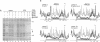Abstract
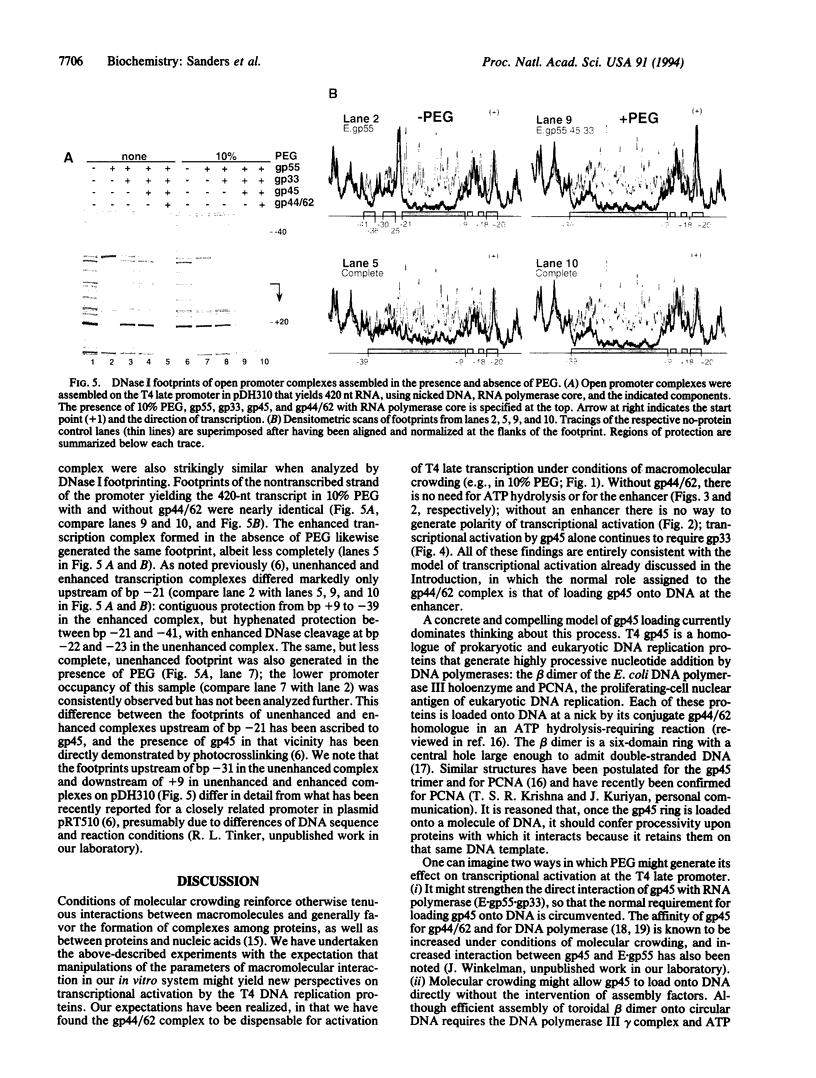
We have used a molecular crowding reagent to define functions in the transcriptional activation of bacteriophage T4 late genes. This activation normally requires the three T4 DNA polymerase accessory proteins encoded by T4 genes 44, 62, and 45 (the gp44/62 complex and gp45), an enhancer-like cis-acting site, an RNA polymerase-bound coactivator, and an unobstructed path along the DNA joining the promoter to the enhancer. We show that molecular crowding eliminates the requirement for the gp44/62 complex and for the enhancer, retains the requirement for gp45 and its coactivator, and generates activated promoter complexes with nearly unchanged DNase I footprints. These experiments identify gp45 as the direct activator of transcription, and the gp44/62 complex as the assembly factor for gp45. They suggest that the enhancer serves as the normal, but not invariably essential, entry site for the gp45 DNA-tracking protein.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Capson T. L., Benkovic S. J., Nossal N. G. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991 Apr 19;65(2):249–258. doi: 10.1016/0092-8674(91)90159-v. [DOI] [PubMed] [Google Scholar]
- Ebright R. H. Transcription activation at Class I CAP-dependent promoters. Mol Microbiol. 1993 May;8(5):797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
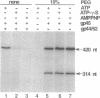
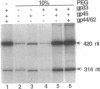
- Herendeen D. R., Kassavetis G. A., Barry J., Alberts B. M., Geiduschek E. P. Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science. 1989 Sep 1;245(4921):952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Williams K. P., Kassavetis G. A., Geiduschek E. P. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990 May 4;248(4955):573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993 May;175(9):2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Jarvis T. C., Ring D. M., Daube S. S., von Hippel P. H. "Macromolecular crowding": thermodynamic consequences for protein-protein interactions within the T4 DNA replication complex. J Biol Chem. 1990 Sep 5;265(25):15160–15167. [PubMed] [Google Scholar]
- Joazeiro C. A., Kassavetis G. A., Geiduschek E. P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994 Apr;14(4):2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., O'Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993 Dec 20;234(4):915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- LaDuca R. J., Crute J. J., McHenry C. S., Bambara R. A. The beta subunit of the Escherichia coli DNA polymerase III holoenzyme interacts functionally with the catalytic core in the absence of other subunits. J Biol Chem. 1986 Jun 5;261(16):7550–7557. [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20034–20044. [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. The T4 DNA polymerase accessory proteins form an ATP-dependent complex on a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20024–20033. [PubMed] [Google Scholar]
- O'Donnell M., Studwell P. S. Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps. J Biol Chem. 1990 Jan 15;265(2):1179–1187. [PubMed] [Google Scholar]
- Onrust R., Stukenberg P. T., O'Donnell M. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J Biol Chem. 1991 Nov 15;266(32):21681–21686. [PubMed] [Google Scholar]
- Piperno J. R., Kallen R. G., Alberta B. M. Analysis of a T4 DNA replication protein complex. Studies of the DNA recognition site for T4 gene 44/62 and 45 protein-catalyzed ATP hydrolysis. J Biol Chem. 1978 Jul 25;253(14):5180–5185. [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., von Hippel P. H. Assembly of a functional replication complex without ATP hydrolysis: a direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A., Tsaneva I. R., West S. C., Benson C. J., Yu X., Egelman E. H. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Tinker R. L., Williams K. P., Kassavetis G. A., Geiduschek E. P. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell. 1994 Apr 22;77(2):225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- Williams K. P., Kassavetis G. A., Geiduschek E. P. Interactions of the bacteriophage T4 gene 55 product with Escherichia coli RNA polymerase. Competition with Escherichia coli sigma 70 and release from late T4 transcription complexes following initiation. J Biol Chem. 1987 Sep 5;262(25):12365–12371. [PubMed] [Google Scholar]
- Williams K. P., Müller R., Rüger W., Geiduschek E. P. Overproduced bacteriophage T4 gene 33 protein binds RNA polymerase. J Bacteriol. 1989 Jun;171(6):3579–3582. doi: 10.1128/jb.171.6.3579-3582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman J. W., Kassavetis G. A., Geiduschek E. P. Molecular genetic analysis of a prokaryotic transcriptional coactivator: functional domains of the bacteriophage T4 gene 33 protein. J Bacteriol. 1994 Feb;176(4):1164–1171. doi: 10.1128/jb.176.4.1164-1171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Minton A. P. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]