Abstract
Objective
To identify additional diabetes susceptibility markers in the MHC that could be responsible for the differential diabetogenicity of different HLA-DR3 CEHs.
Research Design and Methods
High resolution SNP genotyping of the MHC was performed in 15 T1D patients and 39 non-diabetic controls, homozygous for DR3-DQ2 and with one copy of the A*30-B*18-MICA*4-F1C30-DRB1*0301-DQB1*0201-DPB1*0202 HLA haplotype. Significantly associated SNPs were replicated in an independent sample of 554 T1D patients and 841 controls without HLA matching. Electrophoretic mobility shift assay was used to demonstrate a functional effect of an associated SNP.
Results
Seven SNPs showed evidence of association in the initial discovery experiment. Upon replication, only rs419434 (upstream HLA-DOA gene) remained significant. A functional variant (rs432375) in complete LD with rs419434 was shown to affect USF-1 binding and could be responsible for the association signal in the region.
Conclusions
We have identified a novel susceptibility locus within the MHC with a modest contribution to T1D (OR= 1.93; CI: 1.52-2-44; p=10−8) that is independent of HLA-DRB1 locus.
Keywords: Type 1 diabetes, Major Histocompatibility Complex, association study, conserved extended haplotype, HLA-DOA
Introduction
The largest contribution to the genetic predisposition to type 1 diabetes (T1D) maps to the HLA class II region on the MHC, and a strong association of DR3-DQ2 and DR4-DQ8 haplotypes has been very consistently replicated across all Caucasian population groups. It is also widely accepted that the MHC could harbor additional loci that contribute to genetic susceptibility to the disease (1–3). However, the high degree of linkage disequilibrium (LD) within this region has made it difficult to demonstrate that such association signals are truly independent from HLA-DR/DQ risk. In fact, long genomic extensions termed conserved extended haplotypes (CEH) or ancestral haplotypes that have been maintained unaltered during human evolution are characteristic of this genomic region (4). A particular HLA-DR3 CEH (A*30-B*18-MICA*4-F1C30-DRB1*0301-DQB1*0201-DPB1*0202, or B18-DR3) that is relatively common in southern European populations, has been shown to be conserved across a 5 Mb region and to be more diabetogenic than other HLA-DR3 chromosomes (5,6). These findings support the hypothesis that B18-DR3 chromosomes carry additional risk alleles in other susceptibility loci, which are fixed in all B18-DR3 CEHs, but may not be always present in other less-predisposing DR3 chromosomes. Under this hypothesis and in order to search for possible additional T1D susceptibility loci, we have performed high resolution SNP genotyping of the MHC region, comparing HLA-DR3 homozygous T1D patients and non-diabetic individuals carrying one copy of the B18-DR3 CEH, so we can investigate putatively diabetogenic variation at the other HLA-DR3 chromosome. We report evidence for association of a marker close to HLA-DOA gene located at 33.08 Mb that seems to be independent from HLA-DR/DQ, and propose a functional polymorphim at a TFBS as the possible etiological variation underlying this association peak.
Results and discussion
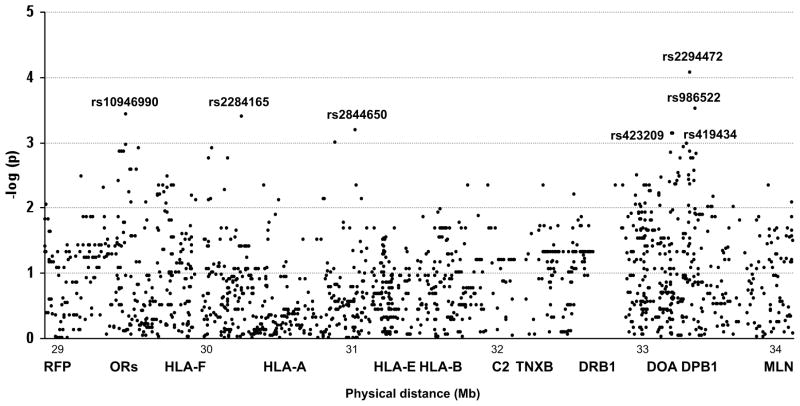
Although during the last years a great amount of effort has been put to the search of additional T1D susceptibility loci in the MHC, results so far have been somewhat controversial (7–10). The discrepancies found among studies could reflect the existence of multiple and unevenly distributed disease predisposing variants, and could also be partly explained by the extreme complexity of the region. The particularly strong LD in the MHC is helpful for the indirect detection of disease associations without necessarily testing the etiological polymorphism, but at the same time, makes it difficult to eliminate secondary signals and to localize independent variants (11). Understanding of the LD structure throughout the MHC, together with the development of new technologies, will be crucial for mapping additional T1D genes in the MHC region. In the present study we have taken advantage of the conservation and diabetogenicity of the B18-DR3 CEH, to identify disease-associated variability while controlling for LD with classical HLA risk variants. We have performed a high resolution SNP genotyping, in which 2,360 SNPs are genotyped along a region of nearly 5 Mb. After strict quality filtering, 2020 SNPs were available for analysis, and 266 SNPs were subsequently discarded because they failed missingness test (Maximum per SNP missing>0.1) or allele frequency test (MAF<0.01). Single-marker case-control association analysis of 1,755 markers showed a significant signal for seven SNPs, (arbitrary p-value threshold 10−4) (figure 1) (Online Appendix, Table 1) and genotypes were confirmed using Taqman assays. Three of the associated SNPs were located telomeric to HLA-B locus at 31.42 Mb, in a region spanning 1.5 Mb. The most telomeric SNP, rs10946990 (29,504,625), is located upstream of OR11A1 gene at 29.50 Mb, outside the classical MHC. The other two SNPs in the region, rs2284165 (30,268,034) and rs2844650 (31,010,512) are located, respectively, in an intron of TRIM26 at 30.26 Mb and upstream of SFTPG at 31.00 Mb. There are several reports describing associations around this extended region including near HLA-A (30.01 Mb) or HLA-B (7) and also in the extended MHC region close to UBD (0.98 Mb) and MASL1 (0.87 Mb) genes (8). The remaining four associated SNPs in our study are located centromeric to the classical T1D loci (HLA-DR/DQ), in a region extending from 33,091,452 to 33,243,940, between HLA-DOA and COL11A2 (33.24 Mb). In this centromeric region, ITPR3 (33.69 Mb) was initially proposed as a susceptibility gene (10) although this association was later revoked due to LD with HLA-DRB1 risk alleles (7,13).
Figure 1.
In order to confirm the association of these seven SNPs with the disease, six of them were genotyped in a larger, independent sample set without HLA-matching. Since rs419434 and rs423209 are in total LD with an r2=1 (both in our population and in the Hapmap CEU data) only rs419434 was genotyped to represent both SNPs. rs419434 was the only SNP for which association with T1D was replicated [p=1×10−8; OR (95% CI)=1.93 (1.52–2.44)] (table 1). This SNP is upstream of HLA-DOA, an HLA class II alpha chain paralog expressed in B cells and involved in the regulation of HLA-DM-mediated peptide loading on MHC class II molecules. HLA-DOA is located approximately 0.5 Mb centromeric from HLA-DRB1, so two tag SNPs, rs2187668 and rs7454108, that are proxies for HLA-DR3/DQ2 and HLA-DR4/DQ8, respectively (14,15) were genotyped and LD between each of them and rs419434 was measured using Haploview (http://www.broad.mit.edu/mpg/haploview/) (16). No evidence for LD between rs419434 and HLA-DR3/DQ2 or with HLA-DR4/DQ8 was observed (r2<0.1), and haplotype-based association analyses of rs419434 risk allele after stratification for the effects of DR3 and DR4 also supported independent contribution of rs419434 to T1D (Table 2).
Table 1.
Results of the replication study of the most significantly associated SNPs, performed in an HLA-unmatched, independent sample set comprising 554 T1D patients and 841 healthy white blood donors of southern European origin, using pre-designed Taqman SNP Genotyping
| SNP | Coordinate | Alleles 1/2 | Allele 1 Frequency (%) | p value | OR (95% CI) | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| rs10946990* | 29504625 | C/T | 25 | 26 | ns | - |
| rs2284165* | 30268034 | G/C | 21 | 18 | ns | - |
| rs2844650* | 31010512 | A/G | 7 | 6 | ns | - |
| rs419434† | 33094727 | A/G | 18 | 10 | 1×10−8 | 1.93 (1.52–2.44) |
| rs2294472 | 33207188 | C/T | 45 | 47 | ns | - |
| rs986522* | 33243940 | C/G | 54 | 51 | ns | - |
Assays (Applied Biosystems, Foster City, CA). Allele frequencies were compared in 2 × 2 contingency tables using Fisher’s exact test with EPI-INFO v.6.0 (Center for Disease Control, Atlanta, GA).
Genotyping was interrupted after analyzing 190 patients vs. 190 controls, because allele frequency did not differ between the two groups.
In complete LD (r2=1) with rs423209.
Table 2.
Results of Mantel Haenszel stratified analysis of rs419434*A, after controlling for the presence of rs2187668*T and rs7454108*G (Proxies for HLA-DR3-DQ2 and DR4-DQ8, respectively) risk variants in two SNP haplotypes, which were constructed using the sliding-window function in PLINK.
|
rs2187668-rs419434 haplotypes
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype | T1D | CTRL | Haplotype | T1D | CTRL | χ2 | MH p-value | OR(95% CI) |
| TA | 104 | 55 | CA | 81 | 119 | 6.05 | 0.0139 | 1.35 (1.06–1.72) |
| TG | 216 | 166 | CG | 707 | 1342 | |||
|
rs7454108-rs419434 haplotypes
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype | T1D | CTRL | Haplotype | T1D | CTRL | χ2 | MH p-value | OR(95% CI) |
| GA | 22 | 15 | AA | 161 | 159 | 29.09 | 10−8 | 1.85 (1.48–2.37) |
| GG | 246 | 127 | AG | 678 | 1380 | |||
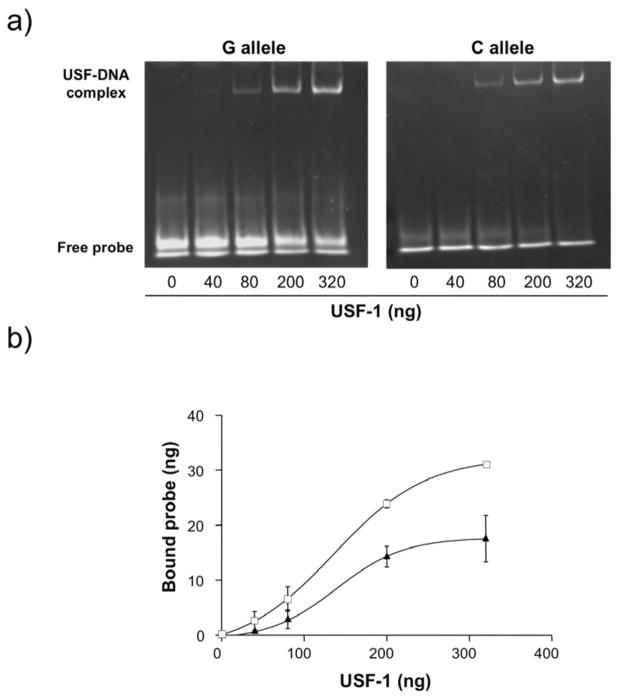
Although it is generally accepted that the degree of LD throughout the MHC is relatively constant until it drops centromeric to HLA-DPB1, there is evidence demonstrating that it fluctuates greatly among MHC subregions and is particularly lower in the classical HLA class II and extended class II regions, where several recombination hotspots have been identified (17,18). In particular, HLA-DOA is flanked by two such recombination hotspots, close to Bromodomain Containing 2 (BRD2) on the telomeric side and to HLA-DPA1 on the centromeric side (19,20) (Online Appendix, figure 1). In an attempt to locate a functional variant that could explain this association signal, we searched within the region of 59 kb between these two recombination hotspots for SNPs in total correlation with rs419434 (r2=1) that were predicted in silico to be regulatory SNPs. Out of the four SNPs that were perfect proxies for rs419434 according to the HapMap CEU data (http://www.hapmap.org) rs432375, that had not been genotyped in our discovery data set, was predicted to disrupt a binding site for Upstream Stimulatory Factor 1 (USF-1) with a match-score >98.0, based on FastSNP (http://fastsnp.ibms.sinica.edu.tw) and MatInspector v.7.7.3 demonstration version (http://www.genomatix.de/products/MatInspector/MatInspector2.html) results. To confirm this theoretical effect, we performed a functional analysis using an Electrophoretic Mobility Shift Assay (EMSA) and observed that the minor allele (allele C) provoked a moderate decrease in the binding capacity of USF-1, that was more pronounced at lower protein concentrations (figure 2). This alteration of binding affinity could possibly affect the level, timing and localization of the gene product, which could in turn have an effect in disease pathogenesis. For instance, SNPs in intronic or intergenic regulatory sequences that are binding sites for transcription factor RUNX1 have been associated with different autoimmune diseases, including systemic lupus erythematosus (21) and rheumatoid arthritis (22). We subsequently genotyped rs432375 in our sample set using a custom Taqman Allelic Discrimination Assay (Applied Biosystems), designed using Primer Express V3.0 software (Applied Biosystems). Due to the complexity of the region, allele-specific probes were designed with a degenerate base (Online Appendix, Supplementary methods). Our results revealed that this SNP was not in total LD with rs419434 (r2=0.92) in our population, but was still significantly associated with the disease [p=3×10−7; OR (95% CI) =1.78 (1.42–2.25)]. The presence of another SNP (rs11935463) only two bases away from rs432375 complicated the genotyping assay design, making it necessary to include four oligonucleotide probes in the assay mix, and might influence the performance of the Taqman assay. Although samples were genotyped in duplicate and only concordant replicates were included in the association analysis, results are less robust than those obtained for rs419434, and the few inconsistencies observed between the two associated SNPs could be due in part to genotyping errors unsolvable with this methodology.
Figure 2.
In the present study we present the association with T1D of a marker close to HLA-DOA that seems to be independent from HLA class II-conferred risk. Further studies in other populations will clarify whether this polymorphism is associated with T1D in different populations or if it is restricted to southern European populations. We also propose a functional variant that provokes a subtle alteration in the binding of transcription factor USF-1 as the underlying etiological culprit, but again, further analyses are necessary to confirm whether this disruption has an effect in gene expression in vivo, of HLA-DOA (cis effect) or other genes (trans effect). An attractive hypothesis would link a defect in HLA-DOA regulatory elements with an enhanced cellular immune response, based on data suggesting that overexpression of HLA-DOA correlates with a reduction in antigen presentation to T cells (23). This exciting idea needs experimental confirmation before it can be accepted. Finally, we have successfully employed a CEH-matching strategy to control for the major effects of HLA loci and to detect modest contribution to T1D risk. Our growing knowledge of the MHC structure will make similar approaches with other CEH possible, in order to investigate whether other minor T1D-susceptibility loci are present in the MHC.
Supplementary Material
Acknowledgments
This work was partially funded by CIBER de Diabetes y Enfermedades metabólicas (CIBERDEM) and Research Project grants 05/2291 from Instituto de Salud Carlos III (LC) and 2005/111039 from the Basque Department of Health (AMA). IS and AC-R are predoctoral fellows supported by grants from the University of the Basque Country and the Spanish Ministry of Education, respectively. JRB is co-funded by the I3SNS Program of the Spanish Ministry of Health (CES05/036).
References
- 1.Johansson S, Lie BA, Todd JA, Pociot F, Nerup J, Cambon-Thomsen A, et al. Evidence of at least two type 1 diabetes susceptibility genes distinct from HLA-DQB1, -DQA1 and -DRB1. Genes Immun. 2003;4:46–53. doi: 10.1038/sj.gene.6363917. [DOI] [PubMed] [Google Scholar]
- 2.Nejentsev S, Gombos Z, Laine AP, Veijola R, Knip M, Simell O, et al. Non-class II HLA gene associated with type 1 diabetes maps to the 240-kb region near HLA-B. Diabetes. 2000;49:2217–2221. doi: 10.2337/diabetes.49.12.2217. [DOI] [PubMed] [Google Scholar]
- 3.Koeleman BV, De Groot KN, Van Der Slik AR, Roep BO, Giphart MJ. Association between D6S2223 and type 1 diabetes independent of HLA class II in Dutch families. Diabetologia. 2002;45:598–599. doi: 10.1007/s00125-001-0725-1. [DOI] [PubMed] [Google Scholar]
- 4.Yunis EJ, Larsen CE, Fernandez-Viña M, Awdeh ZL, Romero T, Hansen JA, et al. Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue antigens. 2003;62:1–20. doi: 10.1034/j.1399-0039.2003.00098.x. [DOI] [PubMed] [Google Scholar]
- 5.Cambon-de Mouzon A, Ohayon E, Hauptmann G, Sevin A, Abbal M, Sommer E, et al. HLA-A, B, C, DR antigens, Bf, C4 and glyoxalase I (GLO) polymorphisms in French Basques with insulin-dependent diabetes mellitus (IDDM) Tissue Antigens. 1982;19:366–79. doi: 10.1111/j.1399-0039.1982.tb01463.x. [DOI] [PubMed] [Google Scholar]
- 6.Bilbao JR, Calvo B, Aransay AM, Martin-Pagola A, Perez de Nanclares G, Aly TA, et al. Conserved extended haplotypes discriminate HLA-DR3-homozygous Basque patients with type 1 diabetes mellitus and celiac disease. Genes Immun. 2006;7:550–554. doi: 10.1038/sj.gene.6364328. [DOI] [PubMed] [Google Scholar]
- 7.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;45:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aly TA, Baschal EE, Jahromi MM, Fernando MS, Babu SR, Fingerlin TE, et al. Analysis of single nucleotide polymorphisms identifies major type 1A diabetes locus telomeric of the major histocompatibility complex. Diabetes. 2008;57:770–776. doi: 10.2337/db07-0900. [DOI] [PubMed] [Google Scholar]
- 9.Husain Z, Kelly MA, Eisenbarth GS, Pugliese A, Awdeh ZL, Larsen CE, et al. The MHC type 1 diabetes susceptibility gene is centromeric to HLA-DQB1. J Autoimmun. 2008;30:266–272. doi: 10.1016/j.jaut.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, et al. Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. Am J Hum Genet. 2006;79:614–627. doi: 10.1086/507876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lie BA, Thorsby E. Several genes in the extended human MHC contribute to predisposition to autoimmune diseases. Curr Opin Immunol. 2005;17:526–531. doi: 10.1016/j.coi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu HQ, Marchand L, Szymborski A, Grabs R, Polychronakos C. The association between type 1 diabetes and the ITPR3 gene polymorphism due to linkage disequilibrium with HLA class II. Genes Immun. 2008;9:264–266. doi: 10.1038/gene.2008.12. [DOI] [PubMed] [Google Scholar]
- 14.Monsuur AJ, de Bakker PIW, Zhernakova A, Pinto D, Verduijn W, Romanos J, et al. Effective detection of human leukocyte antigen risk alleles in celiac disease using tag single nucleotide polymorphisms. Plos One. 2008;3:1–6. doi: 10.1371/journal.pone.0002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker JM, Triolo TM, Aly TA, Baschal EE, Babu SR, Kretowski A, et al. Two single nucleotide polymorphisms identify the highest-risk diabetes HLA genotype: potential for rapid screening. Diabetes. 2008;57:3152–3155. doi: 10.2337/db08-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 19.Zavattari P, Deidda E, Whalen M, Lampis R, Mulargia A, Loddo M, et al. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet. 2000;9:2947–2957. doi: 10.1093/hmg/9.20.2947. [DOI] [PubMed] [Google Scholar]
- 20.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 21.Prokunina L, Castillejo-López C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32:666–669. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 22.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003;35:341–348. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]
- 23.Fallas JL, Tobin HM, Lou O, Guo D, Sant’Angelo DB, Denzin LK. Ectopic Expression of HLA-DO in Mouse Dendritic Cells Diminishes MHC Class II Antigen Presentation. J Immunol. 2004;173:1549–1560. doi: 10.4049/jimmunol.173.3.1549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.