, we plotted the fractions for causative genes by age (Figure 3, B and C). SRNS resulting from recessive genes manifests in early childhood (NPHS1, LAMB2, and PLCE1), whereas mutations in the dominant genes INF2 and TRPC6 are more frequent in early adulthood (Figure 3, B and C). Mutations in WT1 show a biphasic distribution with a first peak at 4–12 months and a second peak for age of onset beyond 18 years. These findings are compatible with the notion that mutations in recessive disease genes are found more frequently in early-onset disease, whereas mutations in dominant genes more frequently cause adult-onset disease.26 Genes involved in the coenzyme Q10 biosynthesis pathway (COQ2, COQ6, PDSS2, and ADCK4) were mutated in about 1% of SRNS cases.
Renal Histologic Findings
A renal biopsy was performed in 337 of 544 individuals, in whom we detected the disease-causing mutation (Supplemental Figure 1A). Compared to children older than 7 years (>90%), renal biopsy was less frequently done in infants (37.8%), most likely due to higher complication risk of the procedure. In the different age groups, different biopsy pattern were seen: In infants diffuse mesangial sclerosis (DMS) was a frequent finding (26.9%), whereas FSGS was seen in >90% of individuals with proteinuria onset at 7 to 25 years (Supplemental Figure 1A). In individuals with disease-causing mutations detected in the early manifesting genes WT1, PLCE1, LAMB2, and NPHS1, DMS was more frequent (23.1%, 17.8%, 13.6%, and 4.9% respectively), whereas mutations in genes with a later age of onset never led to DMS (Supplemental Figure 1B). This is compatible with the notion that DMS represents a developmental glomerular phenotype.26 For individuals with an NPHS1 mutation, kidney biopsy was least frequently performed (32.6%) (Supplemental Figure 1B), most likely because of the high risk of percutaneous renal biopsy in patients with congenital-onset SRNS.
Genotype-Phenotype Correlations
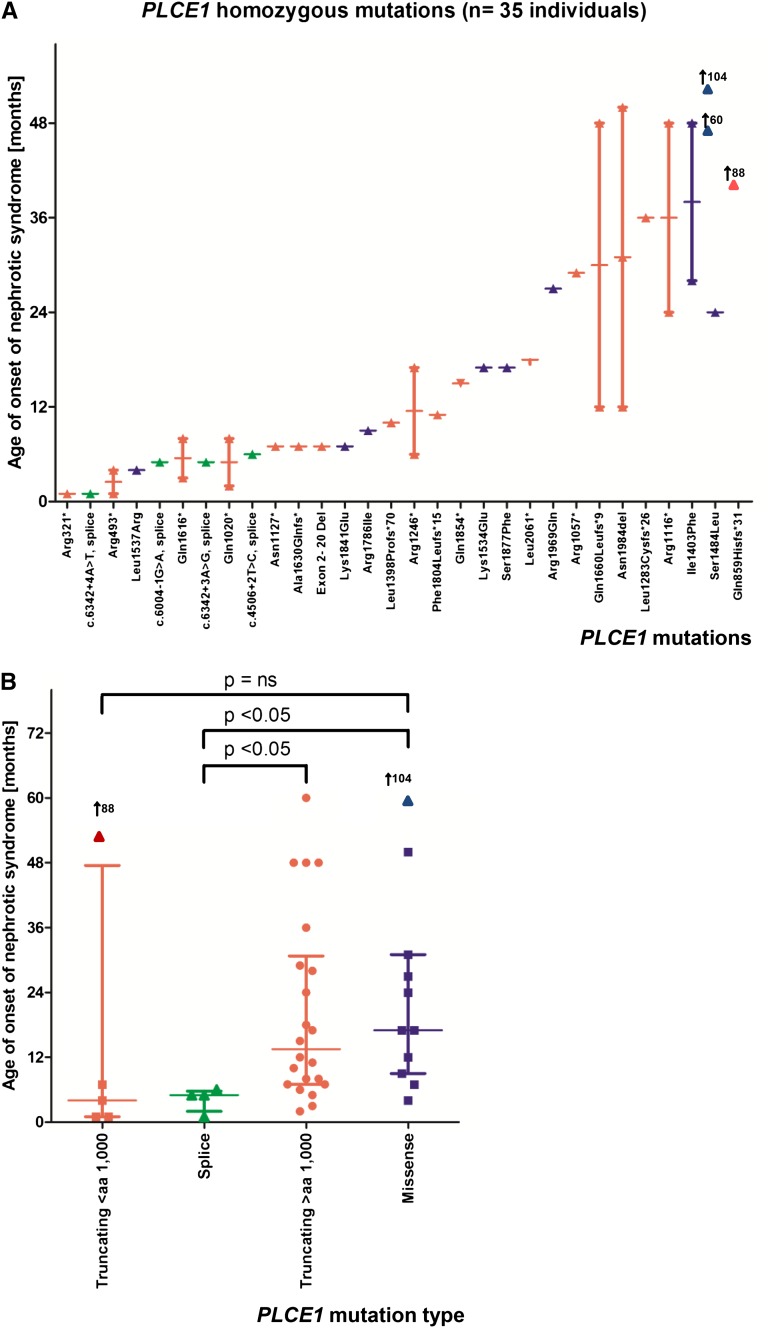
To examine genotype-phenotype correlations, we specifically evaluated the 4 most frequently mutated genes in our cohort: NPHS2, NPHS1, PLCE1, and LAMB2. For PLCE1 mutations, we detected a genotype-phenotype correlation, which has not been described previously. The age of onset was significantly earlier for splice site mutations compared with C-terminal truncating mutations (after amino acid residue 1000) or missense mutations (Figure 4).
Figure 4.
Distribution for age of onset of NS in 35 individuals with homozygous mutations in PLCE1. (A) Homozygous PLCE1 mutations are represented by 30 different alleles. The x-axis indicates the mutations sorted by median age of onset. The y-axis indicates the age of onset of SRNS. Symbols are colored red, green, or blue if the allele is truncating, splice, or missense, respectively. (B) Age of onset of SRNS with causative mutations detected in PLCE1, grouped by type of mutation. The x-axis indicates the type of mutations (truncating mutations before amino acid (aa) residue 1000 (N-terminal), truncating mutations after aa residue 1000 (C-terminal), splice and missense mutations. The y-axis indicates the age of onset of proteinuria. Symbols are blue for missense, green for splice, and red for stop and frameshift. Note that the age of onset was significantly different between splice site mutations versus C-terminal truncating mutations (P<0.05), or splice site versus missense mutations (P<0.05). P values are from two-tailed t test. Arrows represent position of outlier values.
Because homozygous mutations in recessive genes represent the “pure” functional action of gene loss of function, we examined genotype-phenotype correlations for homozygous NPHS2 mutations (Supplemental Figure 2). There was no correlation regarding age of onset for truncating mutations versus missense mutations. However, age of onset was significantly different in the presence of the European founder mutation Arg138Gln (median, 17 months) compared with the milder alleles Leu312Val (median, 96 months) or Val180Met (median, 97.5 months) (Supplemental Figure 2). Because the phenotypic severity of SRNS in individuals with recessive compound heterozygous mutations depends on the “milder” mutation, we ranked compound heterozygous mutations in NPHS2 (Supplemental Figure 3) according to their severity (if present homozygously) from mild (later onset) to severe (earlier onset) (Supplemental Figure 2). The European founder mutation Arg138Gln11,27 occurred more frequently in the early-onset group (group A in Supplemental Figure 3). The allele Arg229Gln was more frequent (8 of 11 times) in the later-onset group (group B). Arg229Gln was recently found to cause SRNS only in compound heterozygosity with specific other alleles.28 We confirmed that Arg229Gln occurred with second alleles that were recently described to be compatible with causing SRNS in combination with Arg229Gln28 (Supplemental Figure 3). In NPHS1, no genotype-phenotype correlation was found for homozygous alleles (Supplemental Figure 4). In LAMB2 we found that N-terminal truncating mutations manifest before 2 months of life, whereas C-terminal truncating mutations manifest after 2 month of life (Supplemental Figure 5). The genotype-phenotype correlations described will be important in assessing the natural course of disease in SRNS and for the management of renal replacement therapy over time.
Frequency of Mutations and Consanguinity
The causative mutation was detected in 184 of 372 (49.5%) consanguineous families but in only 327 of 1308 (25%) of nonconsanguineous families (Supplemental Table 1). In the 8 largest centers the rate of detection of causative mutations ranged from 45.2% in the center with the highest rate of consanguinity (71.4%, Saudi-Arabia) to 14.3% in the center with the lowest rate of consanguinity (0%, Ann Arbor, Michigan) (Supplemental Figure 6). We found a positive correlation between the detection rate of the disease-causing mutation and the rate of consanguinity in the selected centers (R2=0.9414). This finding reflects the fact that consanguinity represents a predilection factor for recessive disease causes.
Allelic Distribution in the 8 Largest Centers
Recessive alleles that were present in the 8 largest centers ≥5 times (18 different alleles) were considered as “frequent alleles” (total n=125 families and 223 mutations) and inspected for their distribution between the 8 different largest contributing centers (Supplemental Figure 7). The previously published founder alleles for NPHS2: Arg138Gln (European founder),11 Pro118Leu (Turkish founder),12 Val180Met (North African founder),11 p.Arg138* (Israeli/Arab founder),11 Gly140Aspfs*41 (Southern European founder)11 and Val260Glu (Comoros and the Sultanate of Oman founder)12 were confirmed in our cohort as predominant in their regions of origin (Supplemental Figure 7).27 We additionally identified two novel Egyptian founder alleles in NPHS2: Met1? (12%; 14 of 116 alleles) and Asn199Lysfs*14 (4.3%; 5 of 116 alleles) in Egypt. In NPHS1 we detected novel founder mutations for the following alleles: Arg367Cys in India (15.2%; 14 of 116 alleles), and Val1084Glyfs*12 in Saudi Arabia (15.4%; 8 of 52 alleles). We also discovered a founder allele for SMARCAL1 Arg586Trp in India (8.7%; 4 of 46 alleles) and for COQ6 Ala353Asp in Turkey (5.9%; 6 of 102 alleles) (Supplemental Figure 7). These findings will help establish genotype-phenotype correlations in clinical settings of distinct locations around the world.
Discussion
In the largest international cohort thus far studied for monogenic causes of SRNS, we present data on mutation analysis in—the 27 genes that, if mutated, cause single-gene forms of SRNS. To render the large number of exon sequences possible, we applied a high-throughput, barcoded exon-sequencing technique that we recently developed with consecutive next-generation sequencing. We report that in more than one quarter of all families with SRNS manifesting before age 25 years, the disease is caused by single-gene mutations in 1 of 21 SRNS genes. Specifically, we found that (1) a surprisingly high fraction of SRNS manifesting before 25 years of life is caused by single-gene mutations (29.5%); (2) the fraction of detecting single-gene causation is inversely correlated to the age of manifestation; (3) specific genotype-phenotype correlations exist for PLCE1 mutations; and (4) certain founder mutations prevail in specific geographic regions. Our approach of sequencing 27 genes was very cost-efficient, resulting in cost of $26 per patient. Our data demonstrate the heuristic power of mutation analysis in SRNS. The findings will guide diagnostic expectations regarding the frequency and nature of causative mutations in many regions of the world.
Identification of causative single-gene mutations may have therapeutic consequences in some cases. For instance, we have shown that whereas single-gene causes of SRNS were previously found in 26% of SRNS cases,2 they were never detected in 120 cases with SSNS,5 strongly suggesting that individuals with single-gene causes of SRNS will not respond to steroid treatment.29 This finding has since been confirmed by another group27 and was again confirmed for 185 individuals with SSNS in this study. Identification of the causative mutation may reveal that a potential therapy is available for some rare single-gene causes of SRNS. For examples, if a mutation in a gene of coenzyme Q10 biosynthesis (COQ2, COQ6, ADCK4, or PDSS2) is detected, experimental treatment with coenzyme Q10 may be indicated.20,30 Likewise, two patients with recessive mutations in PLCE1 responded fully to treatment with steroids or cyclosporine A.18 Finally, individuals with mutations of CUBN may be amenable to treatment with vitamin B12, and individuals with ARHGDIA may theoretically be responsive to the eplerenone treatment.25
In summary, detection of single-gene causes of SRNS will strongly inform practice, diagnostics, understanding of pathogenesis, and treatment of SRNS for the following reasons: (1) Unnecessary initiation or extension of steroid treatment can be avoided if a single-gene cause of SRNS is found; (2) if causative mutations are detected in a gene of coenzyme Q10 biosynthesis pathway (COQ2, COQ6, ADCK4, or PDSS2) treatment with coenzyme Q10 can be attempted20,24; (3) for clinical trials, mutation detection allows etiologic stratification by causative gene in patient cohorts, and thus the small number of patients with a rare disease such as SRNS can be optimally used by stratifying patients according to the etiologic criteria of each causative gene mutation; (4) mutation detection allows definition of correlations between genotype and phenotype as well as between genotype and treatment response; and (5) identification of causative mutations in known single-gene causes of SRNS will help to rapidly define cohorts without mutations, in which additional unknown causative genes can be discovered using whole exome sequencing.25
Concise Methods
Study Participants
Between May 2003 and May 2013 we obtained blood samples, pedigree information, and clinical information from an international cohort of 1783 families (2016 affected individuals) with SRNS following informed consent (www.renalgenes.org) (Figure 1). Study participants did not include patients from Russia, China, sub-Saharan Africa, or Pacific Rim countries. The Institutional Review Boards of the University of Michigan and the Boston Children’s Hospital approved the study. Pediatric nephrologists diagnosed SRNS on the basis of standardized clinical and renal histologic criteria.31 Renal pathologists evaluated renal biopsy specimens. We obtained clinical data using a standardized questionnaire (http://www.renalgenes.org). In the total cohort of 1783 families with SRNS, consanguinity was known to be present in 372 (20.9%) families. The cohort consisted of 187 families with multiple affected cases and 1596 families with single affected cases. The median age of onset of proteinuria was 41 months (3.4 years), with a range of 0–756 months (0–63 years). The cohort had a 1–1.3 female-to-male ratio (Supplemental Table 1). As a control group we included 185 individuals with SSNS based on standardized clinical criteria. Recruitment was performed in the same centers in a consecutive manner.31
Gene Selection, Targeted Exon Sequencing, and Bioinformatics
From 2003 through 2013, when the two most frequent single-gene causes of SRNS were known, we performed exon sequencing in NPHS2 and WT1 (exons 8 and 9) by PCR and Sanger sequencing in our total cohort of 1783 SRNS families.5,9,11,32–37 253 families of these 1783 families, with age of onset <1 year (congenital nephrotic syndrome), were additionally sequenced for mutations in NPHS1.9,32,38–41 In August 2013, when 27 single-gene causes of SRNS were published, the same 1783 families with SRNS were examined for these 27 reported SRNS genes (Supplemental Table 2). We therefore used a strategy of microfluidic multiplex PCR and next-generation sequencing that we recently developed.14,16 The panel included 21 genes with a recessive mode of inheritance (NPHS2, NPHS1, PLCE1, LAMB2, SMARCAL1, COQ6, ITGA3, MYO1E, COQ2, CUBN, ADCK4, DGKE, PDSS2, ARHGDIA, CD2AP, CFH, ITGB4, NEIL1, PTPRO/GLEPP1, SCARB2, and MEFV) and 6 genes with a dominant mode of inheritance (WT1, INF2, TRPC6, ARHGAP24, ACTN4 and LMX1B) (Supplemental Table 2). For these 27 genes, we designed 612 target-specific primer pairs to cover all 512 coding exons and intron/exon boundaries (primer information is available upon request). Amplicon sizes ranged from 129 bp to 342 bp (median, 207 bp). Targeted amplification using barcoded microfluidic multiplex PCR (Fluidigm) together with next-generation sequencing was done as previously described.14,15 Next-generation sequencing was performed on an Illumina HiSeq2000 (1×150 bp single-end, 8 lanes) and an Illumina MiSeq V2 instrument (2×250 bp, paired-end). The next-generation sequencing data were aligned and variants detected via CLC Genomics Workbench 4.9 software. Alignment of resulting reads to reference sequence revealed a mean exon coverage of 735, with 95.4% of the targeted coding regions covered at least 5-fold and 94.5% covered at least 10-fold. No coverage was found in 17 of 612 targeted exons (2.8%). Variants were prefiltered on the basis of quality parameters as described elsewhere.14,15 All mutations were confirmed by Sanger sequencing using genomic DNA samples. Segregation analysis was performed whenever parental DNA was available.
Sensitivity of Mutation Detection
To calculate the sensitivity of mutation detection we included 26 DNA samples with 30 known mutations as positive controls. Overall, 29 of the 30 mutations were redetected (sensitivity, 96.7%). The 1 missing mutation was due to an amplicon failure of NPHS2 exon 1 resulting from a GC-rich region within the target sequence, as previously described.16
Variant Analysis Criteria
We considered variants in the 27 known SRNS to be disease-causing according to strict criteria. We therefore distinguished between criteria for recessive and dominant genes. We considered variants in recessive genes to be disease-causing if two alleles were found in the same individual that fulfilled at least one of the following criteria:
At least one allele truncating allele (stop, abrogation of start or stop, obligatory splice site, or frameshift); or
At least one allele reported in the Human Gene Mutation Database (http://www.hgmd.org/) as disease-causing.
For individuals the second allele needs to meet at least one of the following criteria:
Missense mutation exhibits high evolutionary conservation (Supplemental Table 2); or
Two of three prediction scores classify the allele as disease-causing: SIFT (http://sift.jcvi.org), Mutation Taster (http://www.mutationtaster.org), PolyPhen-2 prediction Humvar >0.9 (http://genetics.bwh.harvard.edu/pph2); or
Loss of function of the identified allele is supported by functional data
For recessive genes two such variants had to be present “in trans.”
Exclusion criteria: Except for the NPHS2 allele, p.Arg229Gln27,28 variants were not assumed to be causative if minor allelic frequency was >1% in genotyping data of the National Heart, Lung, and Blood Institute GO Exome Sequencing Project Exome (http://evs.gs.washington.edu/EVS/). Furthermore, variants were excluded if they did not segregate with the affected status in family members.
For dominant genes we considered variants as disease causing if one allele fulfilled at least one of the following criteria:
Truncating mutation (stop, abrogation of start or stop, obligatory splice site, and frameshift); or
Variant was reported in the Human Gene Mutation Database (http://www.hgmd.org/) as disease causing; or
Missense mutation, if at least conserved from Homo sapiens to Danio rerio; or
Two of three prediction scores classify the allele as disease causing: SIFT (http://sift.jcvi.org), Mutation Taster (http://www.mutationtaster.org), Polyphen 2 prediction Humvar >0.9 (http://genetics.bwh.harvard.edu/pph2); or
loss/gain of function of the identified allele is supported by functional data; or
Full segregation exists in the affected status for ≥7 affected family members.
Exclusion criteria: Variants were excluded if the allele occurred at least once heterozygously or homozygously in the EVS server (http://evs.gs.washington.edu/EVS/) or if the allele did not segregate with the affected status in the family.
Supplementary Material
Acknowledgments
The authors thank the affected individuals and their families for participation in this study and the study coordinators, Leslie Spaneas and Brittany Fisher.
This work was supported by a grant from the National Institutes of Health to F.H. (RC4-DK076683) and the NephCure Foundation. F.H. is an investigator of the Howard Hughes Medical Institute and the Warren E. Grupe Professor. F.H. is an Associate of the Manton Center for Rare Diseases.
Contributing members of the SRNS study group are: H.A. Repetto (Buenos Aires, Argentina), P. MacTaggart (Brisbane, Australia), L. Johnstone (Clayton, Australia), S. Alexander and E. Hodson (Sydney, Australia), C. Mache (Graz, Austria), T.C. Jungraithmayr (Innsbruck, Austria), C. Aufricht and K, Arbeiter (Vienna, Austria), M. Lilova (Sofia, Bulgaria), C. Sweeny (Hamilton, Ontario, Canada), G. Filler (Ottawa, Canada), C. Licht (Toronto, Ontario, Canada), S.Y. Chan (Hong Kong, China), S. Skalova (Hradec Kralove, Czech Republic), T. Seeman (Prague, Czech Republic), M. Nuutinen (Oulu, Finnland), C. Antignac (Paris, France), S. Briese (Berlin, Germany), U. Querfeld (Berlin, Germany), I. Franke (Bonn, Germany), H. Bachmann (Bremen, Germany), M. Kirschstein (Celle, Germany), L.T. Weber, B. Hoppe, B.B. Beck, S. Habbig (Cologne, Germany), B. Mayer (Dresden, Germany), R. Büscher, R. Mallmann, and U. Vester (Essen, Germany), K. Latta (Frankfurt, Germany), M. Pohl and K. Häffner (Freiburg, Germany), L. Patzer (Halle, Germany), T. Henne, L. Pape, M. Schiffer, and A. Schwarz (Hannover, Germany), D. Kiepe, F. Schäfer, B. Tönshoff (Heidelberg, Germany), G. Rönnefarth (Jena, Germany), J.S. Strehlau (Leipzig, Germany), M. Schumacher (Lübeck, Germany), R. Beetz (Mainz, Germany), G. Klaus (Marburg, Germany), H. Fehrenbach (Memmingen, Germany), E. Kuwertz-Broeking, M. Konrad, and A. Schulze-Everding (Muenster, Germany), M.R. Benz, M. Griebel, and J. Hoefele (Munich, Germany), J. Muscheistes (Rostock, Germany), M. Wigger (Rostock, Germany), M. Bald and H. Leichter (Stuttgart, Germany), G. Reusz (Budapest, Hungary), D. Louis (Chandigarh, India), P. Senguttuvan, R. Aravind, R. Padmaraj, and M. Manorajan (Chennai, India), S. Nampoothiri (Cochin, India), J.S.Sharma (Maharashtra, India), A. Bagga and S. Choudry (New Delhi, India), R. Schreiber (Be'er Sheva, Israel), Y. Frishberg (Jerusalem, Israel), R. Cleper and G. Chenin (Tel Aviv, Israel), A. Gianviti (Rome, Italy), F. Al-Kandari (Al Adaliya, Kuwait), V. Tasic (Skopje, Macedonia), G.S. Ch'ng and W.T. Keng (Kuala Lumpur, Malaysia), A. Bokenkamp (Amsterdam, The Netherlands), S. Hashmi and K.N. Moorani (Karachi, Pakistan), F. Anacleto (Manila, Philippines), A. Wasilewska (Bialystok, Poland), D. Drożdż (Krakow, Poland), M. Mizerska-Wasiak (Warsaw, Poland), M. Szczpanska (Zabre, Poland), A.N. Tsygin (Moscow, Russia), S.M. El Desoky (Jeddah, Saudi Arabia), M. Albalwi (Riyadh, Saudi Arabia), R. Bogdanovic, A. Peco-Antić (Belgrade, Serbia), F. Burkhalter and C. Rudin (Basel, Switzerland), A. Pasch (Bern, Switzerland), T.J. Neuhaus (Luzern, Switzerland), U. Odermatt (Luzern, Switzerland), D. Marx-Berger (St. Gallen, Switzerland), G. Laube and G. Spartà (Zurich, Switzerland), I. Attrach (Aleppo, Syria), P. Tanpaiboon (Chiang Mai, Thailand), A. Noyan (Adana, Turkey), S.A. Bakkaloglu, N. Besbas, F. Gok, F. Ozaltin, and A. Bakkaloglu (Ankara, Turkey), E. Comak and A. Gür-Güven (Antalya, Turkey), A. Nayir (Istanbul, Turkey), A. Berdeli, S. Mir, E. Serdaroglu, and A. Soylu (Izmir, Turkey), I. Dursun (Kaysen, Turkey), D.V. Milford and L. Kerecuk (Birmingham, United Kingdom), M.A. Saleem (Bristol, United Kingdom), R.S. Trompeter (London, United Kingdom), J. Sayer (Newcastle upon Tyne, United Kingdom), J. Scheinman (Kansas City, Kansas), M. Bitzer and D.S. Gipson (Ann Arbor, Michigan), C. Hanevold (Augusta, Georgia), G. Gorman (Baltimore, Maryland), M. Pollak (Boston, Massachusetts), C. Langman (Chicago, Illinois), B.P. Dixon (Cincinnati, Ohio), M.A. Cadnapaphornchai (Denver, Colorado), G. Hidalgo (Detroit, Michigan), R.A. Gbadegesin (Durham, North Carolina), C. Silva (Hartford, Connecticut), S. Andreoli (Indianapolis, Indiana), A.D. Torres (Kalamazoo, Michigan), I. Roberti (Livingston, New Jersey), R. Ettenger and N. Kearsley (Los Angeles, California), O. Mansoor (Miami, Florida), A. Singh (New Brunswick, New Jersey), V. Feygina (New York City, New York), F.J. Kaskel (New York City, New York), B. Kaplan and S. Muneeruddin (Philadelphia, Pennsylvania), S. Hsieh (Phoenix, Arizona), V. Chadha (Richmond, Virginia), E.G. Wood (St. Louis, Missouri), R. Weiss (Valhalla, New York), and A. Zolotnistskaya (Valhalla, New York).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014050489/-/DCSupplemental.
References
- 1.Honeycutt AA, Segel JE, Zhuo X, Hoerger TJ, Imai K, Williams D: Medical costs of CKD in the Medicare population. J Am Soc Nephrol 24: 1478–1483, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA: Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr Transplant 11: 366–373, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Cheong HI, Han HW, Park HW, Ha IS, Han KS, Lee HS, Kim SJ, Choi Y. Early recurrent nephrotic syndrome after renal transplantation in children with focal segmental glomerulosclerosis. Nephrol Dial Transplant 15: 78-81, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Senggutuvan P, Cameron JS, Hartley RB, Rigden S, Chantler C, Haycock G, Williams DG, Ogg C, Koffman G: Recurrence of focal segmental glomerulosclerosis in transplanted kidneys: Analysis of incidence and risk factors in 59 allografts. Pediatr Nephrol 4: 21–28, 1990 [DOI] [PubMed] [Google Scholar]
- 5.Ruf RG, Lichtenberger A, Karle SM, Haas JP, Anacleto FE, Schultheiss M, Zalewski I, Imm A, Ruf EM, Mucha B, Bagga A, Neuhaus T, Fuchshuber A, Bakkaloglu A, Hildebrandt F, Arbeitsgemeinschaft Für Pädiatrische Nephrologie Study Group : Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol 15: 722–732, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Höcker B, Knüppel T, Waldherr R, Schaefer F, Weber S, Tönshoff B: Recurrence of proteinuria 10 years post-transplant in NPHS2-associated focal segmental glomerulosclerosis after conversion from cyclosporin A to sirolimus. Pediatr Nephrol 21: 1476–1479, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Somlo S, Mundel P: Getting a foothold in nephrotic syndrome. Nat Genet 24: 333–335, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Wiggins RC: The spectrum of podocytopathies: A unifying view of glomerular diseases. Kidney Int 71: 1205–1214, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Hinkes BG, Mucha B, Vlangos CN, Gbadegesin R, Liu J, Hasselbacher K, Hangan D, Ozaltin F, Zenker M, Hildebrandt F, Arbeitsgemeinschaft für Paediatrische Nephrologie Study Group : Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics 119: e907–e919, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Karle SM, Uetz B, Ronner V, Glaeser L, Hildebrandt F, Fuchshuber A: Novel mutations in NPHS2 detected in both familial and sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol 13: 388–393, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C: NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24: 349–354, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Weber S, Gribouval O, Esquivel EL, Morinière V, Tête MJ, Legendre C, Niaudet P, Antignac C: NPHS2 mutation analysis shows genetic heterogeneity of steroid-resistant nephrotic syndrome and low post-transplant recurrence. Kidney Int 66: 571–579, 2004 [DOI] [PubMed] [Google Scholar]
- 13.McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA, RADAR the UK SRNS Study Group : Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 8: 637–648, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbritter J, Diaz K, Chaki M, Porath JD, Tarrier B, Fu C, Innis JL, Allen SJ, Lyons RH, Stefanidis CJ, Omran H, Soliman NA, Otto EA: High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet 49: 756–767, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Halbritter J, Porath JD, Diaz KA, Braun DA, Kohl S, Chaki M, Allen SJ, Soliman NA, Hildebrandt F, Otto EA, GPN Study Group : Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet 132: 865–884, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovric S, Fang H, Vega-Warner V, Sadowski CE, Gee HY, Halbritter J, Ashraf S, Saisawat P, Soliman NA, Kari JA, Otto EA, Hildebrandt F, Nephrotic Syndrome Study Group : Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 9: 1109–1116, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasselbacher K, Wiggins RC, Matejas V, Hinkes BG, Mucha B, Hoskins BE, Ozaltin F, Nürnberg G, Becker C, Hangan D, Pohl M, Kuwertz-Bröking E, Griebel M, Schumacher V, Royer-Pokora B, Bakkaloglu A, Nürnberg P, Zenker M, Hildebrandt F: Recessive missense mutations in LAMB2 expand the clinical spectrum of LAMB2-associated disorders. Kidney Int 70: 1008–1012, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nürnberg G, Garg P, Verma R, Chaib H, Hoskins BE, Ashraf S, Becker C, Hennies HC, Goyal M, Wharram BL, Schachter AD, Mudumana S, Drummond I, Kerjaschki D, Waldherr R, Dietrich A, Ozaltin F, Bakkaloglu A, Cleper R, Basel-Vanagaite L, Pohl M, Griebel M, Tsygin AN, Soylu A, Müller D, Sorli CS, Bunney TD, Katan M, Liu J, Attanasio M, O’toole JF, Hasselbacher K, Mucha B, Otto EA, Airik R, Kispert A, Kelley GG, Smrcka AV, Gudermann T, Holzman LB, Nürnberg P, Hildebrandt F: Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet 38: 1397–1405, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Gbadegesin R, Hinkes BG, Hoskins BE, Vlangos CN, Heeringa SF, Liu J, Loirat C, Ozaltin F, Hashmi S, Ulmer F, Cleper R, Ettenger R, Antignac C, Wiggins RC, Zenker M, Hildebrandt F. Mutations in PLCE1 are a major cause of isolated diffuse mesangial sclerosis (IDMS). Nephrol Dial Transplant 23: 1291–1297, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, Xie LX, Salviati L, Hurd TW, Vega-Warner V, Killen PD, Raphael Y, Ashraf S, Ovunc B, Schoeb DS, McLaughlin HM, Airik R, Vlangos CN, Gbadegesin R, Hinkes B, Saisawat P, Trevisson E, Doimo M, Casarin A, Pertegato V, Giorgi G, Prokisch H, Rötig A, Nürnberg G, Becker C, Wang S, Ozaltin F, Topaloglu R, Bakkaloglu A, Bakkaloglu SA, Müller D, Beissert A, Mir S, Berdeli A, Varpizen S, Zenker M, Matejas V, Santos-Ocaña C, Navas P, Kusakabe T, Kispert A, Akman S, Soliman NA, Krick S, Mundel P, Reiser J, Nürnberg P, Clarke CF, Wiggins RC, Faul C, Hildebrandt F: COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 121: 2013–2024, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heeringa SF, Möller CC, Du J, Yue L, Hinkes B, Chernin G, Vlangos CN, Hoyer PF, Reiser J, Hildebrandt F: A novel TRPC6 mutation that causes childhood FSGS. PLoS ONE 4: e7771, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Has C, Spartà G, Kiritsi D, Weibel L, Moeller A, Vega-Warner V, Waters A, He Y, Anikster Y, Esser P, Straub BK, Hausser I, Bockenhauer D, Dekel B, Hildebrandt F, Bruckner-Tuderman L, Laube GF: Integrin α3 mutations with kidney, lung, and skin disease. N Engl J Med 366: 1508–1514, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovunc B, Otto EA, Vega-Warner V, Saisawat P, Ashraf S, Ramaswami G, Fathy HM, Schoeb D, Chernin G, Lyons RH, Yilmaz E, Hildebrandt F: Exome sequencing reveals cubilin mutation as a single-gene cause of proteinuria. J Am Soc Nephrol 22: 1815–1820, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashraf S, Gee HY, Woerner S, Xie LX, Vega-Warner V, Lovric S, Fang H, Song X, Cattran DC, Avila-Casado C, Paterson AD, Nitschké P, Bole-Feysot C, Cochat P, Esteve-Rudd J, Haberberger B, Allen SJ, Zhou W, Airik R, Otto EA, Barua M, Al-Hamed MH, Kari JA, Evans J, Bierzynska A, Saleem MA, Böckenhauer D, Kleta R, El Desoky S, Hacihamdioglu DO, Gok F, Washburn J, Wiggins RC, Choi M, Lifton RP, Levy S, Han Z, Salviati L, Prokisch H, Williams DS, Pollak M, Clarke CF, Pei Y, Antignac C, Hildebrandt F: ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest 123: 5179–5189, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gee HY, Saisawat P, Ashraf S, Hurd TW, Vega-Warner V, Fang H, Beck BB, Gribouval O, Zhou W, Diaz KA, Natarajan S, Wiggins RC, Lovric S, Chernin G, Schoeb DS, Ovunc B, Frishberg Y, Soliman NA, Fathy HM, Goebel H, Hoefele J, Weber LT, Innis JW, Faul C, Han Z, Washburn J, Antignac C, Levy S, Otto EA, Hildebrandt F: ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest 123: 3243–3253, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hildebrandt F, Heeringa SF: Specific podocin mutations determine age of onset of nephrotic syndrome all the way into adult life. Kidney Int 75: 669–671, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Bouchireb K, Boyer O, Gribouval O, Nevo F, Huynh-Cong E, Morinière V, Campait R, Ars E, Brackman D, Dantal J, Eckart P, Gigante M, Lipska BS, Liutkus A, Megarbane A, Mohsin N, Ozaltin F, Saleem MA, Schaefer F, Soulami K, Torra R, Garcelon N, Mollet G, Dahan K, Antignac C: NPHS2 mutations in steroid-resistant nephrotic syndrome: A mutation update and the associated phenotypic spectrum. Hum Mutat 35: 178–186, 2014 [DOI] [PubMed] [Google Scholar]
- 28.Tory K, Menyhárd DK, Woerner S, Nevo F, Gribouval O, Kerti A, Stráner P, Arrondel C, Cong EH, Tulassay T, Mollet G, Perczel A, Antignac C: Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 46: 299–304, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Niaudet P, Broyer M: Management of steroid-responsive nephrotic syndrome. Pediatr Nephrol 14: 770–771, 2000 [DOI] [PubMed] [Google Scholar]
- 30.Montini G, Malaventura C, Salviati L: Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med 358: 2849–2850, 2008 [DOI] [PubMed] [Google Scholar]
- 31.ISKDC : Primary nephrotic syndrome in children: Clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int 20: 765–771, 1981 [DOI] [PubMed] [Google Scholar]
- 32.Schultheiss M, Ruf RG, Mucha BE, Wiggins R, Fuchshuber A, Lichtenberger A, Hildebrandt F: No evidence for genotype/phenotype correlation in NPHS1 and NPHS2 mutations. Pediatr Nephrol 19: 1340–1348, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Hinkes B, Vlangos C, Heeringa S, Mucha B, Gbadegesin R, Liu J, Hasselbacher K, Ozaltin F, Hildebrandt F, APN Study Group : Specific podocin mutations correlate with age of onset in steroid-resistant nephrotic syndrome. J Am Soc Nephrol 19: 365–371, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruf RG, Schultheiss M, Lichtenberger A, Karle SM, Zalewski I, Mucha B, Everding AS, Neuhaus T, Patzer L, Plank C, Haas JP, Ozaltin F, Imm A, Fuchshuber A, Bakkaloglu A, Hildebrandt F, APN Study Group : Prevalence of WT1 mutations in a large cohort of patients with steroid-resistant and steroid-sensitive nephrotic syndrome. Kidney Int 66: 564–570, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Mucha B, Ozaltin F, Hinkes BG, Hasselbacher K, Ruf RG, Schultheiss M, Hangan D, Hoskins BE, Everding AS, Bogdanovic R, Seeman T, Hoppe B, Hildebrandt F, Members of the APN Study Group : Mutations in the Wilms’ tumor 1 gene cause isolated steroid resistant nephrotic syndrome and occur in exons 8 and 9. Pediatr Res 59: 325–331, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Chernin G, Vega-Warner V, Schoeb DS, Heeringa SF, Ovunc B, Saisawat P, Cleper R, Ozaltin F, Hildebrandt F, Members of the GPN Study Group : Genotype/phenotype correlation in nephrotic syndrome caused by WT1 mutations. Clin J Am Soc Nephrol 5: 1655–1662, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeanpierre C, Denamur E, Henry I, Cabanis MO, Luce S, Cécille A, Elion J, Peuchmaur M, Loirat C, Niaudet P, Gubler MC, Junien C: Identification of constitutional WT1 mutations, in patients with isolated diffuse mesangial sclerosis, and analysis of genotype/phenotype correlations by use of a computerized mutation database. Am J Hum Genet 62: 824–833, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heeringa SF, Vlangos CN, Chernin G, Hinkes B, Gbadegesin R, Liu J, Hoskins BE, Ozaltin F, Hildebrandt F. Thirteen novel NPHS1 mutations in a large cohort of children with congenital nephrotic syndrome. Nephrol Dial Transplant 23: 3527–3533, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoeb DS, Chernin G, Heeringa SF, Matejas V, Held S, Vega-Warner V, Bockenhauer D, Vlangos CN, Moorani KN, Neuhaus TJ, Kari JA, MacDonald J, Saisawat P, Ashraf S, Ovunc B, Zenker M, Hildebrandt F. Nineteen novel NPHS1 mutations in a worldwide cohort of patients with congenital nephrotic syndrome (CNS). Nephrol Dial Transplant 25: 2970–2976, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ovunc B, Ashraf S, Vega-Warner V, Bockenhauer D, Elshakhs NA, Joseph M, Hildebrandt F, Gesellschaft für Pädiatrische Nephrologie (GPN) Study Group : Mutation analysis of NPHS1 in a worldwide cohort of congenital nephrotic syndrome patients. Nephron Clin Pract 120: c139–c146, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kestilä M, Männikkö M, Holmberg C, Gyapay G, Weissenbach J, Savolainen ER, Peltonen L, Tryggvason K: Congenital nephrotic syndrome of the Finnish type maps to the long arm of chromosome 19. Am J Hum Genet 54: 757–764, 1994 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.