Abstract
Memory CD8 T cells generated after acute viral infections or live vaccines can persist for extended periods, in some instances for life, and play an important role in protective immunity. This long-lived immunity is achieved in part through cytokine-mediated homeostatic proliferation of memory T cells while maintaining the acquired capacity for rapid recall of effector cytokines and cytolytic molecules. The ability of memory CD8 T cells to retain their acquired properties, including their ability to remain poised to recall effector functions, is a truly impressive feat given that these acquired properties can be maintained for decades without exposure to cognate antigen. Here, we discuss general mechanisms for acquisition and maintenance of transcriptional programs in memory CD8 T cells and the potential role of epigenetic programming in maintaining the phenotypic and functional heterogeneity of cellular subsets among the pool of memory cells.
Introduction and context
It is now well established that memory CD8 T cells generated from an acute viral infection acquire the ability to persist in the absence of antigen [1–3]. The realization that memory CD8 T cells undergo antigen-independent homeostasis while retaining the ability to rapidly recall effector functions upon antigen re-encounter was a major conceptual advance for the field. In the years following this observation, efforts by many labs to dissect the mechanisms that instill memory T cells with their long-lived nature have progressively transformed the concept of T cell memory into therapeutic applications to treat or prevent disease and have paved the way for vaccine efforts focused on generating long-lived T cell immunity [4–7]. We often describe the cardinal properties of memory T cells as being their ability to undergo interleukin-15 (IL-15)- and IL-7-dependent self-renewal and survival in the absence of antigen, an ability to reside in non-lymphoid tissues to survey for antigen, and the heightened capacity to recall effector functions upon antigen encounter [8–11]. However, recent investigation of the cellular heterogeneity within the pool of memory T cells has revealed that these generalizable attributes of T cell memory are actually the result of a collection of subsets of cells with distinct phenotypic and functional properties (Figure 1). It is now evident that protective CD8 T cell immunity against a given pathogen is achieved by the collective efforts of each of these subsets. Although the discovery and dissection of the functional differences of memory subsets have significantly advanced our basic understanding of the cellular and molecular mechanisms controlling their development, many important questions remain regarding the plasticity of these subsets and their role in long-lived immunity. Here, we examine phenotypic and functional characteristics of memory CD8 T cell subsets and discuss current issues regarding the plasticity versus stability of acquired transcriptional programs after memory differentiation.
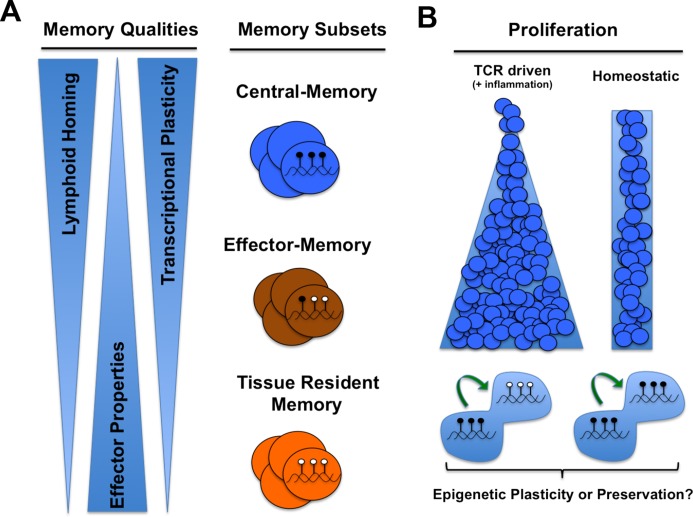
Figure 1. Memory CD8 T cell differentiation and plasticity.
(A) The pool of memory CD8 T cells consists of cells with varying degrees of effector function, proliferative capacity, and distinct tissue homing properties. The total population of memory cells has been broadly divided into subsets classified as central-memory, effector-memory, and tissue-resident memory. Central-memory CD8 T cells are restricted primarily to lymphoid tissues, whereas effector-memory CD8 T cells are found in circulation and non-lymphoid tissues. Tissue-resident memory CD8 T cells are retained at sites of pathogen entry. Upon re-infection of the host, central-memory and effector-memory cells further differentiate and contribute to the pool of secondary effector cells. During a recall response, tissue-resident memory CD8 T cells serve as a vanguard against pathogens, recruiting secondary effector cells differentiated from central-memory and effector-memory CD8 T cells. The memory subsets’ collective abilities of self-renewal, persistence in non-lymphoid tissue, and rapid differentiation to effector cells upon re-infection provide the host with protective immunological memory. Memory CD8 T cells have acquired epigenetic programs that are distinct from those of naïve and effector cells. These programs are coupled to the long-lived maintenance of memory qualities. (B) It remains to be determined whether each of the memory subsets has distinct epigenetic programs and whether the epigenetic programs can be further modified during antigen- or cytokine-driven proliferation or both. The permissive versus repressive epigenetic programs in the cartoon schematic are denoted as filled (methylated DNA) or open (unmethylated DNA) lollipops, respectively.
Major recent advances
Memory subsets
Protective T cell immunity is achieved in part by partitioning the pool of memory CD8 T cells into subsets of cells with distinct tissue homing, self-renewal, and effector recall potentials. The first functional description of memory subsets came from Sallusto and colleagues [12] when they parsed memory cells into cellular subsets with distinct phenotypic properties. These subsets became classically known as effector-memory (Tem) and central-memory (Tcm) T cells. After the initial characterization of human Tem and Tcm memory subsets, mouse model systems amenable to tracking primary immune responses in lymphoid and non-lymphoid tissues were used to better define the proliferative and trafficking properties of T cell memory subsets [13]. From these studies emerged the model that the pool of memory CD8 T cells can be subdivided into two subsets: Tem and Tcm. Downregulation of the lymphoid homing molecules CD62L and CCR7 in the Tem subset of cells limits their ability to reside in the lymph node, allowing them to circulate and home to non-lymphoid tissues. Additionally, the Tem subset of cells remain poised to provide immediate effector functions. The Tcm subset of cells express CD62L and CCR7, restricting their homing to lymphoid tissues. It is believed that the Tcm subset of cells serve as a self-renewing source for the total pool of memory cells. Recent investigations of memory and effector functions of human CD8 T cells subsets have identified a new subset of memory T cells that have naïve-phenotypic qualities (as well as many naïve gene expression programs) but that possess the ability to undergo IL-7 and IL-15 homeostatic proliferation. This subset, now referred to as Tscm because of its many stem cell-like qualities, has the potential to give rise to multiple memory subsets and subsequently yield an effector recall response [14]. During investigation of the various memory subsets, it became apparent that an additional subset of memory CD8 T cells that entered peripheral tissues were inhibited from recirculating. Subsequently, a series of adoptive transfer and parabiosis studies demonstrated that a distinct subset of memory CD8 T cells reside in mucosal tissues, remaining at the site of pathogen entry [15–18]. Memory T cells with restricted egress from peripheral sites of pathogen entry and exposure have become broadly defined as tissue-resident memory cells (Trm). These studies have shaped our current view of CD8 T cell memory differentiation and raised key conceptual questions (highlighted in Table 1). For instance, with the latest description of the Trm and Tscm cell subsets, a question receiving considerable attention is whether the functional differences between the memory subsets are maintained by signals from the local tissue environment or whether they are maintained by cell-intrinsic mechanisms that instill them with distinct proliferative and tissue-homing potential [17,19]. Recent functional, phenotypic, gene expression, and epigenetic profiling studies of effector and memory T cell subsets have provided insights into question of memory fate stability.
Table 1. Open questions in CD8 T cell memory.
|
|
|
|
|
|
Memory gene expression profiles
The core principle that memory cells remain poised to recall effector functions has been a conceptual driver for the field. Since this concept was established, a major focus has been to identify the cellular and molecular mechanisms that allow resting memory T cells to retain their antiviral properties. Broadly, retention of tissue-specific (environment-driven changes to the cells) and lineage-specific properties acquired during cellular differentiation is often maintained by heritable changes in gene regulation. Such stability in transcriptional programming is achieved by select expression of transcription factors, as well as changes in epigenetic programming that coordinate chromatin accessibility by either restricting or allowing transcription factors access to specific regions of chromatin. Initial genome-wide analyses of gene expression patterns in antigen-specific CD8 T cells differentiating in response to an acute viral infection revealed that the progressive change in transcriptional regulation for thousands of genes was also coupled to changes in expression of key transcription factors [20]. Several ensuing gene expression profiling studies provided further evidence that the mechanism for maintenance of acquired antiviral properties in long-lived memory T cells is mediated in large part by changes in transcriptional regulation [20–22]. Adoptive transfer and cell-fate tracking experiments have reinforced the idea that many of the gene expression programs acquired in memory CD8 T cells are stably maintained in the absence of T cell receptor (TCR) or inflammation [23–25].
Dozens of transcription factors that promote memory CD8 T cell differentiation have now been identified (Table 2). Although it is clear that these transcription factors are critical for the generation of memory cells, less is known about their role in the maintenance of subset-specific acquired gene expression programs during homeostasis. Indeed, several knockout studies of transcription factors have revealed that, although deletion of the transcription factor had minimal impact on the quantity of the memory pool, the expression programs of the knockout cells were strikingly skewed toward effector versus central-memory phenotype. Specifically, Rutishauser and colleagues [26] found that conditional deletion of blimp-1 resulted in heightened re-expression of CD62L and CD127 during the conversion of antigen-specific effector CD8 T cells into memory cells, indicating that blimp-1 is used to promote transcriptional programs of effector-memory cells. Likewise, Kallies and colleagues [27] found that blimp-1 was critical for poising the memory cells for a rapid recall of effector functions [26]. Deletion of many transcription factors (such as blimp-1) in CD8 T cells has been reported to mediate memory T cell homeostasis or regulate (or both) the expression of gene expression programs now associated with specific subsets (Table 2). For many of these factors, it remains to be determined whether their impact on memory CD8 T cell function is directly due to their regulation in gene expression programming after memory differentiation and establishment of the memory population. Indeed, because the pool of memory CD8 T cells is a composite of many phenotypically distinct cellular subsets, significant efforts are now focused on identifying transcription factors that play a role in the acquisition and maintenance of phenotypic and functional differences among the memory cell subsets.
Table 2. Transcription factors of memory CD8 T cell differentiation.
| Protein name | Reference(s) |
|---|---|
| Klf2 | [43] |
| Bcl6 | [44] |
| Stat5 | [45,46] |
| Bcl6b (BAZF) | [47] |
| Tbet | [48] |
| Eomes | [48] |
| Id2 | [49,51] |
| Id3 | [50] |
| Elf4 | [51] |
| Blimp-1 | [26,27] |
| Tcf1 | [52] |
| Ets1 | [53] |
| Stat3 | [54,55] |
| Foxo3 | [56] |
| Foxo1 | [57] |
| Irf4 | [58] |
This is a partial list of transcription factors known to regulate memory CD8 T cell differentiation.
Additional insights into gene regulation stability have come from studies investigating the changes in phenotype and function of resting memory CD8 T cells following multiple rounds of acute infection. Using a heterologous prime and boost strategy, Masopust and colleagues [28] demonstrated that, relative to primary memory CD8 T cells, secondary and tertiary memory CD8 T cells were enriched for effector-like qualities and showed increased localization to non-lymphoid tissues. This study demonstrated that iterative rounds of antigen exposure progressively enriched the memory pool with cells having lower expression (slower re-expression) of CD62L and CD127 and higher granzyme B expression, indicating that the repetitive (or prolonged) stimulation of the cells promotes development of the Tem population [28]. The issue of gene regulation stability was assessed more directly in a similar study investigating gene expression programs in secondary, tertiary, and quaternary memory CD8 T cells. The authors found that, with each additional boost, a subset of gene expression programs were progressively modified in the successive pool of memory CD8 T cells [29]. However, the authors also observed that a core memory gene expression program was preserved in the pool of memory CD8 T cells even after multiple rounds of stimulation [29]. Together, these prime-boost studies demonstrated that the quantity of vaccine-generated memory T cells can be increased by boosting strategies and have important implications for generating novel T cell-based vaccines. In addition to the practical application of these findings to vaccine design, these studies provided important insight into the mechanisms that regulate the generation of memory subsets. Given that the phenotypic differences used to delineate the memory subsets were progressively modified upon subsequent exposure to antigen during the prime-boost protocols, it raises the question of whether the subsets are committed to their respective fate or whether they retain some level of transcriptional plasticity.
Whereas it is clear that the individual memory subsets enriched in different tissues have distinct functional differences, it was unclear until recently whether these differences were a reflection of the environment in which they reside (lymphoid versus non-lymphoid) or whether they are dictated by cell-intrinsic gene expression programs acquired during previous antigenic and inflammatory stimuli. To assess the role of the local tissue environment in maintaining memory subset functions, Wakim and colleagues [30] recently performed a series of adoptive transfer experiments transplanting memory subsets into lymphoid versus non-lymphoid tissues and then examined the recall response of these cells. Trm cells taken from the brain, transferred into the spleen (via intravenous adoptive transfer), and then re-exposed to antigen were still restricted in their ability to proliferate [30]. Likewise, memory CD8 T cells taken from the spleen and transferred into the brain retained a heightened proliferative potential [31]. These data demonstrate that differentiation of brain Trm cells is coupled to acquisition of a cell-intrinsic program that restricts their ability to undergo antigen-driven proliferation (Figure 1). In contrast to studies of brain Trm, a study by Masopust and colleagues [18] revealed that Trm cells taken from the gut and transferred back into circulation were in fact capable of developing into effector and multiple memory subsets upon antigen re-encounter, suggesting that the cell-extrinsic signals from the local tissue environment also contribute to subset-specific memory qualities.
To better understand the influence that the local tissue environment has on Trm transcriptional regulation, Wakim and colleagues [31] analyzed the gene expression profile of brain Trm cells versus the conventional memory T cell subsets. Their analysis revealed that brain Trm cells acquired distinct gene expression programs of several transcription factors previously reported to control memory differentiation, including Tcf1 and Eomes [31] (Table 2). Similarly, Skon and colleagues [32] reported that downregulation of the transcription factor Klf2 is critical for programming antigen-specific CD8 T cells with the expression of homing molecules that control the ability of the cell to enter circulation versus maintain residency in non-lymphoid tissue. Taken together, the prime-boost studies and analysis of the Trm indicate that stability of gene expression programs is coupled to cell-intrinsic changes in transcription factor expression and that the relative plasticity of the programming is sensitive to duration of TCR and co-receptor stimulation.
Epigenetic programming
Many studies have demonstrated that acquired gene expression programs regulating effector and memory qualities can be maintained in the absence of TCR signaling. Most efforts to understand the stability of acquired gene expression programs centered on investigating transcription factors that are specifically associated with effector and memory stages of differentiation. Recently, though, some of the focus has shifted to investigating epigenetic modifications to histones and DNA as a mechanism to maintain chromatin accessibility for transcription factors in resting memory cells. Epigenetic enzymes, including DNA methyltransferases and histone modifiers, work in concert to coordinate genome-wide epigenetic programs that provide gene-specific chromatin accessibility. These enzymes include DNA methyltransferase that provide new modifications (de novo methylation) as well as methyltransferases that propagate the newly acquired program from parental cell to daughter cell during division (maintenance methylation). Additionally, a variety of enzymes are used to modify multiple histone amino acids. Depending on the combination of modifications to histone amino acid, these marks can result in repressive or permissive chromatin states, commonly referred to as the histone code.
Global epigenetic correlates of poised functions in memory T cells were initially assessed by Araki and colleagues [33] by performing genome-wide histone H3K4me3 and H3K27me3 ChIP-sequencing (ChIP-seq) analysis of polyclonal human memory CD8 T cell subsets. The authors observed that H3K4me3 histone modifications positively associated with expressed genes in memory cells but that H3K27me3 modifications, known to be coupled to transcriptional repression, negatively correlated with expressed genes. Considering the combination of gene expression data and ChIP-seq data, the authors were able to organize the transcriptional states of the genes into the following categories in memory T cells: (a) active (expressed and associated with the H3K4me3 histone modification), (b) repressed (not expressed and associated with the H3K27me3 histone modification), and (c) poised (not expressed but associated with H3K4me3-positive histone marks), and (d) bivalent (not expressed and associated with both H3K4me3 and H3K27me3 marks). Their findings were the first to demonstrate at a genome-wide level that the acquired changes in gene expression in memory CD8 T cells are coupled with changes in permissive and repressive epigenetic programs [33]. Recently, Russ and colleagues [34] used a mouse model of acute viral infection to track changes in histone modifications in an antigen-specific population of cells undergoing effector and memory differentiation. The authors demonstrated that the coordinate regulation of genes that contribute to a common function of the cell was coupled to distinct histone modification applied broadly to the genes coordinately expressed at different stages of differentiation [34]. Since the analysis of epigenetic programs in the mouse antigen-specific CD8 T cells was performed on the total pool of effector and memory cells, it remains to be determined whether the epigenetic programs identified in the human memory CD8 T cell subsets are present in the corresponding mouse memory CD8 T cell subsets.
Stability of acquired transcriptional programs is often reinforced by changes in DNA methylation programming. This concept was recently applied to the acquired effector functions in CD8 T cells in a study that analyzed genome-wide changes in CpG DNA methylation in antigen-specific CD8 T cells at the naïve and effector stage of differentiation [35]. Using the lymphocytic choriomeningitis virus (LCMV) model system of acute viral infection in mice, Scharer and colleagues [35] measured the DNA methylation programs in LCMV-specific naïve and effector CD8 T cells. Data from this study demonstrated that effector genes (such as those that encode granzymes B and K) go from a methylated state in naïve cells to a demethylated state in effector CD8 T cells. Notably, it was observed that differentially methylated regions between naïve and effector CD8 T cells were enriched for binding sites of transcription factors that regulate the naïve versus effector cell state [35]. Together, these studies demonstrate that naïve, effector, and memory CD8 T cells each have distinct epigenetic programs that are coupled to the transcriptional and functional capacities of the cells. In resting memory cells, active/permissive epigenetic marks likely facilitate the ability of the cells to rapidly recall effector gene re-expression, whereas repressive epigenetic marks play a role in restricting re-expression of genes that are irrelevant or detrimental (or both) to the function of the cell.
Collectively, the genome-wide profiling studies of histone and DNA epigenetic programs are providing new insights into the fate commitment and recall potential of memory cells. Considering these studies, one might predict that Tcm cells, having the highest degree of cell plasticity and lowest degree of committed effector functions, may retain the highest degree of poised epigenetic programs at genes related to fate-commitment and lymphoid tissue retention. Additionally, these cells may retain an ability to undergo further modification of the epigenetic programs during antigen-driven proliferation or homeostasis (Figure 1) (the following logic may apply to the newly defined Tscm subset of cells). In contrast, one might predict that Trm cells may retain not only poised/active epigenetic programs at genes required for effector function and trafficking/retention to peripheral tissues but also repressive epigenetic programs that restrict genes related to proliferation and developmental pluripotency.
Several studies have provided convincing evidence that transcriptional plasticity of memory cells is inversely correlated with the strength and duration of antigen exposure [29,36]. This raises the possibility that memory cells also possess an ability to undergo epigenetic reprogramming, and this may also be dependent upon their stimulation history (Figure 1). Recently, we explored the plasticity of the DNA methylation in functional memory and exhausted CD8 T cells by using mouse and human model systems of acute and chronic viral infection. Tracking changes in DNA methylation at the PD-1 locus in CD8 T cells undergoing effector and memory differentiation during acute viral infection of both mice and humans, we observed that antigen-specific effector CD8 T cells transiently demethylate the PD-1 promoter consistent with upregulation of PD-1 gene expression. Upon viral clearance, the antigen-specific CD8 T cells progressively reacquired a methylated promoter as the cells further differentiated into functional memory cells (Figure 2) [37]. Similar to effector cells from acutely infected mice and humans, the PD-1 promoter was demethylated in antigen-specific CD8 T cells at the early stages of chronic LCMV infection. As expected, the locus remained demethylated in LCMV-specific CD8 T cells as they became functionally exhausted during the sustained stimulation at the later stage of the infection. Importantly, though, the PD-1 locus remained completely demethylated in virus-specific CD8 T cells even after control of the chronic LCMV infection. Therefore, unlike functional memory CD8 T cells, which are able to reacquire the repressive methylation program, these data indicated that CD8 T cells with prolonged exposure to antigen were refractory to acquiring a methylation program at the PD-1 locus. To further examine the stability of the demethylation program at the PD-1 locus, we measured the PD-1 methylation program in HIV-specific CD8 T cells from individuals who naturally controlled HIV infection (elite controllers) or who had achieved viral control to undetectable levels via highly active antiretroviral therapy (HAART). Strikingly, HIV-specific CD8 T cells either from the elite controllers that had achieved viral control for more than a decade or from HAART-treated individuals retained a demethylated PD-1 locus (Figure 2) [37,38]. These data support the concept that prolonged exposure of CD8 T cells to antigen reinforces acquired epigenetic programs. These results have significant implications for therapeutic strategies that attempt to rest exhausted CD8 T cells to rejuvenate their effector potential, as they suggest that stable epigenetic programs may maintain an exhausted state even after long periods of rest. Furthermore, these data highlight a major unanswered question in the field: can the transcriptional program of exhausted T cells be stably reprogrammed during therapies that transiently block inhibitory receptor signaling? The answer to this question will provide much-needed mechanistic insight into whether rejuvenating therapies have the potential to induce long-lived immunity from a pool of exhausted T cells. As such, future studies are needed to test the stability of epigenetic modifications in the generation of functional and exhausted memory CD8 T cell subsets and the maintenance of their subset-specific functions during homeostasis and recall responses.
Figure 2. Reinforced DNA methylation programming during chronic infection.
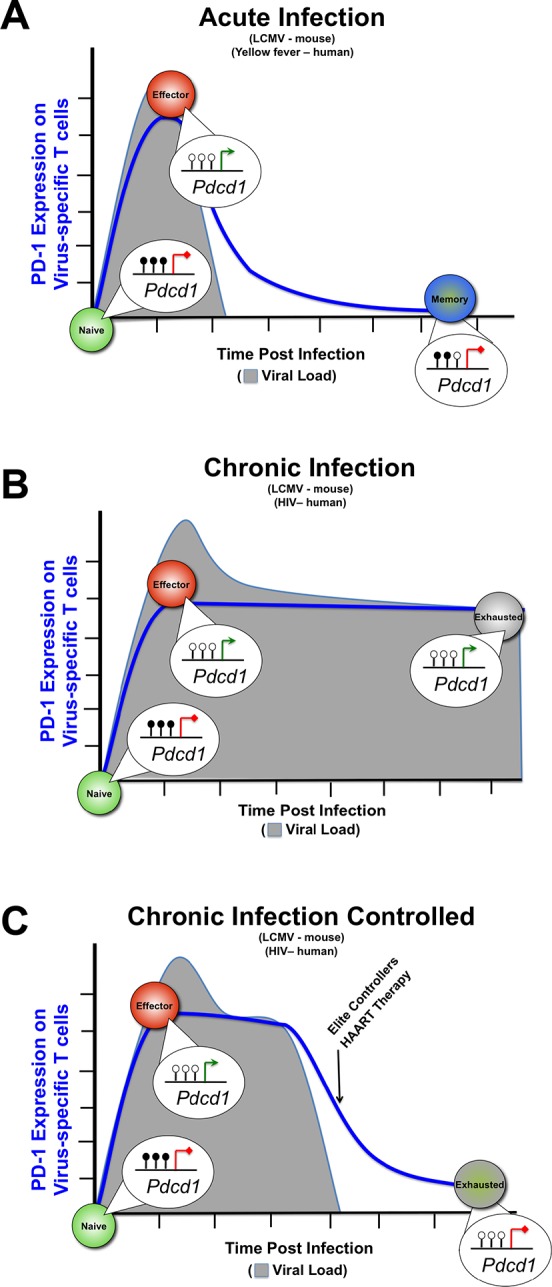
Naïve CD8 T cells have a fully methylated PD-1 (Pdcd1) promoter, but upon antigen exposure and differentiation into effector CD8 T cells, the PD-1 locus becomes demethylated. (A) During acute antigen exposure, antigen-specific CD8 T cells that survive to the memory stage of the immune response re-acquire a methylated PD-1 promoter. (B) During the persistent antigen exposure of a chronic infection, the antigen-specific CD8 T cells retain a demethylated PD-1 locus. (C) After control of the chronic pathogen, either by natural mechanisms such as those that occur during elite control of HIV infection or by therapeutic intervention, the antigen-specific CD8 T cells retain a demethylated locus. In the cartoon schematic, methylation at the PD-1 locus (pdcd1) is represented by filled lollipops, and demethylated locus is represented by an open lollipop.
Concluding remarks
Over the past two decades, studies investigating the stability of memory CD8 T cell phenotype and function during repetitive or continuous antigen exposure (or both) have demonstrated that prolonged TCR signaling (as well as accompanying inflammation) progressively erodes away at the functional T cell properties known to contribute to long-lived immunity. Efforts to dissect these mechanisms have demonstrated that many of the differences between functional and exhausted memory T cells occur at the transcriptional level and have provided key insights into the mechanisms that maintain the functional and exhausted state. These primary studies have played a significant role in the development of novel therapies to treat chronic infections and cancer by identifying mechanisms for reactivating effector functions from cells once thought to be fully committed to the non-functional state of T cell exhaustion. These findings also highlight the importance of research efforts focused on the fundamentals of CD8 T cell memory, as they have resulted in tangible advances in human health.
Our recent appreciation that protective immunological T cell memory is the result of the collective efforts of several distinct memory subsets has now focused our attention on investigating the mechanisms for maintenance of acquired subset-specific functions. Broadly, epigenetic regulation has emerged as a mechanism for a cell to retain tissue- and gene-specific transcriptional regulation and thereby maintain cellular specialization. Strong evidence now supports the concept that changes in epigenetic programming in memory CD8 T cells are coupled with the phenotype and function of memory CD8 T cells. It remains to be determined whether these acquired epigenetic programs can be further modified during memory cell homeostatic self-renewal or following a recall response. Additionally, recent exciting and provocative studies tracking the lineage and memory potential of single CD8 T cells have raised questions regarding the stochastic versus fixed fate of cells during their development into memory-subsets and has focused our attention on changes in gene regulation at a single-cell level [39–42]. Future studies focused on the stability of acquired gene expression programs of memory subsets will likely shed light on the questions surrounding fate commitment and the mechanism for long-lived maintenance of the memory pool. As the mechanisms that dictate stable versus pliable gene regulation become better defined, so comes the ability to manipulate “committed” memory cells, paving the way for therapies that use cellular reprogramming.
Acknowledgments
This work was supported by National Institutes of Health (NIH) funding (R01 AI30048, P01 AI056299, and HHSN266200700006C) to Rafi Ahmed, NIH funding (F32 AI096709) to J. Scott Hale, and NIH funding (R01 AI114442) to Ben Youngblood.
Abbreviations
- ChIP-seq
ChIP-sequencing
- HAART
highly active antiretroviral therapy
- IL
interleukin
- LCMV
lymphocytic choriomeningitis virus
- PD-1
Pdcd1
- Tcm
central-memory T cell
- TCR
T cell receptor
- Tem
effector-memory T cell
- Trm
tissue resident memory T cell
- Tscm
stem cell-like memory T cell
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/7/38
Contributor Information
Ben Youngblood, Email: benjamin.youngblood@stjude.org.
Rafi Ahmed, Email: rahmed@emory.edu.
References
- 1.Lau LL, Jamieson BD, Somasundaram T, Ahmed R. Cytotoxic T-cell memory without antigen. Nature. 1994;369:648–52. doi: 10.1038/369648a0. [DOI] [PubMed] [Google Scholar]
- 2.Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, Ahmed R. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science (New York, NY) 1999;286:1377–81. doi: 10.1126/science.286.5443.1377. [DOI] [PubMed] [Google Scholar]
- 3.Polic B, Kunkel D, Scheffold A, Rajewsky K. How alpha beta T cells deal with induced TCR alpha ablation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8744–9. doi: 10.1073/pnas.141218898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim Y, Veloso de Santana, Marlon G, Rakasz E, Capuano S, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak M, Haase AT, Lifson JD, Allen TM, Watkins DI. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129–33. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717962625
- 5.Ha S, Mueller SN, Wherry EJ, Barber DL, Aubert RD, Sharpe AH, Freeman GJ, Ahmed R. Enhancing therapeutic vaccination by blocking PD-1-mediated inhibitory signals during chronic infection. The Journal of experimental medicine. 2008;205:543–55. doi: 10.1084/jem.20071949. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1104204
- 6.Zhang Z, Jin B, Zhang J, Xu B, Wang H, Shi M, Wherry EJ, Lau George KK, Wang F. Dynamic decrease in PD-1 expression correlates with HBV-specific memory CD8 T-cell development in acute self-limited hepatitis B patients. Journal of hepatology. 2009;50:1163–73. doi: 10.1016/j.jhep.2009.01.026. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/722867436
- 7.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–63. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldrath AW, Sivakumar PV, Glaccum M, Kennedy MK, Bevan MJ, Benoist C, Mathis D, Butz EA. Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. The Journal of experimental medicine. 2002;195:1515–22. doi: 10.1084/jem.20020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–9. doi: 10.1016/S1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 10.Jameson SC. Maintaining the norm: T-cell homeostasis. Nature reviews. Immunology. 2002;2:547–56. doi: 10.1038/nri853. [DOI] [PubMed] [Google Scholar]
- 11.Lefrançois L, Masopust D. T cell immunity in lymphoid and non-lymphoid tissues. Current opinion in immunology. 2002;14:503–8. doi: 10.1016/S0952-7915(02)00360-6. [DOI] [PubMed] [Google Scholar]
- 12.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718581300
- 13.Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science (New York, NY) 2001;291:2413–7. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 14.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, Wang E, Douek DC, Price DA, June CH, Marincola FM, Roederer M, Restifo NP. A human memory T cell subset with stem cell-like properties. Nature medicine. 2011;17:1290–7. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13306998
- 15.Klonowski KD, Williams KJ, Marzo AL, Blair DA, Lingenheld EG, Lefrançois L. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20:551–62. doi: 10.1016/S1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1019156
- 16.Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, Carbone FR, Mueller SN. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 2011;477:216–9. doi: 10.1038/nature10339. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/13183958
- 17.Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, Vezys V, Masopust D. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. Journal of immunology (Baltimore, Md.: 1950) 2012;188:4866–75. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/715197800
- 18.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. Journal of immunology (Baltimore, Md.: 1950) 2006;176:2079–83. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/9220
- 19.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annual review of immunology. 2013;31:137–61. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 20.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–51. doi: 10.1016/S0092-8674(02)01139-X. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1011444
- 21.Wherry EJ, Ha S, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–84. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan Margaret FC. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. Journal of immunology (Baltimore, Md.: 1950) 2005;175:5895–903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 23.Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian Ulrich H, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nature immunology. 2003;4:225–34. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1012300
- 24.Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrançois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nature immunology. 2005;6:793–9. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1027008
- 25.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593–7. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 26.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/720541022
- 27.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–95. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718106509
- 28.Masopust D, Ha S, Vezys V, Ahmed R. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. Journal of immunology (Baltimore, Md.: 1950) 2006;177:831–9. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- 29.Wirth TC, Xue H, Rai D, Sabel JT, Bair T, Harty JT, Badovinac VP. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010;33:128–40. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/7192959
- 30.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17872–9. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/6781956
- 31.Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. Journal of immunology (Baltimore, Md.: 1950) 2012;189:3462–71. doi: 10.4049/jimmunol.1201305. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/719452533
- 32.Skon CN, Lee J, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nature immunology. 2013;14:1285–93. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718155698
- 33.Araki Y, Wang Z, Zang C, Wood WH, Schones D, Cui K, Roh T, Lhotsky B, Wersto RP, Peng W, Becker KG, Zhao K, Weng N. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–25. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/720541047
- 34.Russ BE, Olshanksy M, Smallwood HS, Li J, Denton AE, Prier JE, Stock AT, Croom HA, Cullen JG, Nguyen Michelle LT, Rowe S, Olson MR, Finkelstein DB, Kelso A, Thomas PG, Speed TP, Rao S, Turner SJ. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41:853–65. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/725308442
- 35.Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. Journal of immunology (Baltimore, Md.: 1950) 2013;191:3419–29. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. Journal of virology. 2012;86:8161–70. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718413490
- 37.Youngblood B, Oestreich KJ, Ha S, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, Boss JM, Ahmed R. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400–12. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13356030
- 38.Youngblood B, Noto A, Porichis F, Akondy RS, Ndhlovu ZM, Austin JW, Bordi R, Procopio FA, Miura T, Allen TM, Sidney J, Sette A, Walker BD, Ahmed R, Boss JM, Sékaly R, Kaufmann DE. Cutting edge: Prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. Journal of immunology (Baltimore, Md.: 1950) 2013;191:540–4. doi: 10.4049/jimmunol.1203161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tubo NJ, Pagán AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–96. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718009707
- 40.Plumlee CR, Sheridan BS, Cicek BB, Lefrançois L. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity. 2013;39:347–56. doi: 10.1016/j.immuni.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718071429
- 41.Gerlach C, Rohr JC, Perié L, van Rooij N, van Heijst Jeroen WJ, Velds A, Urbanus J, Naik SH, Jacobs H, Beltman JB, de Boer Rob J, Schumacher Ton NM. Heterogeneous differentiation patterns of individual CD8+ T cells. Science (New York, NY) 2013;340:635–9. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/717989435
- 42.Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Gräf P, Verschoor A, Schiemann M, Höfer T, Busch DH. Disparate individual fates compose robust CD8+ T cell immunity. Science (New York, NY) 2013;340:630–5. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 43.Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Krüppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. Journal of immunology (Baltimore, Md.: 1950) 1999;163:3662–7. [PubMed] [Google Scholar]
- 44.Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nature immunology. 2002;3:558–63. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 45.Kelly J, Spolski R, Imada K, Bollenbacher J, Lee S, Leonard WJ. A role for Stat5 in CD8+ T cell homeostasis. Journal of immunology (Baltimore, Md.: 1950) 2003;170:210–7. doi: 10.4049/jimmunol.170.1.210. [DOI] [PubMed] [Google Scholar]
- 46.Burchill MA, Goetz CA, Prlic M, O'Neil JJ, Harmon IR, Bensinger SJ, Turka LA, Brennan P, Jameson SC, Farrar MA. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. Journal of immunology (Baltimore, Md.: 1950) 2003;171:5853–64. doi: 10.4049/jimmunol.171.11.5853. [DOI] [PubMed] [Google Scholar]
- 47.Manders PM, Hunter PJ, Telaranta AI, Carr JM, Marshall JL, Carrasco M, Murakami Y, Palmowski MJ, Cerundolo V, Kaech SM, Ahmed R, Fearon DT. BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7418–25. doi: 10.1073/pnas.0501585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature immunology. 2005;6:1236–44. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 49.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nature immunology. 2006;7:1317–25. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1050653
- 50.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nature immunology. 2011;12:1221–9. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13675956
- 51.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nature immunology. 2009;10:618–26. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X, Yu S, Zhao D, Harty JT, Badovinac VP, Xue H. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–40. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/6936956
- 53.Grenningloh R, Tai T, Frahm N, Hongo TC, Chicoine AT, Brander C, Kaufmann DE, Ho I. Ets-1 maintains IL-7 receptor expression in peripheral T cells. Journal of immunology (Baltimore, Md.: 1950) 2011;186:969–76. doi: 10.4049/jimmunol.1002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, Milner JD. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–18. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13395965
- 55.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13409018
- 56.Sullivan JA, Kim EH, Plisch EH, Peng SL, Suresh M. FOXO3 regulates CD8 T cell memory by T cell-intrinsic mechanisms. PLoS pathogens. 2012;8:e1002533. doi: 10.1371/journal.ppat.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao RR, Li Q, Gubbels Bupp Melanie R, Shrikant PA. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. 2012;36:374–87. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, Brehm MA, Swain SL, Welsh RM, Berg LJ. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. Journal of immunology (Baltimore, Md.: 1950) 2014;192:5881–93. doi: 10.4049/jimmunol.1303187. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718391914