Abstract
Conventional T (Tcon) cells and Foxp3+ T-regulatory (Treg) cells are thought to have differing metabolic requirements, but little is known of mitochondrial functions within these cell populations in vivo. In murine studies, we found that activation of both Tcon and Treg cells led to myocyte enhancer factor 2 (Mef2)-induced expression of genes important to oxidative phosphorylation (OXPHOS). Inhibition of OXPHOS impaired both Tcon and Treg cell function compared to wild-type cells but disproportionally affected Treg cells. Deletion of Pgc1α or Sirt3, which are key regulators of OXPHOS, abrogated Treg-dependent suppressive function and impaired allograft survival. Mef2 is inhibited by histone/protein deacetylase-9 (Hdac9), and Hdac9 deletion increased Treg suppressive function. Hdac9−/− Treg showed increased expression of Pgc1α and Sirt3, and improved mitochondrial respiration, compared to wild-type Treg cells. Our data show that key OXPHOS regulators are required for optimal Treg function and Treg-dependent allograft acceptance. These findings provide a novel approach to increase Treg function and give insights into the fundamental mechanisms by which mitochondrial energy metabolism regulates immune cell functions in vivo.—Beier, U. H., Angelin, A., Akimova, T., Wang, L., Liu, Y., Xiao, H., Koike, M. A., Hancock, S. A., Bhatti, T. R., Han, R., Jiao, J., Veasey, S. C., Sims, C. A., Baur, J. A., Wallace, D. C., Hancock, W. W. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival.
Keywords: histone deacetylase, immunity, immunometabolism, immunoregulation, transplant survival
Understanding the complex network of signaling pathways that guides immune responses is essential to the development of new immunotherapies. Recently, the field of immunometabolism has gained prominence, given that metabolic products can directly affect immune signaling and that the nutrient environment may favor some immune cells over others according to differing metabolic needs (1–3). Proliferating conventional T (Tcon) cells rely primarily on glycolysis even under aerobic conditions in a Warburg-like manner typical of tumor cells. This reliance on glycolysis is largely attributed to the need to preserve fatty acids and proteins for proliferation and cytokine production and independence from oxygen in hypoxic environments (4–8), as well as utilizing glyceraldehyde phosphate dehydrogenase (GAPDH), which can interfere with IFN-γ transcription when not engaged in glycolysis (9). In contrast to proliferating Tcon cells, CD8+ memory T cells rely mainly on oxidative phosphorylation (OXPHOS) for energy production (10, 11).
OXPHOS is also thought important for energy production by Foxp3+ T-regulatory (Treg) cells (7, 8, 12, 13), a subset of T cells key to maintaining immune homeostasis and suppressing immune responses (14). Modulation of Treg numbers or function is currently of considerable therapeutic interest (15). Increasing Treg function could prove beneficial in autoimmune diseases and after transplantation (16), whereas inhibiting Treg function may promote protective host antitumor immunity (17). Altering cellular metabolism or the host metabolic environment could influence immune function and cell differentiation, and, for example, promote or inhibit Treg differentiation (4). Medical interventions aimed at changing cellular energy metabolism toward OXPHOS have long been linked to some degree of immunosuppression. For example, patients on ketogenic diets for seizure prevention anecdotally been noted to experience alleviation of allergic disease and increased susceptibility to minor illness (18). In addition, both a ketogenic diet and metformin, which activates AMPK by decreasing ATP levels (19), reduce inflammation in murine experimental autoimmune encephalomyelitis (20, 21). Likewise, augmenting the activity of pyruvate dehydrogenase, which promotes the conversion of pyruvate into acetate and thereby supports OXPHOS, leads to increased Foxp3+ Treg formation (22). In contrast, inhibiting fatty acid oxidation could be useful in cancer treatment, as it interferes with Treg function (7). However, the development of such therapeutic strategies will require further studies, especially with regards to the regulatory mechanisms that govern T cell metabolism and function.
In this report, we sought to investigate the metabolic properties of Tcon and Treg cells, and to assess the roles of key metabolic regulators in their functions. Using metabolic and functional assays, we analyzed the immune phenotypes of mice lacking regulator genes essential to OXPHOS metabolism. We identified key regulators of energy metabolism in Tregs and showed that they were essential for Treg suppressive function and Treg-dependent allograft acceptance. Our findings provide novel insights into T cell biology and identify new therapeutic options for interventions aimed at altering Treg function.
MATERIALS AND METHODS
Animal studies
We purchased BALB/c, C57BL/6, B6/Rag1−/−, and fl-Pgc1α mice (The Jackson Laboratory, Bar Harbor, ME, USA), and obtained YFP-Foxp3cre (23), Hdac9−/− (histone/protein deacetylase-9) (24), and Sirt3−/− (25) mice from their developers. Mice housed under specific-pathogen-free conditions were studied using protocols approved by the Institutional Animal Care and Use Committees of the Children’s Hospital of Philadelphia and the University of Pennsylvania. We transplanted BALB/c hearts into B6/Rag1−/− recipients (26). Allograft recipients were adoptively transferred intraperitoneal with 1 × 106 wild-type (WT) Tcon cells plus 5 × 105 Treg (WT, fl-Pgc1α/Foxp3cre or Sirt3−/−) (Supplemental Fig. 1). Allograft survival was assessed by palpation, and rejection was confirmed by histology.
Antibodies, media, and small molecules
For flow cytometry, we purchased mAbs to murine CD4 (allophycocyanin [APC]-eFluor 780, phycoerythrin [PE]), CD8 (PE/Cy7), CD25 (APC), CD62L (APC/Cy7), CD44 (PE), and Foxp3 (APC) from eBioscience (San Diego, CA, USA), as well as CD4 (Pacific Blue) from BD Pharmingen (San Diego, CA, USA). For mitochondrial staining, we purchased 10-nonyl acridine orange (NAO; Invitrogen, Carlsbad, CA, USA) and MitoSOX Red as well as LIVE/DEAD Fixable Dead Cell Stain Kit, aqua (Life Technologies, Carlsbad, CA). For immunoblotting, we purchased antibodies to Sirt3, Pgc1α, AMPKα, phospho-AMPKα (Thr172), and β-actin from Cell Signaling Technology (Danvers, MA, USA). We also purchased MitoProfile (Abcam, Cambridge, MA, USA) and Foxp3 (eBioscience) Abs. For standard T cell culture medium, we used RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 µg/ml), and 55 nM β2-mercaptoethanol. For low-glucose media, we used glucose-free RMPI-1640 (#11875) and glucose-free dialyzed FBS (#26400-036) from Gibco (Carlsbad, CA, USA). We purchased EX-527 (Tocris Bioscience, Bristol, United Kingdom), rotenone, and suberoylanilide hydroxamic acid (SAHA; Sigma-Aldrich, St. Louis, MO, USA); these were dissolved in DMSO and diluted in PBS. Equally diluted DMSO alone was used as a control.
Cell isolation and flow cytometry
Spleen and peripheral lymph nodes were collected and processed to single-cell suspensions of lymphocytes. We used magnetic beads (Miltenyi Biotec, San Diego, CA, USA) for isolation of Tcon (CD4+CD25–), Treg (CD4+CD25+), and antigen presenting cells (CD90.2–). We confirmed equal purity postisolation by flow cytometry (Supplemental Fig. 2). Cells of interest were analyzed using surface markers, and for Foxp3 staining, surface marker–stained cells were fixed, permeabilized, and labeled with Foxp3-specific mAb (27). Flow cytometry data were captured using Cyan (Dako, Glostrup, Denmark) and analyzed using the FlowJo 9.5.3 (Treestar) software. Pooled histogram data are shown as percentage of maximum, which is a normalization of overlaid data and represents the number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells.
T cell assays
For Treg suppression assays, purified Tcon cells were labeled with carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes, Eugene, OR, USA) and stimulated with irradiated antigen-presenting cells plus CD3ε mAb (1 µg/ml; BD Pharmingen). After 72 hours, proliferation of Tcon cells was determined by flow cytometric analysis of CFSE dilution, followed by normalization of relative suppression and calculation of area under the curve (28). For conversion to Foxp3+ Tregs, Tcon cells were incubated for 3 to 5 days with CD3ε/CD28 mAb beads, plus TGF-β (3 ng/ml) and IL-2 (25 U/ml), and analyzed by flow cytometry for Foxp3+ induced Treg (iTreg) (29).
Bioenergetic analyses
We measured T cell bioenergetic functions—oxygen consumption rate (OCR) and extracellular acidification rate (ECAR)—using the XF24 Analyzer (Seahorse Biosciences, North Billerica, MA, USA). In brief, XF24 24-well plates were coated using Cell-Tak (BD Biosciences, San Jose, CA, USA) as described in the Seahorse protocol. Isolated T cells were plated at a concentration of 1 × 106 cells/100 μl XF assay medium in 100 μl unbuffered XF assay medium–modified DMEM (#100965) at pH 7.4, supplemented with 5 mM glucose, 2 mM glutamate, and 1 mM sodium pyruvate, then incubated for 10 min at 37°C without CO2. To enhance cell adherence, plates were spun at room temperature for 10 min at 500 rpm, and an additional 570 μl of medium was added per well. Four baseline measurements of OCR and ECAR were taken; then oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), rotenone, and antimycin A were injected sequentially through ports of the Seahorse Flux Pak cartridges to reach the final concentrations of 1.25, 0.5, 1, and 1.8 µM, respectively. Three readings were taken after each sequential injection. Instrumental background was measured in separate control wells using the same conditions without biologic material.
High-resolution respirometry
OCR of forebrain homogenized tissue was determined at 37°C, with an Oxygraph-2K for high-resolution respirometry (Oroboros, Innsbruck, Austria), in a closed chamber with a magnetic stirring. The OCRs were calculated as the time derivative trace (DatLab software for data acquisition and analysis; Oroboros). The respiration medium consisted of a MiR05 buffer, which contains 110 mM sucrose, 60 mM potassium lactobionate, 0.5 mM ethylene glycol tetraacetic acid, 1 g/L essentially fatty acid–free bovine serum albumin, 3 mM MgCl2, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES adjusted to pH 7.1 at 37°C. Catalase (C9322; Sigma-Aldrich) was added, at 280 IU/ml, to the MiR05 buffer to form the MiR06 buffer. Subsequently, 2 ml MIR06 buffer were placed into the oxygraph chamber and stirred at 750 rpm and equilibrated to room air for 30 to 40 minutes until a stable signal was obtained for calibration at air saturation. Next, forebrain tissue (500 mg) was homogenized in 2.5 ml MIR06 in a glass tube and Teflon pestle on ice. The chambers were closed, and 20 µl homogenate was inserted into each chamber. The respirometric states were induced by addition of 2 mM malate, 5 mM pyruvate, and 10 mM glutamate, followed in succession by 2 mM ADP Mg2+, 10 µM cytochrome c, 10 mM succinate, 2 µM FCCP, 0.5 µM rotenone, 5 mM malonic acid, and 2.5 µM antimycin A. The corresponding oxygen concentration was calculated from the digitally recorded barometric pressure and the oxygen solubility at 37°C. Instrumental background flux was measured in separate controls using the same medium without biologic material.
Reactive oxygen species production in T cells using MitoSOX Red staining
In a preliminary series of experiments, we observed numerous limitations of MitoSOX staining that required further experimental steps to obtain reliable data. First, we noted the presence of MitoSOX Red signal from dead and apoptotic cells due to release of the fluorophore from the mitochondria and binding to nuclear DNA, as described previously (30). Furthermore, we noted that the MitoSOX Red signal was decreased in standard Dulbecco PBS (without Ca2+ and Mg2+), suggesting that the presence of these divalent ions was necessary for correct evaluation of mitochondria reactive oxygen species (ROS) production. Using timed of evaluations (every 15 minutes up to 1 hour), we found a significant increase in the MitoSOX Red signal at room temperature if cells were previously stained for superficial markers, such as CD4 and CD25, using standard 4°C buffers. Similarly, if the final washing step was performed at 37°C, the MitoSOX Red signal gradually decreased when the cells reached room temperature. Therefore, the results of any serial evaluations of cells could be compromised when the same order of samples is used. To overcome these limitations, we developed a modified MitoSOX Red staining protocol specific for T cells. First, we used the aqua Live/Dead Fixable Dead Cell Stain Kit to exclude nonspecific ROS signal from dead and apoptotic cells, then washed them 2 times. Then cells were stained with CD4 and CD25 mAbs (BioLegend, San Diego, CA, USA), or with CD4 mAb if YFP-Foxp3cre (23) mice were used. After washing, we incubated cells with 5 μM MitoSOX Red in HBSS (Gibco), which contains Ca2+ and Mg2+, for 10 minutes at 37°C. Cells were then washed twice, first with prewarmed 37°C HBSS and second with room-temperature HBSS, and left for 10 to 15 minutes in the dark at room temperature to ensure that all cells reached room temperature before evaluation by flow cytometry. All samples were evaluated twice in forward (sample 1, sample 2, […], sample n) and backward order (sample n, […], sample 2, sample 1) to ensure consistent observations.
Histology and immunohistochemistry
Sections of cardiac allografts were fixed in 10% neutral buffered formalin and embedded in paraffin. Hematoxylin and eosin– and trichrome-stained sections (4 µm) were reviewed by a pathologist (TRB) blinded to the treatment conditions to judge inflammation, myocyte necrosis, and fibrosis. Additional sections for immunohistochemistry were stained with primary antibodies for CD3 (Dako A0452; 1:100) with DAB (3,3′-diaminobenzidine) (Dako Cytomation) for antigen detection.
Electron microscopy
Purified cells of interest were centrifuged and then fixed in 2.5% glutaraldehyde in 0.1 M Sorenson phosphate buffer (SPB). After fixation, pellets were treated as follows: 3 × 10 minute changes of 0.1 M SPB adjusted to 425 mOsm/L with sucrose followed by fixation in 1% osmium tetroxide in 0.1 M SPB for 1.5 hour; washed in deionized water (3 × 10 minutes) followed by en bloc staining with 2% uranyl acetate for 30 minutes; dehydration in acetone; and infiltration and embedding with increasing concentrations of Spurr resin in acetone. Ultrastructural images were visualized with a Philips EM208S transmission electron microscope by a pathologist blinded to the experimental conditions (TRB). The number and morphologic characteristics of mitochondria present in each cell (24 per sample, ×11,000–22,000 magnification) were recorded. Morphologic changes to include vacuolar change, fusion, and elongation were graded on a scale from 0 to 3 if the findings were seen in 0, 1% to 30%, 30% to 60%, and >60% of the mitochondria within the cell, respectively. Cells without intact nuclei were excluded from analysis to minimize the inclusion of changes resulting from preservation artifacts or cellular degeneration.
RNA isolation, quantitative PCR, and Western blot analysis
RNA was extracted using RNeasy kits (Qiagen, Germantown, MD, USA), and RNA integrity and quantity were analyzed by photometry (DU640; Beckman Coulter, Brea, CA, USA). Reverse transcription, quantitative PCR (qPCR), and Western blot analysis were performed as previously reported (31, 32), with the exception of MitoProfile antibody staining, for which the step of boiling the samples was omitted. Primers were purchased from Applied Biosystems (Foster City, CA, USA).
Microarrays
Microarray experiments were performed using whole-mouse-genome oligoarrays (Mouse430a; Affymetrix, Santa Clara, CA, USA), and array data were analyzed using Mayday 2.12 software (33). Array data were subjected to robust multiarray average normalization. For low-stringency screening of differential expression, fold changes of up- and down-regulated genes were calculated, and significance assessed by significance analysis of microarrays using 100 permutations and false discovery rate (FDR) <0.5, and data with >1.2× differential expression were included in the analysis. Data underwent z-score transformation for heat-map display.
Data analysis
Data were analyzed by GraphPad Prism 5.0d software. Normally distributed data were displayed as means ± sem. Measurements between 2 groups were done by the Student t test if normally distributed or the Mann-Whitney U test if otherwise. Likewise, groups of 3 or more were analyzed by 1-way ANOVA if normally distributed, or the Kruskal-Wallis test if not normally distributed. Survival was calculated by a log-rank (Mantel-Cox) test.
Accession numbers
We deposited our data in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE36095.
RESULTS
OXPHOS is important for T cell proliferation
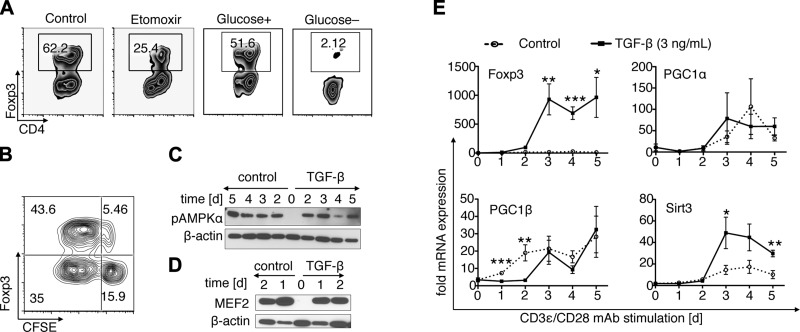
We first examined the importance of OXPHOS for Tcon and Treg function. Naive Tcon cells had reduced ability to transform into iTreg cells if treated with etomoxir, a compound known to directly inhibit fatty acid β-oxidation (Fig. 1A). We, like others (7), noted that limiting glucose in the culture medium reduced Tcon cell proliferation (data not shown), but a complete lack of glucose was even more deleterious to iTreg formation than etomoxir treatment, completely preventing iTreg development (Fig. 1A). Foxp3 induction was especially prominent in highly proliferating T cells (Fig. 1B). These findings indicated that glucose is a key requirement in the early phase of iTreg conversion, just as it is for Tcon cell proliferation. Because proliferating Tcon cells require glucose to allow a Warburg-like metabolism (6, 8), we questioned if and when iTreg would undergo a switch from glycolysis to OXPHOS. We therefore assessed the expression of key OXPHOS regulators during iTreg development. We expected that Tcon stimulation using CD3ε/CD28 mAb would not activate OXPHOS regulators, given the preference of Tcon cells for glycolysis. Likewise, we anticipated that polarizing conditions promoting iTreg development would eventually induce OXPHOS regulators during conversion. However, surprisingly, both conditions led to increased transcription of Pgc1α, Pgc1β, and Sirt3, as well as the phosphorylation of AMPKα, suggesting that both Tcon proliferation and iTreg conversion promote OXPHOS regulation (Fig. 1C–E). In addition, myocyte enhancer factor 2 (Mef2), a coactivator of Pgc1α and a target of class IIa histone/protein deacetylase (HDAC) repression, was up-regulated within 24 hours of T cell costimulation (Fig. 1D). Hence, both glycolysis and OXPHOS are involved in iTreg conversion and Tcon cell proliferation.
Figure 1.
T cell stimulation and Treg induction activate OXPHOS. A) C57BL/6 CD4+CD25–Foxp3– Tcon cells were cultured for 5 days under polarizing conditions using CD3/CD28 mAb-coated beads, plus TGF-β (3 ng/ml), and treated with 200 μM etomoxir or vehicle. Etomoxir treatment decreased development of Foxp3+ iTregs. Complete elimination of glucose, using glucose-free medium with dialyzed FBS, also abolished iTreg conversion. B) iTreg induction of CFSE-labeled Tcon at day 3 showed Foxp3 expression to be prominent within the dividing Tcon cell subset. c–e) C57BL/6 Tcon cells were stimulated using CD3/CD28 mAb-coated beads with or without added TGF-β (2 ng/ml), and protein (C and D) and RNA (E) samples were obtained each day. Both stimulated Tcon and iTreg show up-regulation of Mef2 and phosphorylation of AMPKα. Subsequently, Sirt3, Pgc1α, and Pgc1β were up-regulated regardless of whether the T cells were stimulated to become Foxp3+ iTreg by addition of TGF-β. Data are representative of 3 or 4 independent experiments, and pooled data are shown in (e). *P < 0.05, **P < 0.01, ***P < 0.001.
OXPHOS is essential for Treg suppressive function
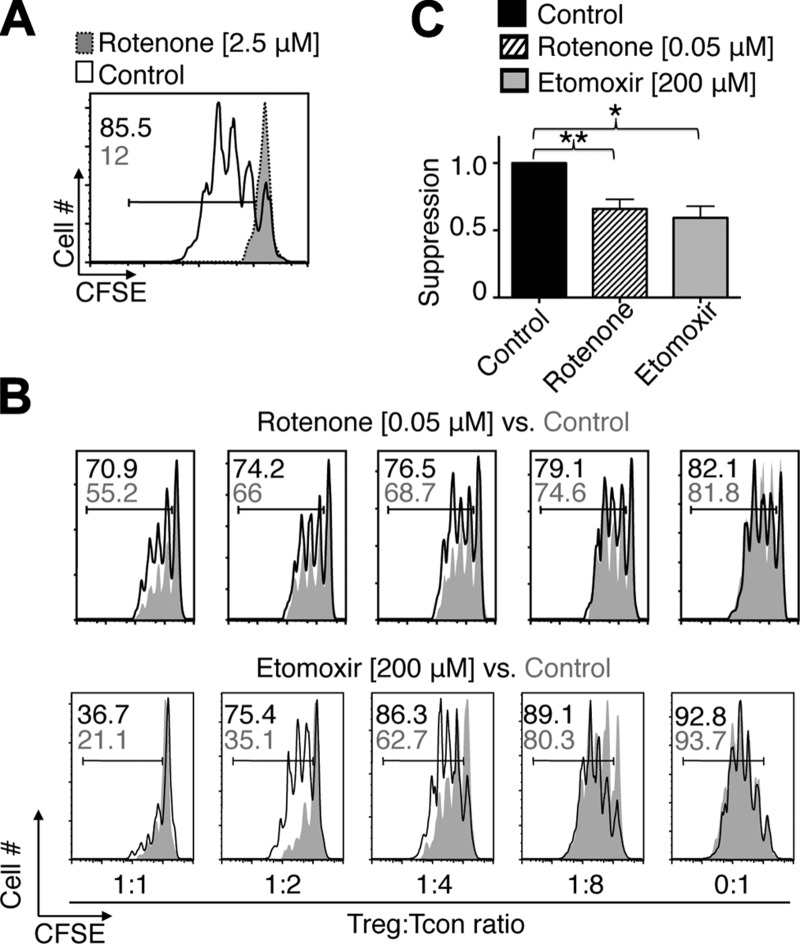
Because OXPHOS can occur in both Tcon and Treg cells, we sought to determine the relative dependence of either cell type on OXPHOS. We targeted complex I of the mitochondrial respiratory chain by adding rotenone to CD3ε/CD28 mAb-stimulated Tcon cells. Rotenone (2.5 μM) halted all proliferation of CD3ε/CD28 mAb-stimulated Tcon cells. Similar to Tcon cells stimulated in the absence of glucose, rotenone-treated Tcon cells remained alive but did not proliferate (Fig. 2A). These findings indicate that mitochondrial respiration is just as important for Tcon proliferation as the presence of glucose. Next, we tested whether OXPHOS was important for both Tcon and Treg cell function by undertaking Treg suppression assays in the presence of OXPHOS inhibitors. To avoid toxicity to proliferating Tcon cells, we tested several concentrations of rotenone. At 0.05 μM, Tcon cells proliferated freely, whereas Treg suppressive function was still markedly impaired (Fig. 2B, top; Fig. 2C). Likewise, inhibiting fatty acid β-oxidation by the addition of low doses of etomoxir (e.g., 200 μM) had only minor effects on Tcon cell proliferation, but markedly decreased Treg suppressive function (Fig. 2B, bottom; Fig. 2C). These data suggest that Tregs are more vulnerable than Tcon cells to OXPHOS inhibition, underscoring the unique metabolic features of Treg cells.
Figure 2.
Marked dependence of Treg vs. Tcon cells on OXPHOS. A) Tcon cells were labeled with CFSE and activated with CD3 mAb plus APC for 72 hours. Addition of 2.5 μM rotenone, an inhibitor of complex I of the mitochondrial respiratory chain, prevented T cell proliferation. B and C) C57BL/6 Tregs were added to CFSE-labeled Tcon cells stimulated as in A. B) Percentage of Tcon cell proliferation is indicated. Less Tcon cell proliferation indicates increased Treg suppression. Treatment with 200 μM etomoxir and 50 nM rotenone weakened Treg suppressive function without affecting Tcon cell proliferation; higher doses impaired Tcon cell proliferation as well. Data are representative of 3 or 4 independent experiments. C) Pooled rotenone (n = 7) and etomoxir (n = 4) treated Treg suppression data normalized to vehicle control. Paired Student’s t test, *P < 0.05, **P < 0.01.
Foxp3+ Treg have greater mitochondrial mass and higher ROS production than Tcon cells
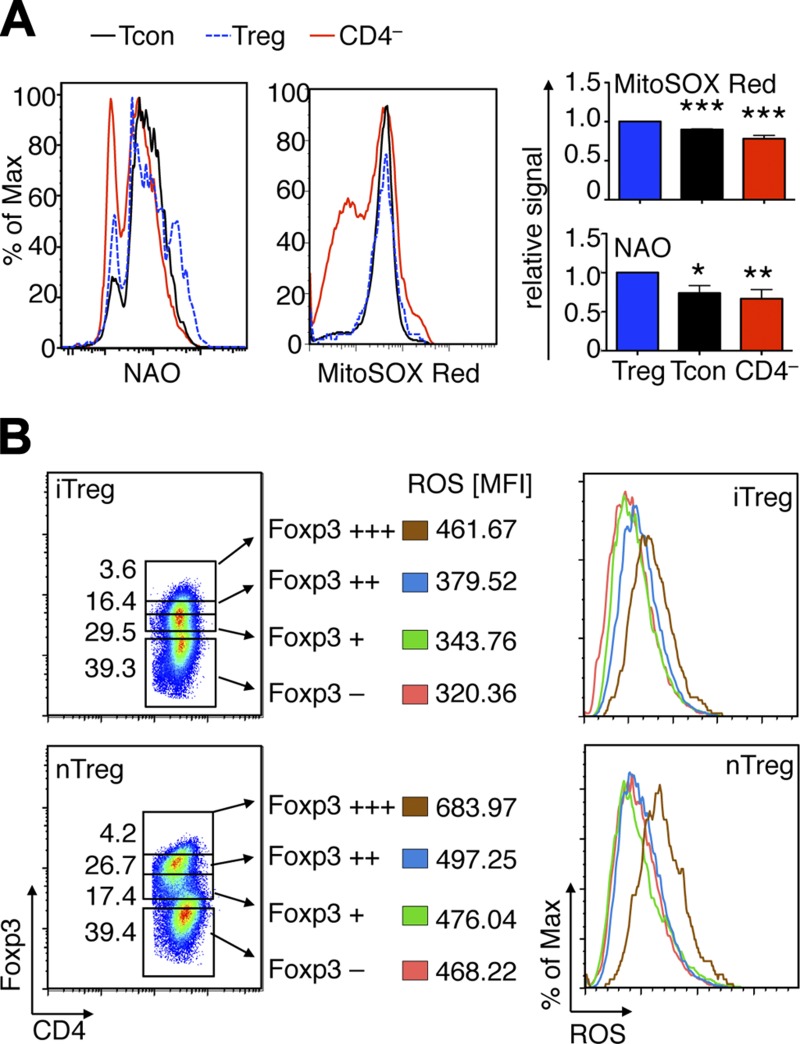
To begin to explore the mitochondrial biology of Foxp3+ Treg cells, we stained freshly isolated splenocytes from C57BL/6 mice for mitochondrial mass (NAO) and ROS production (MitoSOX Red), along with T cell surface markers. Consistent with their increased susceptibility to OXPHOS inhibition, naturally occurring CD4+CD25hi Treg (nTreg) had higher mitochondrial mass and greater ROS production than Tcon (CD4+CD25–) or non–T cells (Fig. 3A). Next, we assessed if higher ROS production was linked to Foxp3 expression. Because Foxp3 staining requires cell fixation and permeabilization, which is incompatible with the assessment of ROS production in live cells, we analyzed both nTregs and iTregs developed using cells from YPF+ Foxp3cre mice. This allowed simultaneous tracking of Foxp3 (via YFP detection) and assessment of ROS production without cell fixation. We found a close correlation between Foxp3 expression and ROS production in both iTreg and nTreg cells (Fig. 3B. Together, these data show that Foxp3+ Tregs have a higher mitochondrial mass at rest and produce more ROS under resting and stimulated conditions than Tcon cells.
Figure 3.
Treg cells have greater mitochondrial mass and ROS production than Tcon cells. A) Flow plots and quantitative data from C57BL/6 splenocytes. Treg (CD4+CD25hi) have higher mitochondrial mass per cell (NAO, n = 6) and, under resting conditions, the highest production of ROS (MitoSOX Red, n = 5), compared with Tcon (CD4+CD25–) or non-CD4 cells. D) Foxp3 and ROS production correlate in iTregs and in nTreg. CD4+Foxp3– Tcon from YFP-Foxp3cre mice were stimulated for 3 days with CD3/CD28 mAb and IL-2 (25 U/ml) alone, or with addition of TGF-β (3 ng/ml) to promote iTreg conversion. Foxp3+ iTreg show higher ROS production than T cells that remained Foxp3–. In both iTreg and nTreg, Foxp3 expression was correlated with ROS production. Percentage of maximum (% of max) shows normalization of overlaid data and represents number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Loss of Sirt3 and Pgc1α weakens Treg function
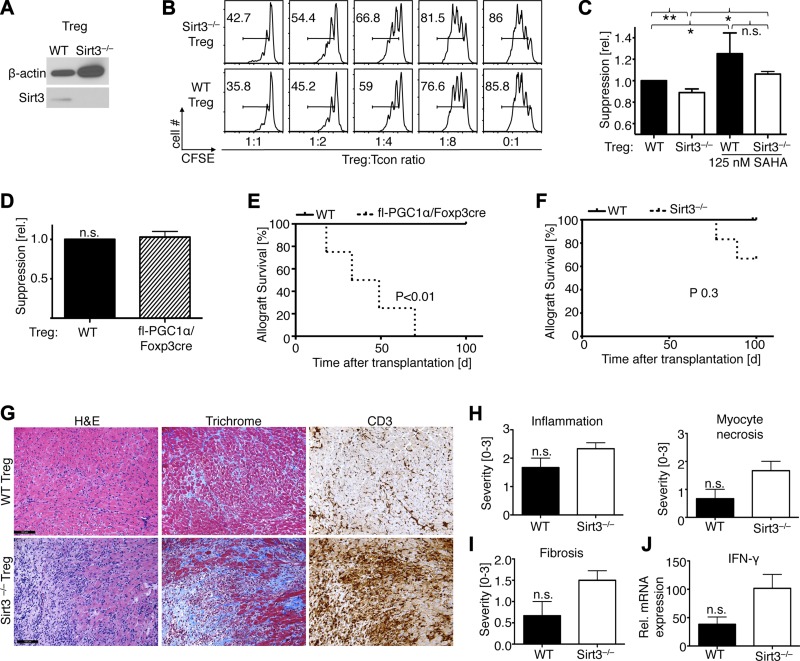
Both Sirt3 and Pgc1α are essential for physiologic mitochondrial activity (34, 35), and if Treg cells are especially dependent on mitochondrial activity, then loss of Sirt3 and Pgc1α might affect Treg function. Isolated Sirt3−/− Foxp3+ Tregs (Fig. 4A) had markedly impaired in vitro suppressive function compared to WT controls (Fig. 4B), though Treg function was still improved by exposure to an HDAC inhibitor, SAHA (Fig. 4C). By contrast, deletion of Pgc1α in Foxp3+ Tregs did not affect their suppressive function in vitro (Fig. 4D). Deletion of Sirt3 or Pgc1α did not affect the macroscopic or microscopic appearance of lymphoid organs compared to age-matched WT controls (data not shown). Likewise, proportions of CD4+ and CD8+ T cells, CD25+ and CD44+CD62L– T cells, and CD4+ Foxp3+ Tregs in thymus, spleen, and lymph nodes were similar, as were Tcon proliferation and iTreg conversion (data not shown). Next, we assessed Treg suppressive function in vivo using an adoptive transfer model of cardiac allograft rejection (Supplemental Fig. 1). BALB/c (H-2d) hearts were transplanted into B6/Rag1−/− (H-2b), and to induce rejection, recipients were injected with 1 × 106 WT Tcon cells (H-2b). Cotransfer of 5 × 105 WT Tregs (H-2b) suppressed WT Tcon proliferation and promoted long-term allograft survival, whereas cotransfer of an equal number of Tregs lacking Pgc1α led to acute allograft rejection (Fig. 4E). Use of Sirt3−/− Tregs had less dramatic effects, although one third of mice rejected their cardiac allografts (Fig. 4F), and hearts still beating at 100 days after transplantation showed increased chronic graft injury (Fig. 4G–J). Taken together, loss of either Pgc1α or Sirt3 weakened Treg suppressive function in vitro or in vivo, indicating their importance in Treg cell function.
Figure 4.
Effects of Sirt3 or Pgc1α deletion on Treg function. A) Lack of Sirt3 protein expression in Sirt3−/− Tregs. B) Comparison of Sirt3−/− and WT Treg showed that Sirt3−/− Tregs had reduced ability to suppress the proliferation of WT Tcon cells in vitro; percentage of CFSE-labeled proliferating Tcon cells is shown. C) Pooled data from 3 independent Treg suppression assays, alone or after Treg treatment with an HDAC inhibitor (SAHA). D) Pooled data from 3 independent Treg suppression assays comparing WT and fl-Pgc1α/Foxp3cre Treg suppressive function showed no differences in vitro. E) B6/Rag1−/− cardiac allograft recipients were adoptively transferred with Tcon cells plus WT or fl-Pgc1α/Foxp3cre Tregs at a 2:1 ratio; P value indicates Mantel-Cox. Use of Pgc1α−/− Tregs led to a significant reduction in allograft survival (n = 4 per group). F) Modest impairment of allograft survival with Sirt3−/− Tregs (n = 6 Sirt3−/− and n = 3 control Treg recipients). G) Compared to grafts from mice receiving WT Tregs, hearts still beating at 100 days after transplantation in mice receiving Sirt3−/− Tregs had increased inflammation, fibrosis, and T cell infiltration (scale bar = 100 μm). H and I) Quantification of hematoxylin and eosin (H) and trichrome (I) data show a trend to more severe inflammation, tissue injury, and fibrosis in recipients of Sirt3−/− Tregs. J) qPCR showed a trend to increased INF-γ production in recipients of Sirt3−/− Tregs. H–J) Data were normalized to native Balb/c hearts. *P < 0.05, **P < 0.01.
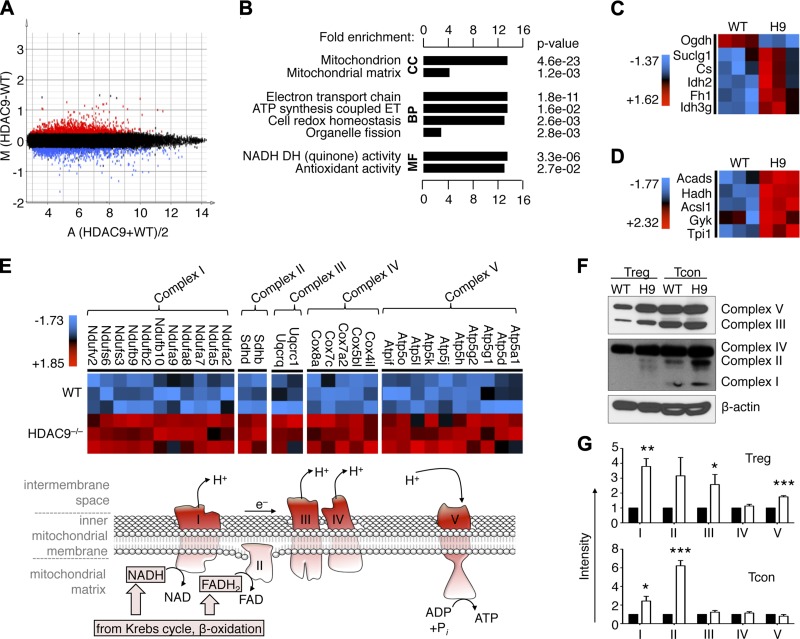
Hdac9 deletion promotes Treg gene transcriptional changes and OXPHOS activity
Our finding of up-regulation of Mef2 after T cell stimulation, and the important role of OXPHOS in promoting optimal Treg function, led us to hypothesize that these findings might be mechanistically linked. Deletion of Hdac9, which normally suppresses Mef2 activity, increases Foxp3+ Treg development and Treg suppressive function (26, 29), and so loss of Hdac9 might aid Treg function by augmenting mitochondrial respiration. By screening microarray data from Tregs of Hdac9−/− and WT mice using nonstringent statistical criteria, we identified patterns of altered gene expression (Fig. 5A). Using DAVID functional annotation clustering to identify biologically relevant groups of genes with altered expression (36), we noted that Hdac9 loss led to a broad up-regulation of genes involved in mitochondrial function and energy metabolism (Fig. 5B). Specifically, we identified increased expression of key transcripts involved in the Krebs cycle (Fig. 5C), fatty acid β-oxidation (Fig. 5D), and mitochondrial respiratory complexes (Fig. 5E). Together, these findings led us to hypothesize that loss of Hdac9 might produce a state of optimized OXPHOS. To assess if the alterations in mitochondrial gene expression translated into increased protein production, we isolated CD4+CD25+ Treg with equal purity (Supplemental Fig. 2), and we noted elevated mitochondrial complex protein expression in Treg and Tcon lacking Hdac9 (Fig. 5F, G). Hence, Hdac9 deletion in Treg cells appears to not only increases the expression of key OXPHOS genes, as noted by microarray, but also augment mitochondrial respiration by increasing the expression of mitochondrial complex proteins.
Figure 5.
Hdac9 deletion increases expression in Tregs of genes favoring OXPHOS. A) Plot of differentially expressed genes showing 1023 probes increased (red) and 889 probes decreased (blue) in Hdac9−/− vs. WT Tregs (20% differential expression, significance analysis of microarrays using 100 permutations and FDR < 0.5). B) DAVID functional annotation clustering of probes with increased expression in Hdac9−/− Tregs with >4 raw scores by Student’s t test (P < 0.05, FDR < 0.3, n = 613). Krebs cycle (C) and fatty acid degradation genes (D) are activated in Hdac9−/− Tregs. E) Up-regulation of mitochondrial complexes I to V in Hdac9−/− Tregs, and schematic illustrating the hypothesis of favored mitochondrial respiration in Hdac9−/− Tregs; electron donors NADH and FADH2 from, e.g., the Krebs cycle and β-oxidation feed into the electron transport chain (complex I to IV), leading to a proton (H+) gradient, which drives ATP production (complex V). C–E) Data shown after z-score transformation. F and G) Western blot analysis showing representative (F) and densitometry quantified (G) increased mitochondrial complex (C1 to C5) protein expression in Hdac9−/− Tregs and Tcon relative to WT controls (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test). Abbreviations: Acads, acyl-CoA DH; Acsl1, acyl-CoA synthetase long-chain family member 1; Cox, cytochrome c oxidase; CS, citrate synthase; DH, dehydrogenase; ET, electron transport; FH, fumarase; Gyk, glycerol kinase; Hadh, hydroxyacyl-CoA DH; Idh1, isocitrate DH1; Nduf, NADH-ubiquinone oxidoreductase; Ogdh, α-ketoglutarate DH; Sdh, succinate DH; Suclg1, succinyl-CoA ligase-α; Tpi1, triosephosphate isomerase 1; Uqcr, ubiquinol–cytochrome c reductase.
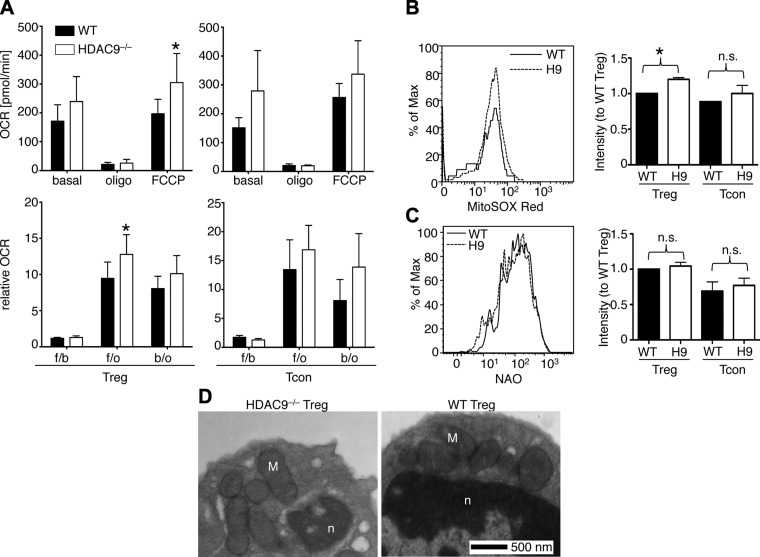
Loss of Hdac9 increases cellular respiration in Treg but not Tcon cells
We assessed if the Hdac9-related differences in gene expression patterns identified by microarray screening were accompanied by biologically relevant differences in OXPHOS metabolism. To quantify mitochondrial respiration in Hdac9−/− T cells, we measured the mitochondrial OCR and ECAR in intact cells using the Seahorse extracellular flux analyzer. Whereas Hdac9−/− Tregs had increased basal and uncoupled (4-(trifluoromethoxy)-phenylhydrazine hydrochloride, FCCP) OCRs, this was not the case for Hdac9−/− vs. WT Tcon cells (Fig. 6A). Likewise, brain and liver tissue homogenates, tested so as to assess effects of Hdac9 deletion beyond that observed in the Treg population, had normal OCR (Supplemental Fig. 3a), though Hdac9−/− T cells had slightly more acidification (Supplemental Fig. 3b). An elevated OCR is indicative of increased mitochondrial respiration, whereas increased acidification implies lactate generation, although this may be masked if cells are also producing ammonia (37). Along with our Seahorse findings of increased OCR in Hdac9−/− Tregs, we noted corresponding higher ROS production by Hdac9−/− Tregs (Fig. 6B), while Hdac9 deletion did not affect mitochondrial mass, shape, or number (Fig. 6C, D; Supplemental Table 1). Collectively, these data indicate increased mitochondrial function in Hdac9−/− Treg cells.
Figure 6.
Hdac9 deletion increases Treg mitochondrial respiratory function. A) OCRs of Tcon (right) and Treg cells (left) from WT or Hdac9−/− Tregs (H9) mice were measured using the Seahorse extracellular flux analyzer as described in text. Basal indicates OCR before any drug addiction; Oligo, OCR after 1.25 μM oligomycin addiction; and FCCP, OCR after 0.5 μM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone addiction. All data were correct for the antimycin A–resistant respiration background (*P < 0.05, n = 6). The OCR ratio was calculated between the following: b/o, basal condition (before any addition) and after oligomycin addition; f/o, after FCCP and oligomycin addition; f/b, after FCCP addition and basal condition. B) ROS production was measured in splenocytes of Hdac9−/− and WT mice by superoxide sensitive fluorescence. Percentage of maximum (% of Max) shows normalization of overlaid data and represents number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells. Treg (CD4+CD25hi) from Hdac9−/− mice had increased ROS, consistent with higher mitochondrial respiration (*P < 0.05, n = 3). C) Mitochondrial mass was measured in WT and Hdac9−/− T cells using NAO (n = 4). D) Representative transmission electron microscopic image of purified Hdac9−/− and WT Tregs shows normal mitochondria without overt pathology. Nucleus and mitochondria are denoted by n and M, respectively; for quantitation, see Supplemental Table 1.
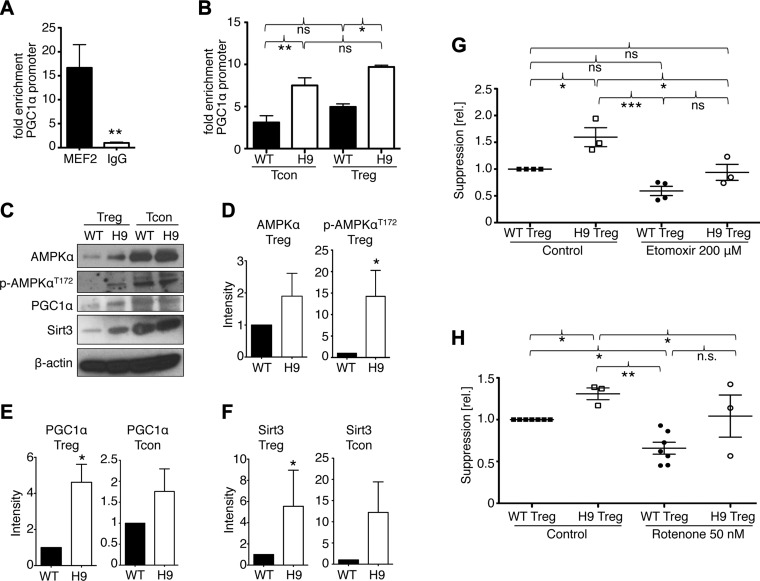
Deletion of Hdac9 boosts Mef2 binding to the Pgc1a promoter and favors OXPHOS
Finding increased OCR in Hdac9−/− Tregs raised the question of how Hdac9 deletion might favor OXPHOS. Given that Mef2 expression increased after Tcon cell stimulation (Fig. 1D), we tested if Mef2 was involved in the control of Pgc1α transcription in T cells. We found that Mef2 was bound to the Pgc1α promoter in Tcon cells stimulated for 24 hours, but not in resting Tcon cells, suggesting that Mef2 may be involved in controlling OXPHOS in T cells (Fig. 7A). Furthermore, the loss of Hdac9 coincided with increased Mef2 binding to the Pgc1α promoter in both Tcon and Treg cells (Fig. 7B). Consistent with increased Mef2-Pgc1α promoter binding, and increased mitochondrial respiration, we found that Treg lacking Hdac9 had higher expression of Pgc1α and Sirt3 proteins, as well as increased AMPKα phosphorylation (Fig. 7C–F; Supplemental Fig. 3c). Because Hdac9 influenced the expression of both Pgc1α and Sirt3, and loss of either protein weakened Treg function, we were interested in the degree to which the enhanced suppressive function of Hdac9−/− Tregs depended on altered mitochondrial function. Therefore, we treated WT and Hdac9−/− Treg with etomoxir (Fig. 7G) or rotenone (Fig. 7H). While inhibition of β-oxidation or mitochondrial respiration decreased the suppressive function of all Tregs, Hdac9−/− Tregs trended toward improved function compared to equally treated WT Treg cells. Likewise, pan-HDAC inhibition was also able to improve Treg function in Sirt3−/− Treg cells (Fig. 4C). Taken together, these data show that Mef2, which is up-regulated upon T cell activation, binds to Pgc1α, and that loss of Hdac9 increases this binding and activates several key components of OXPHOS.
Figure 7.
Hdac9 deletion leads to increased Mef2 binding and expression of OXPHOS regulators. A) Chromatin immunoprecipitation assay of genomic DNA from Tcon, stimulated with CD3ε/CD28 mAb for 24 hours, showing Mef2 binding to the Pgc1α promoter. B) Similar to A, comparing WT and Hdac9−/− T cells. C) Western blot analysis of WT and Hdac9−/− Treg and Tcon cells, comparing AMPKα, Pgc1α, and Sirt3. D) Densitometry showing increased AMPKα protein and threonine 172 phosphorylation in Hdac9−/− Tregs. Data pooled from 3 independent experiments. E and F) Densitometry showing Pgc1α (E) and Sirt3 (F) expression is increased in Hdac9−/− Tcon and Treg cells. Data pooled from 4 to 6 independent experiments; *P < 0.05. G and H) Pooled data from 3 to 6 independent Treg suppression assays normalized to vehicle control–treated WT Treg. Hdac9−/− Tregs show superior suppressive function compared to WT cells. Inhibition of β-oxidation (G) or complex I of the mitochondrial respiratory chain (H) diminished but did not abolish the advantage of Hdac9−/− Treg suppressive function. *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
Numerous studies have suggested a link between T cell metabolism and immune function; however, many of the mechanisms remain to be investigated. One approach has been to study known regulators of cellular metabolism, such as the mechanistic target of rapamycin (mTOR) or AMPK, with regards to their influence on T cell differentiation and function (38). Indeed, mTOR does play a central role in mediating glycolysis and promoting T cell differentiation (39). Not only does deletion of mTOR bias T cell development toward Treg formation (40), but also inhibition of mTOR through rapamycin can aid Treg expansion (41). These findings are supported by our own studies of human kidney and liver allograft recipients, where we found rapamycin-treated patients had better functioning Tregs than those of calcineurin inhibitor–treated patients (42). Other metabolic pathways that favor Tcon function include MYC and hypoxia inducible factor 1α, which inhibits Foxp3 transcription, and promotes glycolysis (38). A recent study showed that CD4+ T recon cells require the glucose transporter Glut1 for proliferation, while Treg cells do not (43). In contrast, AMPK, an important inhibitor of mTOR signaling, plays a key role in the development of cells that have mainly lipid oxidation–based metabolic programs, such as Treg and CD8+ memory T cells (12, 38).
Together, these studies emphasize the divergent modalities of ATP production between Tcon and Treg (7, 8), with Treg being more OXPHOS oriented, and proliferating Tcon relying on a Warburg-like metabolism. Our own data suggest that this is not such a categorical difference, i.e., there is overlap between Tcon and Treg cell metabolism. Stimulated Tcon cells activate AMPKα and key OXPHOS regulators, such as Pgc1α, Sirt3, and Mef2. In addition, we noted that Tcon cells were susceptible to more severe inhibition of mitochondrial respiration. Together, these data suggest that proliferating Tcon cells are not dependent purely on glycolysis. In support, Chang et al. (9) observed that Tcon cells can still proliferate in glucose-free conditions if galactose is present in the medium, thus enabling OXPHOS but not glycolysis. Cheng et al. (9) showed that even though Tcon cells can engage in OXPHOS, glycolysis was essential to keep GAPDH engaged, as it can otherwise interfere with IFN-γ transcription. However, even though both Tcon and Treg use OXPHOS, our results also suggest that compared to Tcon cells, Tregs are more susceptible to inhibition of OXPHOS, and particularly to partial reductions in OXPHOS.
This difference suggests therapeutic opportunities. For example, in oncology, the use of chimeric antigen receptor–T cell therapy has proven successful in hematologic malignancies (44), but less so in solid tumors. Low glucose levels in the tumor microenvironment may aid in the cancer immune escape by rendering tumor-infiltrating cytotoxic T cells less effective (e.g., by producing less IFN-γ). It is well known that immunosuppressive cells such as Tregs are induced and accumulate in the tumor microenvironment, and pose an obstacle to antitumor immunity (17). We can speculate that the metabolic conditions in the tumor microenvironment promote Treg formation and function, and that interfering with key metabolic pathways important to Treg function could aid in counteracting this obstacle to cancer immunotherapy. Our work identifies Sirt3 and Pgc1α, as well as Mef2, as potential candidate targets for such intervention.
On the other hand, the same pathways may be utilized to augment Treg function. For example, gut commensal bacteria produce short chain fatty acids with Hdac-inhibitory properties that are crucial to the formation of Foxp3+ iTreg (45). We noted that targeting of Hdac9, a known repressor of Mef2, led to increased Pgc1α and Sirt3 expression, AMPKα activation, and mitochondrial respiration. We hypothesized that the lack of Hdac9 inhibition on Mef2, which is present in the Mef2d isoform in T cells (46), might up-regulate Pgc1α (47). Indeed, we found Mef2 binding to the Pgc1α promoter in T cells. Pgc1α, on the other hand, is well known to promote Sirt3 expression (48), and both Pgc1α and Sirt3 are critical for mitochondrial function (34, 35). Thus, inhibiting Hdac9 may preferentially increase Treg metabolism. We have previously shown that genetic and pharmacologic targeting of Hdac9 increases nTreg and iTreg development, Treg suppressive function, and Treg-dependent allograft survival (26), and can prevent or treat autoimmune colitis (29). We have also demonstrated that Hdac9 targeting results in increased acetylation of Foxp3 protein, which is important for Treg suppressive function (26, 31, 49). Alternately, Foxp3 may directly steer Treg toward a relatively OXPHOS-dependent metabolism. As such, more abundant and more acetylated Foxp3 may be responsible for the increased OXPHOS observed in Hdac9−/− Treg cells. We plan to investigate the role of Foxp3 in Treg metabolism in future studies.
In summary, our data show that modulation of OXPHOS tips the Tcon and Treg cell balance away from Treg cells because OXPHOS inhibition leads to loss of their suppressive functions. Activation of Tregs promotes Mef2 expression and binding to Pgc1α, a key regulator of OXPHOS, suggesting a feed forward toward Treg function. Loss of Sirt3 or Pgc1α, as occurs in aging and many metabolic conditions, can weaken Treg suppressive function, whereas deletion of Hdac9, which binds and neutralizes Mef2, improves OXPHOS in Treg cells. Although further studies are required especially to delineate the role of Foxp3 in controlling Treg OXPHOS, our data indicate novel pathways to modulate host T cell responses by modifying mitochondrial energy metabolism in Tregs vs. Tcon cells.
Supplementary Material
Acknowledgments
The work was supported by the U.S. National Institutes of Health (Grants K08AI095353, AI073489, AI095276, NS21325, NS070298, AG24373, and DIC73691), as well as Simons Foundation (Grant 205844).
Glossary
- Acads
acyl-CoA DH
- Acsl1
acyl-CoA synthetase long-chain family member 1
- APC
allophycocyanin
- CFSE
carboxyfluorescein succinimidyl ester
- Cox
cytochrome c oxidase
- CS
citrate synthase
- DAB
3,3′-diaminobenzidine
- DH
dehydrogenase
- ECAR
extracellular acidification rate
- ET
electron transport
- FBS
fetal bovine serum
- FCCP
carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- FDR
false discovery rate
- FH
fumarase
- GAPDH
glyceraldehyde phosphate dehydrogenase
- Gyk
glycerol kinase
- Hadh
hydroxyacyl-CoA DH
- HDAC
histone/protein deacetylase
- Hdac9
histone/protein deacetylase-9
- Idh1
isocitrate DH1
- iTreg
induced Treg
- Mef2
myocyte enhancer factor 2
- mTOR
mechanistic target of rapamycin
- NAO
10-nonyl acridine orange
- Nduf
NADH-ubiquinone oxidoreductase
- nTreg
naturally occurring CD4+CD25hi Treg
- OCR
oxygen consumption rate
- Ogdh
α-ketoglutarate DH
- OXPHOS
oxidative phosphorylation
- PE
phycoerythrin
- qPCR
quantitative PCR
- ROS
reactive oxygen species
- SAHA
suberoyl + anilide + hydroxamic acid
- Sdh
succinate DH
- SPB
Sorenson phosphate buffer
- Suclg1
succinyl-CoA ligase-α
- Tcon
conventional T cells
- Tpi1
triosephosphate isomerase 1
- Treg
T-regulatory cells
- Uqcr
ubiquinol–cytochrome c reductase
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Van der Windt G. J., Pearce E. L. (2012) Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol. Rev. 249, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathis D., Shoelson S. E. (2011) Immunometabolism: an emerging frontier. Nat. Rev. Immunol. 11, 81–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathmell J. C. (2012) Metabolism and autophagy in the immune system: immunometabolism comes of age. Immunol. Rev. 249, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mockler M. B., Conroy M. J., Lysaght J. (2014) Targeting T cell immunometabolism for cancer immunotherapy; understanding the impact of the tumor microenvironment. Front. Oncol. 4, 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox C. J., Hammerman P. S., Thompson C. B. (2005) Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5, 844–852 [DOI] [PubMed] [Google Scholar]
- 6.Marelli-Berg F. M., Fu H., Mauro C. (2012) Molecular mechanisms of metabolic reprogramming in proliferating cells: implications for T-cell-mediated immunity. Immunology 136, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalek R. D., Gerriets V. A., Jacobs S. R., Macintyre A. N., MacIver N. J., Mason E. F., Sullivan S. A., Nichols A. G., Rathmell J. C. (2011) Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce E. L. (2010) Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 22, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C. H., Curtis J. D., Maggi L. B. Jr., Faubert B., Villarino A. V., O’Sullivan D., Huang S. C., van der Windt G. J., Blagih J., Qiu J., Weber J. D., Pearce E. J., Jones R. G., Pearce E. L. (2013) Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce E. L., Walsh M. C., Cejas P. J., Harms G. M., Shen H., Wang L. S., Jones R. G., Choi Y. (2009) Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van der Windt G. J., Everts B., Chang C. H., Curtis J. D., Freitas T. C., Amiel E., Pearce E. J., Pearce E. L. (2012) Mitochondrial respiratory capacity is a critical regulator of CD8+T cell memory development. Immunity 36, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacIver N. J., Michalek R. D., Rathmell J. C. (2013) Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalek R. D., Gerriets V. A., Nichols A. G., Inoue M., Kazmin D., Chang C. Y., Dwyer M. A., Nelson E. R., Pollizzi K. N., Ilkayeva O., Giguere V., Zuercher W. J., Powell J. D., Shinohara M. L., McDonnell D. P., Rathmell J. C. (2011) Estrogen-related receptor-α is a metabolic regulator of effector T-cell activation and differentiation. Proc. Natl. Acad. Sci. USA 108, 18348–18353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feuerer M., Hill J. A., Mathis D., Benoist C. (2009) Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat. Immunol. 10, 689–695 [DOI] [PubMed] [Google Scholar]
- 15.Wang L., de Zoeten E. F., Greene M. I., Hancock W. W. (2009) Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat. Rev. Drug Discov. 8, 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Issa F., Robb R. J., Wood K. J. (2013) The where and when of T cell regulation in transplantation. Trends Immunol. 34, 107–113 [DOI] [PubMed] [Google Scholar]
- 17.Nishikawa H., Sakaguchi S. (2010) Regulatory T cells in tumor immunity. Int. J. Cancer 127, 759–767 [DOI] [PubMed] [Google Scholar]
- 18.McDonald L. (1998) The Ketogenic Diet: A Complete Guide for the Dieter and Practitioner, Vol. 1, Morris Publishing, Austin, TX [Google Scholar]
- 19.Viollet B., Guigas B., Sanz Garcia N., Leclerc J., Foretz M., Andreelli F. (2012) Cellular and molecular mechanisms of metformin: an overview. Clin. Sci. 122, 253–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim do Y., Hao J., Liu R., Turner G., Shi F. D., Rho J. M. (2012) Inflammation-mediated memory dysfunction and effects of a ketogenic diet in a murine model of multiple sclerosis. PLoS ONE 7, e35476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nath N., Khan M., Paintlia M. K., Singh I., Hoda M. N., Giri S. (2009) Metformin attenuated the autoimmune disease of the central nervous system in animal models of multiple sclerosis. J. Immunol. 182, 8005–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostroukhova M., Goplen N., Karim M. Z., Michalec L., Guo L., Liang Q., Alam R. (2012) The role of low-level lactate production in airway inflammation in asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L300–L307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubtsov Y. P., Rasmussen J. P., Chi E. Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W. R. Jr., Muller W., Rudensky A. Y. (2008) Regulatory T cell–derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 [DOI] [PubMed] [Google Scholar]
- 24.Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V. Jr., Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao R., de Zoeten E. F., Ozkaynak E., Chen C., Wang L., Porrett P. M., Li B., Turka L. A., Olson E. N., Greene M. I., Wells A. D., Hancock W. W. (2007) Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299–1307 [DOI] [PubMed] [Google Scholar]
- 27.Tao R., Wang L., Han R., Wang T., Ye Q., Honjo T., Murphy T. L., Murphy K. M., Hancock W. W. (2005) Differential effects of B and T lymphocyte attenuator and programmed death-1 on acceptance of partially versus fully MHC-mismatched cardiac allografts. J. Immunol. 175, 5774–5782 [DOI] [PubMed] [Google Scholar]
- 28.Akimova T., Ge G., Golovina T., Mikheeva T., Wang L., Riley J. L., Hancock W. W. (2010) Histone/protein deacetylase inhibitors increase suppressive functions of human FOXP3+ Tregs. Clin. Immunol. 136, 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Zoeten E. F., Wang L., Sai H., Dillmann W. H., Hancock W. W. (2010) Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 138, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay P., Rajesh M., Haskó G., Hawkins B. J., Madesh M., Pacher P. (2007) Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat. Protoc. 2, 2295–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beier U. H., Wang L., Han R., Akimova T., Liu Y., Hancock W. W. (2012) Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci. Signal. 5, ra45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beier U. H., Wang L., Bhatti T. R., Liu Y., Han R., Ge G., Hancock W. W. (2011) Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol. Cell. Biol. 31, 1022–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battke F., Symons S., Nieselt K. (2010) Mayday—integrative analytics for expression data. BMC Bioinformatics 11, 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 35.Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V. Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 37.Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., Chomicz S., Ferrick D. A. (2007) Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 292, C125–C136 [DOI] [PubMed] [Google Scholar]
- 38.Pollizzi K. N., Powell J. D. (2014) Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat. Rev. Immunol. 14, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chi H. (2012) Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12, 325–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgoffe G. M., Kole T. P., Zheng Y., Zarek P. E., Matthews K. L., Xiao B., Worley P. F., Kozma S. C., Powell J. D. (2009) The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Procaccini C., De Rosa V., Galgani M., Abanni L., Calì G., Porcellini A., Carbone F., Fontana S., Horvath T. L., La Cava A., Matarese G. (2010) An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity 33, 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akimova T., Kamath B. M., Goebel J. W., Meyers K. E., Rand E. B., Hawkins A., Levine M. H., Bucuvalas J. C., Hancock W. W. (2012) Differing effects of rapamycin or calcineurin inhibitor on T-regulatory cells in pediatric liver and kidney transplant recipients. Am. J. Transplant. 12, 3449–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macintyre A. N., Gerriets V. A., Nichols A. G., Michalek R. D., Rudolph M. C., Deoliveira D., Anderson S. M., Abel E. D., Chen B. J., Hale L. P., Rathmell J. C. (2014) The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 20, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maus M. V., Grupp S. A., Porter D. L., June C. H. (2014) Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123, 2625–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J. R., Pfeffer K., Coffer P. J., Rudensky A. Y. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson B. J., Jäck H. M., Lyons G. E. (1998) Characterization of myocyte enhancer factor 2 (MEF2) expression in B and T cells: MEF2C is a B cell–restricted transcription factor in lymphocytes. Mol. Immunol. 35, 445–458 [DOI] [PubMed] [Google Scholar]
- 47.Czubryt M. P., Olson E. N. (2004) Balancing contractility and energy production: the role of myocyte enhancer factor 2 (MEF2) in cardiac hypertrophy. Recent Prog. Horm. Res. 59, 105–124 [DOI] [PubMed] [Google Scholar]
- 48.Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. (2010) Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE 5, e11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beier U. H., Akimova T., Liu Y., Wang L., Hancock W. W. (2011) Histone/protein deacetylases control Foxp3 expression and the heat shock response of T-regulatory cells. Curr. Opin. Immunol. 23, 670–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.