Abstract
Ubiquitin (Ub) and the ubiquitin-like modifier interferon stimulated gene 15 (ISG15) participate in the host defense of viral infections. Viruses, including the Severe Acute Respiratory Syndrome human coronavirus (SARS hCoV), have co-opted Ub/ISG15-conjugation pathways for their own advantage or have evolved effector proteins to counter pro-inflammatory properties of Ub/ISG15-conjugated host proteins. Here, we compare substrate specificities of the papain-like protease (PLpro) from the recently emerged Middle Eastern Respiratory Syndrome (MERS) hCoV to the related protease from SARS, SARS PLpro. Through biochemical assays, we show that similar to SARS PLpro, MERS PLpro is both a deubiquitinating and a deISGylating enzyme. Further analysis of the intrinsic deubiquitinating enzyme (DUB) activity of these viral proteases revealed unique differences between the recognition and cleavage specificities of polyUb chains. First, MERS PLpro shows broad linkage specificity for the cleavage of polyUb chains, while SARS PLpro prefers to cleave Lys48-linked polyUb chains. Second, MERS PLpro cleaves polyUb chains in a “mono-distributive” manner (one Ub at a time), and SARS PLpro prefers to cleave K48-linked poly-Ub chains by sensing a di-Ub moiety as a minimal recognition element using a “di-distributive” cleavage mechanism. The di-distributive cleavage mechanism for SARS PLpro appears to be uncommon among USP-family DUBs, as related USP family members from humans do not display such a mechanism. We propose that these intrinsic enzymatic differences between SARS and MERS PLpro will help identify pro-inflammatory substrates of these viral DUBs and can guide in the design of therapeutics to combat infection by coronaviruses.
Keywords: MERS, SARS, PLpro, deubiquitinating enzymes, ubiquitin, ISG15
INTRODUCTION
The emergence of novel infectious agents of zoonotic origin that can cross species barriers and pose a significant risk to human health is a constant threat today [1]. Over a decade ago, a novel coronavirus caused severe upper respiratory infection in humans and was termed the Severe Acute Respiratory Syndrome human coronavirus (SARS hCoV) [2]. SARS caused a global pandemic and had mortality rate of ~10% [3] due to its high infectivity rate, but was eventually contained by health care professionals through early detection and patient isolation However, to date, there is no specific treatment available for SARS or any other type of coronavirus infection, either in the form of small molecule inhibitors or vaccines [2, 4]. Although the last known human-to-human SARS infection chain was broken in 2003, the Center for Disease Control and Prevention (CDC) has recently classified SARS as a select agent (posing a high risk to human health), owing to the high risk of re-emergence and the lack of available treatment options. Just as SARS began to fade from the public’s memory, a new human coronavirus emerged in 2012, termed Middle Eastern Respiratory Syndrome (MERS) hCoV, which to date has infected close to a thousand individuals with a ~30% mortality rate (http://www.cdc.gov/coronavirus/mers/index.html). MERS hCoV, like SARS hCoV, is a beta coronavirus of zoonotic origin, which is also related to coronaviruses found in bats [5], and recent evidence suggests that MERS hCoV likely utilized dromedary camels as intermediary hosts [6]. MERS hCoV, like SARS hCoV, causes severe respiratory infection, as well as acute renal failure in some cases [3, 7]. MERS is currently an ongoing pandemic, with the first US cases reported in May 2014.
Ubiquitin is a small protein that post-translationally modifies target substrates through a defined enzymatic cascade involving E1-E2-E3 enzymes [8–10]. Conversely, deubiquitinating enzymes (DUBs) cleave ubiquitin from substrates [11, 12]. Ubiquitin can be conjugated to itself via any of its seven lysines, as well as via its N-terminus, leading to different functional consequences for the modified substrate [10]. The most common function of ubiquitination is proteasomal degradation of the substrate, mediated by K48-linked poly-ubiquitin (poly-Ub) chains [13]. ISG15, a ubiquitin-like (Ubl) modifier composed of two ubiquitin-like folds in tandem, is conjugated to substrates by an orthogonal enzymatic cascade [14] and plays a role in tagging newly synthesized proteins during an antiviral response [15, 16]. Topologically ISG15 resembles the open conformation of a linear or a K63-linked di-Ub molecule [17, 18], while K48-linked chains are more dynamic and have context-dependent topologies, depending on their interacting proteins [19].
Both ubiquitin and ISG15 are involved in fighting viral infection, and several viruses have co-opted Ubl conjugation pathways for their own advantage or have evolved effector proteins to counter the pro-inflammatory properties of Ubl-conjugated host proteins [20]. For coronaviruses, one such mechanism relies on dampening the host immune response via the action of viral isopeptidases [21]. Thus, for SARS, a highly promising drug target is one of its two viral processing proteases, the SARS papain-like processing protease (PLpro) [22], a multi-function papain-like cysteine protease [23]. In addition to cleaving the viral polypeptide at defined positions, SARS PLpro can remove ubiquitin and ISG15 from cellular conjugates. These activities are believed to be important for its anti-inflammatory function, although the specific ubiquitin- and/or ISG15-conjugated substrates have yet to be identified. Recently, the equivalent processing protease from the MERS hCoV, termed MERS PLpro, has also been shown to have deubiquitinating and deISG15ylating activities when expressed in cells [24, 25].
Here, we present a biochemical characterization of MERS PLpro and a comparison of its isopeptidase activity with SARS PLpro. We show that similar to SARS PLpro, purified, recombinant MERS PLpro is a dual-function isopeptidase that can cleave ISG15 and ubiquitin conjugates. Using a positional-scanning substrate library screen, we show that MERS PLpro has broad cleavage site specificity in terms of amino acid sequence, a feature it shares with SARS PLpro. Furthermore, we demonstrate MERS PLpro to be a mono-distributive DUB that recognizes and cleaves one ubiquitin unit within a chain. We also demonstrate MERS PLpro to be a broad specificity DUB with respect to Ub-Ub linkages. In contrast to MERS PLpro, we found that the unit of recognition and cleavage for SARS PLpro is a K48-linked di-ubiquitin (di-UbK48) unit within a chain length of three or greater, making it a unique “di-distributive” DUB. This finding allows us to dispel the recent notion that SARS PLpro has a preference for cleaving ISG15 conjugates, as opposed to ubiquitin conjugates, as SARS PLpro possesses the capacity to efficiently cleave both poly-Ub chains and ISG15-conjugates at similar rates. Furthermore, we show that this unique di-distributive mechanism of SARS PLpro can result in the accumulation of di-UbK48 units as well as of mono-Ub conjugated substrates, which could have functional consequences during coronavirus infection. Our study reveals key mechanistic differences between two related coronavirus proteases that may be used in identifying their cellular substrates, and which may aid in inhibitor design strategies. Finally, our study reinforces the need to investigate the comparative advantage that deubiquitination versus deISGylation might confer to coronaviruses.
EXPERIMENTAL
Ubiquitin probe labeling assays
Unless otherwise indicated, DUBs (1 μM) were incubated with an excess of activity-based probes (Ub-PA, monoUb-VME or di-UbK48-VME, 5 μM) for the indicated times (1 min to 1 hr) at 37°C in 20 mM Tris, pH=8.0, 150 mM NaCl and 5 mM DTT. Reactions were terminated with loading sample buffer (4X LDS, Invitrogen), boiled for 5 min at 95°C and analyzed by SDS-PAGE and SYPRO-staining. Gels were imaged on BioRad Gel-Doc imagers.
Ubiquitin chain cleavage assays
Unless otherwise indicated, in a reaction volume of 10 μL, 0.5 μg ubiquitin chains (in 20 mM Tris, pH=8.0, 150 mM NaCl and 5 mM DTT) were cleaved with 2–1000 nM DUB at 37°C for the indicated times in a time-course assay or by 1/5-fold serial dilution of DUBs (starting at 1 μM) for 1 hr at 37°C. Reactions were terminated with loading sample buffer (4X LDS, Invitrogen), boiled for 5 min at 95°C and analyzed by SDS-PAGE and SYPRO-staining. Gels were imaged on BioRad Gel-Doc imagers, quantified using ImageJ, and cropped where indicated.
Ubiquitin- and ISG15-AMC kinetics
To determine apparent kcat/Km for SARS and MERS PLpro, ubiquitin-AMC (amino-methyl-coumarine) and ISG15-AMC were prepared as ½-fold serial dilutions (starting at 20 μM and 12 μM, respectively) in 20 mM Tris, pH=8.0, 150 mM NaCl and 5 mM DTT. SARS PLpro was used at 10 nM, MERS PLpro was used at 50 nM, and the final reaction volume was 20 μl. Substrates and DUBs were pre-incubated at 37°C for 5, min and cleavage of Ubl-AMCs was performed at 37°C using a Spectramax fluorescence plate reader running SoftMax Pro 5 software (Molecular Devices) operated in kinetic mode, in black, flat-bottom 384-well plates (Corning #3575), where free AMC fluorescence was monitored by excitation at 355 nm and emission at 460 nm. Initial linear cleavage rates (Vi) were fitted by the Michealis-Menten equation using Prism software (GraphPad) based on a free AMC standard curve. Where indicated, experiments were performed in duplicate; error bars indicate SEM.
Synthesis of the diverse tetrapeptide-ACC Positional Scanning Substrate Combinatorial Library (PS-SCL)
Synthesis of the PS-SCL for screening of P4, P3 and P2 sub-sites was carried out as described previously [26, 27]. After completing the synthesis and analysis steps, each sub-library was dissolved in biochemical grade DMSO at a concentration of 20 mM and stored at −80°C until use.
Fluorogenic substrate synthesis
The synthesis of individual fluorogenic substrates was carried out using classic solid phase peptide synthesis methods as described previously [28]. Each substrate was purified by HPLC on a Waters M600 solvent delivery module with a Waters M2489 detector system using a semi-preparative Waters Spherisorb S10ODS2 column. The solvent composition was as follows: phase A (water/0.1% TFA) and phase B (acetonitrile/0.1% TFA). The purity of each substrate was confirmed by analytical HPLC using a Waters Spherisorb S5ODS2 column. Finally, the molecular weight of each substrate was confirmed by mass spectrometry analysis. All compounds were at least 95% pure. Individual substrates were dissolved in 20 mM in DMSO and stored in −80°C until use.
Assay of the PS-SCL and individual tetra-peptides
Kinetic assays were performed at 37°C in buffer containing 150 mM NaCl, 6 mM DTT, 20 mM Tris, pH=7.5. Assay conditions for the P2, P3 and P4 libraries were as follows: the reaction volume was 100 μl; the total final substrate mixture concentration was 300 μM; MERS PLpro was assayed at 1 μM. Enzyme was pre-incubated for 30 min at 37°C in buffer, followed by addition into 96-well Corning® plate wells containing each substrate. The reaction was monitored for 60 min in kinetic mode. To calculate initial velocity (Vi), the linear portion of the obtained progress curves was used. Results are presented as an average of at least three experiments. Analysis of the results was based on total relative fluorescence units (RFU) for every sub-library, setting the highest value to 100% and adjusting the other results accordingly. Individual substrates were screened against MERS PLpro at 37°C in buffer containing 150 mM NaCl, 6 mM DTT, 20 mM Tris, pH=7.5 and 1 M of the Hofmeister salt sodium citrate. Enzyme was pre-incubated for 30 min at 37°C before being added to wells containing each substrate. Assay conditions were as follows: 100 μl reaction volume; final substrate concentrations ranged from 500 to 29.3 μM; and MERS PLpro concentration was 1 μM. The concentration of DMSO in the assay was less than 1% (v/v). Release of the ACC fluorophore was monitored continuously with excitation at 355 nm and emission at 460 nm. Initial velocity rates were fitted using GraphPad Prism 6 to the Michealis-Menten curve, determining Vmax and Km.
DUB assay in cell lysates
Lysates from human interferon beta (IFNβ) treated HeLa cells (10 μg total protein per reaction) were incubated with a 1/10-fold serial dilution of 10 μM DUB in 20 μl reaction volumes for 1 hr in the presence of 25 mM DTT or with 0.5 μM DUBs for the time course assay. Reactions were terminated by boiling in SDS loading buffer, and the results were analyzed by SDS-PAGE and western blotting. Blots were developed by chemiluminescence.
RESULTS
MERS PLpro is a broad-specificity isopeptidase
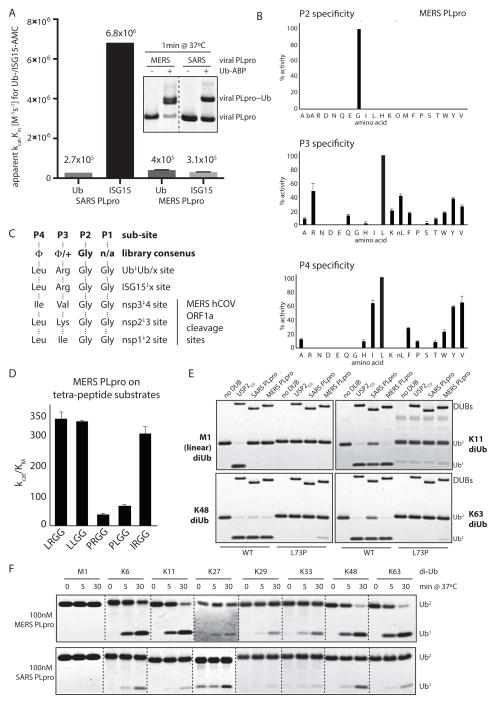
To characterize MERS PLpro, we cloned and expressed the putative MERS PLpro (including its ubiquitin-like (UBL) domain) using domain limits based on alignments with the related SARS PLpro (Supplementary Fig. 1A). First, we tested MERS PLpro by comparing its activity on the fluorogenic model substrates ubiquitin- and ISG15-AMC. Unlike SARS PLpro, which displays a strong preference for cleaving ISG15-AMC over ubiquitin-AMC (Fig. 1A and Supplementary Fig. 1B–C and refs [23, 29, 30]), MERS PLpro did not display a significant preference for either, but processed ISG15- and ubiquitin-AMC similarly when determining overall relative kcat/Km values (Fig. 1A). Additionally, MERS PLpro was more reactive than SARS PLpro towards a ubiquitin activity based probe, Ub-PA, which is used to covalently label active DUBs (Fig 1A, inlet). Overall suggesting divergent preferences of SARS and MERS PLpro towards Ub and ISG15 model substrates.
Figure 1. MERS PLpro is a broad-specificity isopeptidase.
(A) Relative apparent kcat/Km values for SARS and MERS PLpro for Ub- and ISG15-AMC hydrolysis, plotted as compared to SARS PLpro on Ub-AMC. Error bars represent SEM, calculated from Michealis-Menten kinetics as shown in Supplementary Figure 1 B–C). Inlet – SARS and MERS PLpro were labeled by the ubiquitin activity based probe (Ub-PA). (B) P2, P3 and P4 specificity of MERS PLpro. (C) Comparison of the PS-SCL library consensus specificities for P2-P4 with the native cleavage sites used by MERS PLpro in the viral polypeptide. Amino acids are listed as their three letter codes (Φ, hydrophobic; +, positively charged); □ indicates cleavage by MERS PLpro. (D) Comparison of kcat/Km values for MERS PLpro on individual tetra-peptide substrates, that were synthetized based on the PS-SCL results in Figure 1. (E) A panel of wild-type and DUB-resistant (UbL73P) di-ubiquitin chains (M1-, K11-, K48- and K63-linked, displayed in the top left, top right, bottom left and bottom right corners, respectively) assayed with SARS PLpro, MERS PLpro and USP2CD; showing how MERS PLpro can tolerate Pro in P4 in the context of full-length ubiquitin when it is part of a di-ubiquitin chain (F) MERS PLpro (at 100 nM) and SARS PLpro (at 100 nM) were assayed against a panel of di-ubiquitin chains of all 8 Ub-Ub linkages. Dotted lines indicate cropped images from different gels ran side-by-side (see also Supplementary Fig 1F).
Next, to determine the cleavage site specificity of MERS PLpro with regards to the primary amino acid sequence of the cleavage site (P4-P1 residues of the substrate, which correspond to Leu73-Arg74-Gly75-Gly76 in ubiquitin and Leu154-Arg155-Gly156-Gly157 in ISG15), we generated a positional scanning substrate combinatorial library (PS-SCL, Supplementary Fig. 1D) that has been previously used to assay the specificity of SARS PLpro and other DUBs [26]. The results of the PS-SCL screen demonstrated that MERS PLpro has an absolute requirement for Gly in the P2 position, but can accommodate a wide variety of hydrophobic amino acids in both the P3 and P4 positions (Fig. 1B). In the context of this tetra-peptide screen, the most preferred amino acid in P3 was Leu, followed by Arg, Lys and hydrophobic residues. The most preferred amino acid in P4 was also Leu, followed by mostly hydrophobic residues. The library screen results are in line with the three cleavage sites that MERS PLpro processes within the viral polypeptide (Fig. 1C), where the P1 and P2 sites are both Gly, the P3 residues are Ile, Lys or Val, and the P4 is either a Leu or an Ile [25]. Thus, these results indicate that the active site of MERS PLpro appears to be less stringent. When comparing PS-SCL library screen results of other DUBs [26] from the USP-family (which contains both SARS and MERS PLpro), it appears that MERS PLpro exhibits a lesser degree of specificity in P4 than either SARS PLpro or USPS. Its specificity in P3 is broad, yet more stringent than that of SARS PLpro (Supplementary Fig. 1E).
PS-SCL screen reveals relaxed cleavage site specificity for MERS PLpro, allowing cleavage of a DUB-resistant ubiquitin mutant
Notably, to date, no isopeptidase has been shown to accommodate Pro in P4 [27], and the positional library screen suggested that MERS PLpro may be able to cleave substrates containing a Pro in the P4 position (Fig. 1B, bottom panel). To confirm the cleavage of Pro in P4, we generated unique tetra-peptides with defined sequences and determined the rates of cleavage by MERS PLpro (Fig. 1D). The results showed that Pro can be accommodated in P4 for both tetra-peptides tested, although with reduced rates, and that there is little discrimination between Leu and Arg in the P3 position.
In a recent study, we generated a ubiquitin Leu73Pro (UbL73P) mutant that was resistant to cleavage by different families of human DUBs when conjugated to substrates or as a poly-Ub chains [31]. Intrigued by the ability of MERS PLpro to accommodate Pro in P4 (corresponding to UbL73P) we tested whether MERS PLpro can cleave UbL73P. Indeed, we show that in the context of di-ubiquitin (di-Ub) chains, MERS PLpro, but not SARS PLpro or USP2CD, could cleave UbL73P chains, although more weakly when compared to wild-type chains (Fig. 2E). The ability of MERS PLpro to cleave UbL73P was observed for K11-, K48- and K63-linked di-Ub chains, but not for M1-linked (linear) di-Ub chains, suggesting some level of linkage specificity displayed by MERS PLpro for processing ubiquitin chains. Thus, we determined the linkage specificity of MERS PLpro by assaying a panel of di-Ub molecules (all eight linkages [10]) in a time-course assay. Using different concentrations of MERS PLpro, we demonstrate that MERS PLpro is a broad-specificity DUB, displaying cleavage of K6-, K11-, K29-, K33-, K48- and K63-chains, but not linear di-Ub chains (Fig. 1F, top panel; and Supplementary Figure 1F). In contrast, cleavage of di-Ub chains by SARS PLpro – which displays broad linkage specificity in this assay – is not as robust as cleavage by MERS PLpro when comparing the two enzymes at similar enzyme concentrations (Fig. 1F, bottom panel). Neither DUB was able to cleave linear di-Ub or K27-linked di-Ub, although the latter could be structurally quite compact.
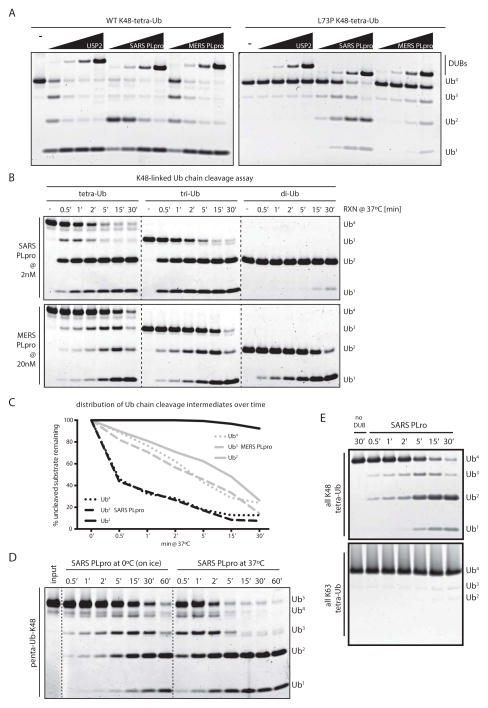
Figure 2. Enhanced activity of SARS PLpro on longer K48-linked ubiquitin chains.
(A) K48-linked wild-type and DUB-resistant (UbL73P) tetra-ubiquitin chains assayed with a 1/5-fold serial-dilution SARS PLpro, MERS PLpro and USP2CD (starting at 2μM DUB), showing how both MERS and SARS PLpro can tolerate Pro in P4 in the context of full-length ubiquitin when it is part of a tetra-ubiquitin chain. (B) K48-linked ubiquitin chains (tetra-, tri- and di-Ub) were cleaved in a time-course assay by SARS PLpro (at 2nM, top panels) and by MERS PLpro (at 20nM, bottom panels); highlighting the different cleavage patterns of the two DUBs. Dotted lines indicate cropped images from three gels, ran side by side. (C) Quantification of cleavage rates by SARS and MERS PLpro from (B). (D) K48-linked penta-ubiquitin chains were cleaved in a time-course assay by SARS PLpro (20nM) at 0°C (on ice) and 37°C, and visualized by SDS-PAGE and SYPRO-staining. Dotted lines are included for clarity. (E) Comparison of tetra-Ub cleavage by SARS PLpro of K48- versus K63-linked chains.
SARS PLpro recognizes a K48-linked tri-Ub as a minimal unit for ubiquitin chain processing
The low activity of SARS PLpro on di-Ub chains compelled us to test its activity on longer ubiquitin chains. Surprisingly, when analyzing K48-linked tetraUb-L73P chains, both MERS and SARS PLpro were able to cleave the mutant ubiquitin chains (Fig. 2A, right panel). The cleavage of UbL73P chains was impaired compared with wild-type chains, however the slowed kinetics allowed visualization of different cleavage intermediates for MERS and SARS PLpro (compare Fig. 2A, left and right panels). The tetra-Ub cleavage pattern for USP2CD and MERS PLpro appeared to resemble a distributive cleavage pattern, as opposed to the SARS PLpro cleavage pattern, which accumulated di-Ub as an intermediate form prior to the appearance of mono-Ub products, during cleavage of UbL73P, suggesting that SARS and MERS PLpro recognize ubiquitin chains in a different manner. Interestingly, a detailed look at the active site clefts of the crystal structures of USP2CD, SARS and MERS PLpro bound to mono-ubiquitin [32–34] suggests a lack of a defined pocket for P4 in MERS PLpro and SARS PLpro, while that pocket is present in USP2CD (Supplementary Fig. 1G), potentially explaining the ability of viral DUBs to accommodate Pro in P4. Thus overall, MERS PLpro is a dual-specificity isopeptidase that cleaves both ISG15- and various ubiquitin conjugates at similar rates. In contrast, SARS PLpro appears to be a poorer DUB on di-Ub substrates, but a superior deISGylase as compared with MERS PLpro. Importantly, the cleavage of wild-type tetra-Ub chains revealed that SARS PLpro can cleave tetra-Ub chains more efficiently than MERS PLpro, whereas the opposite is true for the cleavage of di-Ub chains. These differences suggest that MERS and SARS PLpro are mechanistically distinct in their ability to recognize ISG15 and ubiquitin conjugates.
To further investigate the unique property of SARS PLpro in stabilizing di-Ub cleavage products from a poly-Ub chain, we compared the processing rates for SARS and MERS PLpro on tetra-, tri- and di-Ub substrates (Fig. 2B–C). We show that SARS PLpro (Fig. 2B, top panels and Fig. 2C, black lines) exhibits negligible cleavage of K48-linked di-Ub chains but readily process tetra- and tri-Ub chains. The cleavage-intermediate patterns from the time-course study indicate that SARS PLpro cleaves a tri-Ub chain into a di-Ub and a mono-Ub product. The cleavage of a tetra-Ub chain by SARS PLpro is more complex, as it can either generate a tri-Ub and a mono-Ub product, which can then be further processed into a di-Ub and a mono-Ub, or the tetra-Ub can be cleaved to generate two di-Ub units. Thus, different modes of tetra-Ub cleavage will result in the accumulation of di-Ub (and mono-Ub) along with the transient appearance of the tri-Ub form (Fig. 2B). SARS PLpro therefore appears to preferentially recognize and cleave K48-linked chains in a “di-distributive” manner. This di-distributive cleavage mechanism is best demonstrated when analyzing cleavage of K48-linked penta-Ub chains at lower temperatures (Fig. 2D), allowing visualization of immediate cleavage intermediates. Here, it is visually striking that the initial cleavage products, arising at highly comparable rates from processing a penta-Ub chain, are tri- and di-Ub intermediates. This result strongly indicates that cleavage of a five-member ubiquitin chain can take place stochastically either between the 2nd and 3rd ubiquitin moiety, or between the 3rd and 4th ubiquitin moiety (from either end of the chain), both of which will result in the transient accumulation of tri- and di-Ub cleavage intermediates. The lack of transient accumulation of a single cleavage intermediate, combined with the observation of mono- and tetra-Ub cleavage intermediates suggests a lack of directionality in ubiquitin chain cleavage, strongly suggesting that SARS PLpro is a distributive DUB that processes ubiquitin chains in units of two. Consequently, further cleavage of a di-Ub molecule into monomers appears to be inhibited.
Unlike SARS PLpro, cleavage rates with MERS PLpro (Fig. 2B, bottom panels and Fig. 2C, grey lines) were nearly identical against ubiquitin chains of various lengths, and the patterns of cleavage intermediates demonstrate that MERS PLpro is a (mono)-distributive DUB that cleaves single units of ubiquitin (like the majority of DUBs studied thus far). The di-Ub stabilizing cleavage pattern of SARS PLpro appears to be K48-specific, as no di-Ub intermediates accumulate during cleavage of K63-linked tetra-Ub chains (Fig. 2E). Additionally, K63-linked chains are cleaved poorly by SARS PLpro compared with MERS PLpro (Supplementary Fig. 2A). Both SARS and MERS PLpro failed to cleave linear (M1-linked) tetra-Ub chains, even at equimolar DUB:substrate concentrations (Supplementary Fig. 2B). These results collectively support the idea that MERS PLpro is a broad-specificity mono-distributive DUB, while SARS PLpro is di-distributive and is significantly more active on longer K48-linked ubiquitin chains.
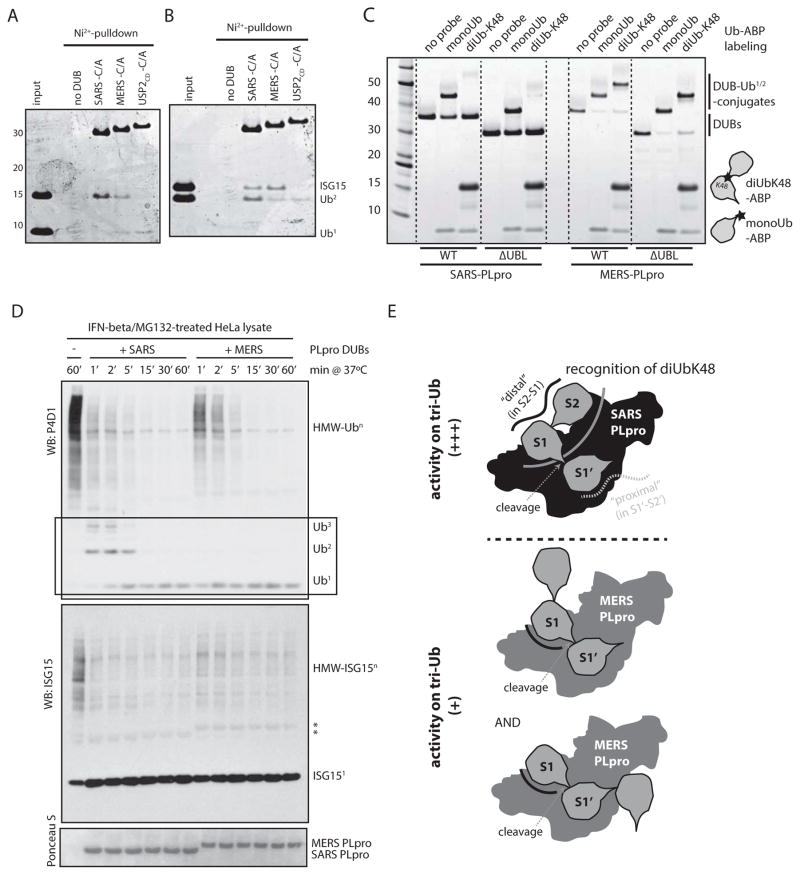
The comparatively poor cleavage activity of SARS PLpro on di-Ub chains could be due the inability of SARS PLpro to recognize and bind di-Ub chains. To test this, we generated recombinant His-tagged catalytic mutant DUBs and used them for pull-down assays by incubating them with different mixtures of Ubl units (Fig. 3A–B). We show that SARS PLpro preferentially binds to K48-linked di-Ub chains over mono-Ub when both were present at equimolar ratio. In contrast, USP2CD preferentially favored mono-Ub over di-Ub, and MERS PLpro bound to both similarly (Fig. 3A). Interestingly, SARS PLpro slightly favored K48-linked di-Ub chains over ISG15 when both were present, whereas the opposite was true for MERS PLpro (Fig. 3B). Thus, it is not the inability of SARS PLpro to recognize di-Ub in order to cleave it, but perhaps the binding of di-Ub by SARS PLpro is mediated by a region that renders the Ub-Ub internal cleavage site inaccessible for cleavage by the active site of the protease.
Figure 3. SARS PLpro is a di-UbK48-binding, tri-UbK48-cleaving DUB.
(A) and (B) Catalytic Cys/Ala mutants of SARS PLpro, MERS PLpro and USP2CD were analyzed for their ability to bind mono- versus K48-di-Ub (A) or ISG15 versus K48-di-Ub (B). (C) Ubiquitin-activity-based probe (ABP) labeling of SARS and MERS PLpro and their ΔUBL mutants. The position of warhead (the reactive group for covalently labeling the active site Cys of the DUB) within the Ub-ABP is indicated by a star. Note the absence of labeling for SARS PLpro by the di-UbK48-ABP. Dotted lines are included for clarity. (D) Time-course cleavage assay of IFNβ-/MG132-treated cell lysates by SARS and MERS PLpro. Box is shown to highlight the different initial cleavage products released from poly-Ub chains by the two viral DUBs. (E) Schematic representation of the model highlighting the different mechanism of cleavage for ubiquitin chains by SARS (in black) and MERS (in grey) PLpro, showing the presence of a unique di-Ub-recognizing surface within SARS PLpro (dark grey curve), which is lost from MERS PLpro. S2, S1 and S1′ indicate the sub-sites occupied by Ub chains when bound across the active site: S2 is the distal-distal Ub in a tri-Ub; S1 is the distal Ub (with its C-terminus in the active site); and S1′ is the proximal Ub (the one cleaved off).
Recent advances in chemical biology have enabled the generation of ubiquitin activity-based probes (ABPs) for DUBs that are linkage-specific, where the warhead used to covalently label the active site Cys of a DUB is positioned in between the two ubiquitin moieties in a di-Ub probe [35, 36]. Using this tool, we tested whether SARS could efficiently bind di-Ub across its active site in the S1-S1′ position (adapting the nomenclature of amino acid positions in protease cleavage sites [37] to positions occupied by the ubiquitin-like protein domains), in order to favor labeling with the ABP; or whether another position is utilized (such as S2-S1), which would prevent reactivity (see model in Fig. 3E). Also, as both SARS and MERS PLpro contain ubiquitin-like (UBL) domains in their N-terminus, we wondered if the UBL domain plays a role in the correct positioning of longer ubiquitin chains for SARS PLpro. Here, we show that while the K48-specific di-Ub ABP (di-UbK48-ABP), as well as a mono-Ub-ABP, efficiently labeled MERS PLpro and its ΔUBL mutant to form a covalently modified DUB (Fig. 3C, right side), the di-UbK48-ABP failed to efficiently label SARS PLpro or its ΔUBL mutant (Fig. 3C, left side). Thus, the lack of di-UbK48-ABP labeling for SARS PLpro is likely due to selective binding of the di-UbK48 chain at a position other than the S1-S1′ position. Therefore, the active site Cys cannot react with the warhead of the ABP. Consistent with Ub-ABP labeling, removal of the UBL-domains of either SARS or MERS PLpro had no effect on their K48-tetra-Ub cleavage rates or the pattern of intermediates observed (Supplementary Fig. 2C–D). Thus, the UBL domains of both MERS and SARS PLpro are dispensable for their ubiquitin chain recognition and processing activities.
To determine whether the different mechanisms for substrate recognition and processing between MERS and SARS PLpro remain observable in the context of multiple cellular substrates, we aimed to create an experimental system that would contain both ISGylated and K48-linked polyubiquitinated substrates where we could simultaneously monitor the appearance of cleavage intermediates of both ubiquitin-like proteins. To this end, we treated HeLa cells with interferon-beta (IFNβ) and the proteasome inhibitor, MG132, to induce stabilization of both ISGylated and K48-linked polyubiquitinated substrates in cells. Next, the lysates were incubated with recombinant viral DUBs in a time-course assay, and the substrate processing patterns were analyzed by western blotting by probing for total cellular ubiquitin and ISG15 conjugates (Fig. 3D). The results indicated that MERS and SARS PLpro are both potent deISGylating enzymes, while SARS PLpro appeared to process higher molecular weight (HMW) ubiquitin conjugates slightly more efficiently. Importantly, in the context of a mixture of cellular substrates, SARS PLpro initially accumulated di-Ub chains as a cleavage product, while MERS PLpro did not (Fig. 3D, black box). Assaying IFNβ-/MG132-treated cell lysates with a serial dilution of viral DUBs (Supplementary Fig. 2E–F) also indicated initial stabilization of di-Ub products for SARS PLpro, overall demonstrating the same mechanistic differences between SARS and MERS PLpro with respect to ubiquitin chain processing in the context of cellular substrates.
In summary, we have demonstrated that SARS PLpro preferentially recognizes di-UbK48 as a unit (possibly in the S2-S1 pockets) and efficiently cleaves the next ubiquitin in the chain (in S1′), resulting in accumulation of di-UbK48 products. These results strongly suggest that the minimal preferred substrate for SARS PLpro is a K48-tri-Ub chain (spanning S2-S1-S1′, see model in Fig. 3E). Importantly, the related MERS PLpro lacks this unique recognition and cleavage preference.
Orientation of ubiquitin chain cleavage by SARS PLpro
Recognition of a di-Ub moiety and cleavage of a third ubiquitin in a tri-Ub chain by SARS PLpro is presumed to be via the S2-S1/S1′ sub-sites (in a “distal” position, see model in Figure 3E, top, shown in black solid line), largely based on the presumed similar recognition of ISG15, where the two ubiquitin-like domains of ISG15 could only occupy those same S2-S1 sub-sites, restricted by the positioning of its C-terminal glycine that is cleaved. Alternatively, recognition of a tri-Ub chain may occur in an S1/S1′-S2′ binding mode (in a “proximal” position, see model in Figure 3E, top, shown in grey dotted line), where, recognition of a di-Ub would take place on the other side of the cleavage site. The results presented thus far cannot differentiate between the two modes of orientation within a tri-Ub chain, given the symmetry in the system and the identical size of the cleavage intermediates. Thus, we sought to determine the orientation of the Ub chain cleavage mechanism by SARS PLpro: to differentiate between “distal” Ub chain processing (S2-S1/S1′) versus a “proximal” (S1/S1′-S2′) mechanism, which would place recognition of ISG15 and the di-UbK48 unit on opposing sides of the SARS active site.
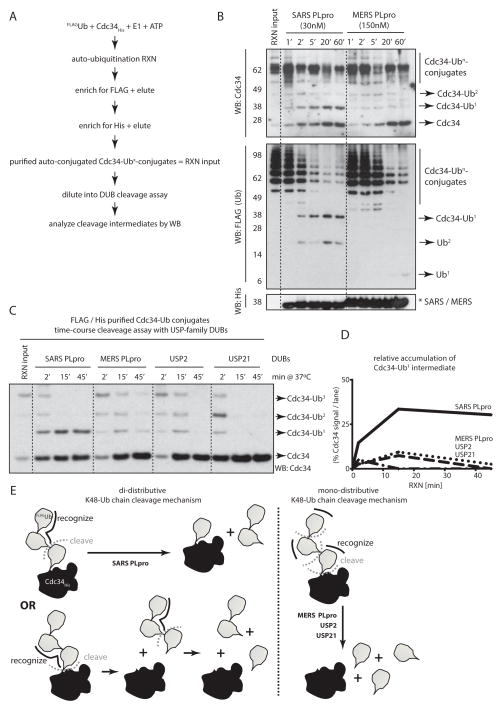
To determine the directionality in the cleavage mechanism, we generated a purified His-tagged substrate conjugated with FLAG-tagged ubiquitin (FLAG-Ub) chains (see Fig. 4A for schematics and Supplementary Fig. 3A). We chose Cdc34 as a model substrate, whose auto-conjugation results in K48-linked ubiquitin chains. When purified His-Cdc34 conjugated with FLAG-Ub chains was cleaved by SARS PLpro in a time-course assay, Cdc34-ubiquitin conjugates of various lengths, as well as free Cdc34, appeared as initial cleavage products (Fig. 4B, compare lanes 1 and 2). Ultimately, two unique product species were stabilized by SARS PLpro: mono-Ub-conjugated Cdc34 and free Cdc34 (Fig. 4B, lanes 5 and 6; and Supplementary Fig. 3B). Cleavage of the same poly-Ub-conjugated Cdc34 by MERS PLpro, on the other hand, did not result in the accumulation of mono-Ub-conjugated Cdc34. In the case of MERS PLpro, even though the initial cleavage intermediates were similar to those of SARS PLpro (various length Cdc34-ubquitin conjugates), the final product was free, fully deconjugated Cdc34 (Fig. 4B, right side). These results are consistent with previous experiments illustrating mechanistic differences between SARS and MERS PLpro, where SARS PLpro stabilized di-UbK48 cleavage products arising from unanchored ubiquitin chains, while MERS PLpro did not. Additionally, these results strongly support a mechanism where recognition of K48-linked ubiquitin chains by SARS PLpro in the context of a conjugated substrate take place via the “distal” approach (via the S2-S1/S1′ sub-sites, Supplementary Fig. 3C), and not by the “proximal” binding mode. In the latter case, the final accumulated cleavage product would have been a di-Ub-conjugated substrate (the two ubiquitin moieties in S1′-S2′, while the conjugated substrate would occupy the S3′ sub-site), which was not observed (Figure 4B and Supplementary Fig. 3B). Furthermore, the appearance of initial cleavage products of various lengths also supports a distributive cleavage mechanism, where there is no discrimination between the positions at which SARS or MERS PLpro cleaves a ubiquitin chain.
Figure 4. SARS PLpro is a unique “distal di-distributive” deubiquitinating enzyme.
(A) Schematics of the purification strategy of poly-Ub-conjugated Cdc34 in order to asses the directionality of cleavage of Ub chains. (B) Time-course cleavage of HisCdc34-poly-UbFLAG conjugates by SARS and MERS PLpro, revealing stabilization of mono-Ub-conjugated Cdc34 as a cleavage product by SARS PLpro and not by MERS PLpro. (C) Time-course cleavage of HisCdc34-poly-UbFLAG conjugates by USP-family DUBs. (D) Quantification of the relative abundance of the HisCdc34-mono-UbFLAG species from the time-course cleavage assay in (C). (E) Schematic representation of the mono-(for MERS PLpro, USP2 and USP21) and the di-distributive cleavage mechanism (for SARS PLpro), indicating the accumulation of mono-Ub-conjugated substrates only in the case of the di-distributive cleavage mechanism displayed by SARS PLpro.
Interestingly, such a “distal, di-distributive” cleavage mechanism of K48-linked chains appears to be unique to SARS PLpro, as two human DUBs from the same USP family (USP2 and USP21) also do not accumulate mono-Ub conjugated Cdc34 as the final cleavage product, but completely deconjugate the substrate (Fig. 4C–D and Supplementary Fig. 3D). Collectively, these results strongly suggest a distributive cleavage mechanism for SARS PLpro (where the di-Ub moiety is recognized in the S2-S1 sub-sites), which may lead to accumulation of mono-Ub-conjugated substrates, depending on the positioning of the cleavage event with respect to the substrate (Fig. 4E).
DISCUSSION
We show here, that although the viral polyprotein processing proteases from the MERS and SARS coronaviruses are both efficient deubiquitinating and deISGylating enzymes, they utilize different mechanisms for substrate recognition and processing. MERS PLpro is mono-distributive deubiquitinating enzyme, while SARS PLpro prefers recognition of a diUbK48 unit, thus it is a distributive deubiquitinating enzyme (see Fig. 4E and Supplementary Fig. 3E and discussion below).
Previously, MERS PLpro has been demonstrated to cleave Ub- and ISG15-conjugates when expressed in cells [24, 25]; here we provide direct in vitro evidence demonstrating that MERS PLpro is a dual-specificity isopeptidase that processes ubiquitin and ISG15conjugates similarly. Our analysis is consistent with a very recent report on the substrate specificity of MERS PLpro [38], where although the authors noted slightly higher processing rates for ISG15-AMC, MERS PLpro was also demonstrated to cleave ubiquitin chains one ubiquitin at a time. We additionally demonstrate that MERS PLpro is a broad-specificity DUB, like most human DUBs from the USP family [11], which cleaves various ubiquitin linkages with similar efficiency, with the exception of linear chains. This lack of specificity for cleaving di-Ub chains is somewhat shared by SARS PLpro, when a di-Ub molecule occupies the S1-S1′ sub-sites. However, given the preference of SARS PLpro to recognize di-UbK48 units (discussed in detail below), the physiological relevance of the broad linkage specificity of SARS PLpro at the S1-S1′ sub-sites remains to be determined.
It is interesting to note that linear Ub chains are not cleaved by either SARS or MERS PLpro, even though the cleavage of a linear di-Ub is proteolytically the same as cleavage of a viral polypeptide at the processing sites, where the protease is cleaving a peptide bond (as opposed to various isopeptide-bond linkages in Ub-Ub dimers). Furthermore, since cleavage of K63-linked ubiquitin chains (which are similar in topology to linear Ub chains [17]) is well tolerated by MERS PLpro, it would be expected that viral DUBs might also cleave linear Ub chains. Interestingly, some human DUBs of the USP family, such as USP7, USP8 and USP11, also cannot tolerate linear Ub chains, even though they are less specific towards isopeptide linkages [39]. One reasonable explanation is that the peptide bond of the Gly-Met junction of a linear Ub is conformationally too rigid for SARS and MERS PLpro (and for some human DUBs) to accommodate, while an iso-peptide bond via a Lys side-chain is far more flexible in conformation. The flexibility of the isopeptide-linkage could also help explain the lack of linkage specificity for ubiquitin-ubiquitin linkages (when occupying S1-S1′ binding sites), suggesting that there could be only minimal contacts with the substrate on the primed side of the cleavage site (S1′ sub-site for ubiquitin chains or the sequence following the viral polypeptide following the cleavage site). On the other hand, in a linear di-Ub, the flexible C-terminus of the distal ubiquitin (in S1) ending in Gly continues immediately into the initiator Met of a rigid β-strand of the proximal ubiquitin (in S1′), potentially leading to a Ub-Ub conformation which is not suitable for binding in the S1′ pocket of viral PLpros. Further structural work on viral DUBs in complex with longer ubiquitin chains – or with viral polyprotein substrates – that span the active site are needed to shed light on the primed side specificity of viral DUBs.
On the other hand, addressing the issue of unprimed side specificity of viral DUBs, using a positional scanning substrate combinatorial library (PS-SCL) we show that, unlike human DUBs, both SARS and MERS PLpro have broad cleavage site specificities in terms of amino acid preference. Both SARS and MERS PLpro appear to have loosely defined pockets in their active site clefts, even though available crystal structures of both SARS and MERS PLpro in complex with mono-Ub [33, 34] indicate an intricate network of hydrogen bonds stabilizing the C-terminus of ubiquitin in their active site clefts. Nevertheless, they are both able to accommodate a hitherto DUB-resistant mutation in ubiquitin [31], suggesting greater flexibility of their active site clefts than human DUBs. Interestingly, the accommodation of Pro in P4 for SARS PLpro is only observable in the context of longer, tetra-Ub chain substrates, suggesting that recognition and processing of poly-Ub chains by SARS PLpro is via a unique mechanism. Whether these differences in viral PLpro active site specifies may be exploited in inhibitor design strategies has yet to be addressed.
Furthermore, we demonstrate mechanistic differences between SARS and MERS PLpro in how they recognize and process polyubiquitin chains (Fig. 4E and Supplementary Fig. 3E): MERS PLpro is a mono-distributive DUB, which cleaves ubiquitin chains one ubiquitin at a time (like the majority of human DUBs), while SARS PLpro is preferentially a di-distributive DUB that recognizes K48-linked di-Ub molecules as its minimal recognition unit and a processes the third ubiquitin in a poly-Ub chain – or removes a di-Ub from a substrate. This unique recognition is possibly mediated by an extended binding surface in SARS PLpro, which is not conserved in MERS PLpro. Traditionally, it has been accepted that SARS PLpro is primarily a deISGylating enzyme, mainly based on its preference for ISG15-AMC over Ub-AMC [21, 23, 30]. We propose that this is an inaccurate description of the activity and function of SARS PLpro, as we here show the cleavage of poly-Ub chains (minimally a tri-Ub or a di-Ub-conjugated substrate) to be highly efficient and comparable to that of ISG15 conjugates. MERS PLpro does not share this mechanism, as it possibly recognizes Ub/ISG15 in the S1 site and cleaves the next Ub in the S1′ site in a broadly linkage-independent manner, with no or minimal contacts in the S2 site (Fig. 4E and Supplementary Fig. 3E).
While our manuscript was in preparation, two very recent papers from the Mesecar and Baker groups demonstrated a similar mechanism of bivalent Ubl recognition for SARS PLpro, which is not shared by MERS PLpro [33, 38]. By modeling and mutational analysis, the authors identified a region in SARS PLpro (Phe70-Leu76) that is responsible for both di-UbK48 and ISG15 recognition in the S2 sub-site, and which is not conserved in MERS PLpro. In their model, Ratia et al. also suggested the S2 interaction to be via the classical Ile44 hydrophobic patch of ubiquitin, which interaction remains to be demonstrated experimentally. In the future, more precise mutations, in S1 or in S2 sub-sites, guided by structural work on ISG15- and di-Ub-bound SARS PLpro complexes, will allow for discrimination between the surfaces involved in ubiquitin vs ISG15 recognition, which will ultimately pave the way for determining the relative contributions of deubiquitination versus deISGylation to coronavirus pathogenesis.
Our study fully supports the Ratia et al. model and further shows that the unique di-distributive cleavage mechanism of SARS PLpro can likely result in the stabilization of not only di-UbK48 species, but also of mono-ubiquitin conjugated substrates that accumulate as remnants following substrate deconjugation by SARS PLpro. These mono-Ub-conjugated substrates are stabilized when the remnant ubiquitin on the substrate occupies the S1′ sub-site during the first cleavage event by SARS PLpro, leaving a poor substrate for SARS PLpro to recognize in subsequent cleavage events. Here, we also show that related USP-family DUBs and MERS PLpro do not share this feature of SARS PLpro to accumulate di-Ub units and mono-Ub-conjugated substrate cleavage products as they are mono-distributive DUBs. Although it is possible that in vivo, SARS PLpro may ultimately remove all ubiquitin molecules from its substrates, it is tempting to speculate that transiently stabilizing mono-Ub-conjugated substrates could have functional consequences during coronavirus infection. However, the exact in vivo substrates of SARS (and MERS) PLpro are currently ill defined, and the biological significance of this mode of di-Ub recognition remains to be elucidated. However, there are examples of human DUBs that appear to function similarly. The Machado-Josephin family cysteine protease DUB, ataxin-3, has been shown to be a poor deubiquitinating enzyme overall, but has been demonstrated to edit longer K48- and K63-mixed chains by recognizing a ubiquitin chain via its UIM domains and cleaving proximal to the recognized elements [40]. Additionally, a recently solved crystal structure of OTUD2 (a human DUB from the OTU family) in complex with a K11-linked di-Ub substrate has revealed an extended Ub (S2) binding surface in OTUD2 responsible for its K11 tri-Ub processing into a mono-Ub and di-Ub unit [41]. The function of these additional ubiquitin-recognizing domains in DUBs and how they contribute to biological pathways driven by each unique DUB has yet to be addressed.
MERS and SARS PLpro are primarily processing proteases that cleave the viral polypeptide at defined positions to release non-structural effector proteins [24, 42]. Interestingly, the positional scanning library screen for both SARS [26] and MERS PLpro (herein) faithfully recapitulated the nature of the sequences cleaved within the viral polypeptides [25], although these cleavage sites are not part of any ubiquitin-like domains in the viral polypeptide. Whether SARS and MERS PLpro have the capability to cleave Φ-Φ/✚-Gly-Gly-motifs (where Φ is a hydrophobic and ✚ is a positively charged amino acid) in exposed loops of human proteins, pointing to potential additional virulence-related non-Ubl substrates in human cells, has yet to be determined. The contribution of SARS PLpro to the infectivity of SARS hCoV and the contribution of MERS PLpro towards MERS hCoV infectivity is well established, as the catalytic activity of these proteases is required for viral polypeptide processing [23]. Thus, both viral DUBs are targets of intense drug development efforts in attempts to inhibit viral propagation. Recently, promising small molecule inhibitors have been developed for SARS hCoV based on their ability to inhibit SARS PLpro; however they do not inhibit MERS PLpro [38], suggesting that finding a common anti-coronaviral drug based on PLpros could be a difficult task. The in vivo testing of such compounds is also hampered by safety concerns using these highly pathogenic BSL-3 level viruses, thus the recent development of a proxy experimental system, using transgenic mice harboring an engineered Sindbis virus (a BSL-2 level organism) expressing SARS and MERS PLpros [43] is an extremely important step.
During viral infection – after viral genome translation and polypeptide expression – the host immune response must be suppressed to ensure timely viral particle assembly to propagate infection. It is accepted that the ubiquitin/ISG15 deconjugating activity of these membrane-bound viral PLpros plays a role in promoting viral replication in the host by deconjugating ISG15- and ubiquitin-conjugated proteins in the infected cell [23], contributing to down-regulation of innate immune functions [44]. However, the identities of the ubiquitin/Ubl-conjugated substrates whose deconjugation by viral PLpros are essential for pathogenicity remain ill defined. Whether they cleave ubiquitin or Ubl conjugates from viral proteins (i.e., to spare them from degradation by the proteasome or remove the ISG15 mark that each newly synthesized protein receives [15]), or cellular substrates (i.e., to dampen the host immune response by deconjugating signaling molecules [21, 25]), or both, within the vicinity of the membrane environment remains unknown, as most studies have been performed using over-expression systems relying on model pro-inflammatory substrates. It is also currently unknown to what extent deISGylation and/or deubiquitination activities contribute to pathogenesis. Thus, identifying viral PLpro mutants that can differentiate between ubiquitin and ISG15 recognition and cleavage are of considerable interest. Interestingly, mutations in PLpros that affect global Ubl-deconjugation, but not viral polypeptide processing, have been recently described [33, 34], which could aid in the development of attenuated viruses for coronavirus vaccine development. It would also be of interest to determine whether the two PLpro enzymes can be functionally swapped between the two viruses to determine whether coronavirus infection merely requires an active processing protease/isopeptidase or if substrate specificity of that protease is a key factor. With reverse genetics systems for coronaviruses in place [45] that have been used to uncover the function of SARS proteins in viral pathogenesis [46], together with the rapid development of the full-length MERS hCoV genome as a bacterial artificial chromosome system [47], it may be possible to answer these questions in the near future.
Supplementary Material
SUMMARY STATEMENT.
We compare processing proteases from two human coronaviruses (SARS and MERS hCoV) with respect to their activities and substrate specificities for ubiquitin-like signaling molecules, ubiquitin and ISG15; and we uncover a unique mode of poly-ubiquitin recognition by the SARS protease.
Acknowledgments
We thank members of the Huang, Lima and Reinberg labs for helpful discussion and reagents and Erin Bekes for editorial support.
FUNDING
M.B. is recipient of an individual post-doctoral NRSA fellowship from the NIGMS [F32GM100598]. This work was also supported the by Foundation for Polish Science and a statutory activity subsidy from the Polish Ministry of Science and Higher Education for the Faculty of Chemistry at Wroclaw University of Technology (to M.D). C.D.L is a Howard Hughes Medical Institute investigator and research in the Huang lab is supported by grants from the NIH [GM084244] and from the ACS [RSG-12-158-DMC].
Footnotes
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
M.B. conceived the study, designed the research, carried out experiments (with the exception of the PS-SCL screen in Fig. 1, which was carried out by W.R. and P.K. under the supervision of M.D.; while M.P.C.M. and H.O. synthetized the di-UbK48VME probe used in Fig. 3C), analyzed the data and wrote the manuscript. C.D.L and T.T.H provided data analysis, experimental guidance and support.
References
- 1.Herfst S, Fouchier R. Epidemiological and genetic investigations of human-to-human transmission of zoonotic influenza viruses. Euro Surveill 2014. 2014;19(25) doi: 10.2807/1560-7917.es2014.19.25.20840. pii=20840. [DOI] [PubMed] [Google Scholar]
- 2.Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Research. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilgenfeld R, Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Research. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baez-Santos YM, Barraza SJ, Wilson MW, Agius MP, Mielech AM, Davis NM, Baker SC, Larsen SD, Mesecar AD. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. Journal of Medicinal Chemistry. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3 doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusken CB, Haagmans BL, Muller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler JF, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch BJ, Rottier P, Osterhaus A, Gortazar-Schmidt C, Drosten C, Koopmans MP. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. The Lancet Infectious Diseases. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danielsson N, Catchpole M. Novel coronavirus associated with severe respiratory disease: case definition and public health measures. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2012;17 doi: 10.2807/ese.17.39.20282-en. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 9.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 10.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 11.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 12.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochstrasser M. Introduction to intracellular protein degradation. Chem Rev. 2009;109:1479–1480. doi: 10.1021/cr900054t. [DOI] [PubMed] [Google Scholar]
- 14.Durfee LA, Huibregtse JM. The ISG15 conjugation system. Methods Mol Biol. 2012;832:141–149. doi: 10.1007/978-1-61779-474-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narasimhan J, Wang M, Fu Z, Klein JM, Haas AL, Kim JJ. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 19.Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mielech AM, Chen Y, Mesecar AD, Baker SC. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Research. 2014 doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. The Journal of General Virology. 2014;95:614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 26.Drag M, Mikolajczyk J, Bekes M, Reyes-Turcu FE, Ellman JA, Wilkinson KD, Salvesen GS. Positional-scanning fluorigenic substrate libraries reveal unexpected specificity determinants of DUBs (deubiquitinating enzymes) Biochem J. 2008;415:367–375. doi: 10.1042/BJ20080779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drag M, Mikolajczyk J, Krishnakumar IM, Huang Z, Salvesen GS. Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem J. 2008;409:461–469. doi: 10.1042/BJ20070940. [DOI] [PubMed] [Google Scholar]
- 28.Kasperkiewicz P, Poreba M, Snipas SJ, Parker H, Winterbourn CC, Salvesen GS, Drag M. Design of ultrasensitive probes for human neutrophil elastase through hybrid combinatorial substrate library profiling. Proc Natl Acad Sci U S A. 2014;111:2518–2523. doi: 10.1073/pnas.1318548111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein science: a publication of the Protein Society. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindner HA, Lytvyn V, Qi H, Lachance P, Ziomek E, Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Archives of Biochemistry and Biophysics. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekes M, Okamoto K, Crist SB, Jones MJ, Chapman JR, Brasher BB, Melandri FD, Ueberheide BM, Denchi EL, Huang TT. DUB-resistant ubiquitin to survey ubiquitination switches in mammalian cells. Cell Reports. 2013;5:826–838. doi: 10.1016/j.celrep.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratia K, Kilianski A, Baez-Santos YM, Baker SC, Mesecar A. Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathogens. 2014;10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey-Elkin BA, Knaap RC, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL. Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression. J Biol Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Liang Q, Gong P, Tencer AH, Zhuang Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem Commun (Camb) 2014;50:216–218. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulder MP, El Oualid F, ter Beek J, Ovaa H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. Chembiochem. 2014;15:946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schechter I, Berger A. On the size of the active site in proteases. I Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 38.Baez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD. Catalytic Function and Substrate Specificity of the PLpro Domain of nsp3 from the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) J Virol. 2014 doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faesen AC, Luna-Vargas MP, Geurink PP, Clerici M, Merkx R, van Dijk WJ, Hameed DS, El Oualid F, Ovaa H, Sixma TK. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, Freund SM, Ovaa H, Komander D. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng X, Agnihothram S, Mielech AM, Nichols DB, Wilson MW, StJohn SE, Larsen SD, Mesecar AD, Lenschow DJ, Baric RS, Baker SC. A chimeric virus-mouse model system for evaluating the function and inhibition of papain-like proteases of emerging coronaviruses. J Virol. 2014;88:11825–11833. doi: 10.1128/JVI.01749-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Research. 2014;115C:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almazan F, Gonzalez JM, Penzes Z, Izeta A, Calvo E, Plana-Duran J, Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci U S A. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almazan F, Sola I, Zuniga S, Marquez-Jurado S, Morales L, Becares M, Enjuanes L. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Research. 2014 doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almazan F, DeDiego ML, Sola I, Zuniga S, Nieto-Torres JL, Marquez-Jurado S, Andres G, Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4:e00650–00613. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.