Abstract
Historically introduced by McConkey to explain the slow mutation rate of highly abundant proteins, weak protein (quinary) interactions are an emergent property of living cells. The protein complexes that result from quinary interactions are transient and thus difficult to study biochemically in vitro. Cross-correlated relaxation-induced polarization transfer-based in-cell nuclear magnetic resonance allows the characterization of protein quinary interactions with atomic resolution inside live prokaryotic and eukaryotic cells. We show that RNAs are an important component of protein quinary interactions. Protein quinary interactions are unique to the target protein and may affect physicochemical properties, protein activity, and interactions with drugs.

The interior of a living cell is an extraordinarily dense environment containing up to 400 mg/mL macromolecules.1 Hydrophobic and hydrophilic interactions are perturbed, and there is limited bulk water. Cytosolic water forms solvent shells on protein surfaces,2 and the distances between macromolecules are shorter than the characteristic Debye screening length of ion charges.3 In the cellular milieu, even very weak interactions may serve as a basis for evolutionary selection.4,5 To provide an explanation for the unexpectedly slow rate of mutation of proteins observed in various types of cells, McConkey postulated that weak, inherently transient interactions that are prevalent in cells play a role in forming a fifth level of protein structural organization, quinary structure.6
Protein quinary interactions are refractory to biochemical analyses.4 The study of these interactions requires in-cell methodologies because isolation techniques can easily disrupt protein complexes with limited thermodynamic stability.4,7 In-cell nuclear magnetic resonance (NMR) of proteins and protein complexes can potentially provide atomic resolution information about quinary interactions, thus opening the door for the study of a new level of structural complexity present in intact cells.8–13 To collect two-dimensional solution in-cell NMR spectra, 15N heteronuclear single-quantum coherence (1H–15N HSQC)14 experiments with 15N-labeled proteins either overexpressed8,15–17 or delivered into cells by using cell-penetrating peptide,9 toxins,18 injections,19 or electroporation techniques20,21 are performed. Peaks in this two-dimensional NMR experiment correspond to the protein backbone amide protons and nitrogens and are exquisitely sensitive to changes in protein structure.
At present, in-cell NMR is seriously limited by the molecular size of cellular complexes and the intrinsic cytosolic viscosity.22–25 Transverse proton relaxation of the in-cell NMR signal, T2, is so short for high-molecular mass complexes that only proteins that are unstructured, and thus have a longer T2 because of internal flexibility, or interact extremely weakly with the cytosol have been studied by using two-dimensional in-cell NMR, essentially leaving quinary interactions of the majority of folded proteins unobservable.22 In vitro solution NMR performed on extensively deuterated supramolecular structures with molecular masses of >100 kDa utilizes cross-correlated relaxation-enhanced polarization transfer-based NMR experiments (CRINEPT) to largely circumvent the limitations of molecular size.26 Relaxation-optimized 15N-edited CRINEPT–heteronuclear multiple-quantum coherence (HMQC)–transverse relaxation-optimized spectroscopy (TROSY) experiments are attractive for in-cell NMR studies because of superior sensitivity to NMR signals and relative insensitivity to unavoidable magnetic field inhomogeneity.27 1H–15N CRINEPT–HMQC–TROSY was used to probe the quinary interactions of proteins that were previously difficult to observe by in-cell NMR, thioredoxin (Trx),28 FKBP,28 adenylate kinase (ADK), and ubiquitin,9 all of which strongly interact with either prokaryotic or eukaryotic cytosol. Reduced proton density (REDPRO) labeling was used to exchange α and β protons of amino acids for deuterons to minimize proton relaxation.29
MATERIALS AND METHODS
Plasmid Construction
DNA encoding full-length FKBP was amplified from pC4EN-F1 (ARIAD Pharmaceuticals) using the oligonucleotides 5′-TTT TTT CCA TGG TGT CTA GAG GAG TGC AGG TGG AAA CC-3′ and 5′-TTT TTT GGT ACC TTA ATA ACT AGT TTC CAG TTT TAG AAG CTC CAC ATC-3′. The gene was ligated into pRSF-1b (Novagen) using the NcoI and KpnI linker sites. The resulting plasmid, pRSF-FKBP, confers kanamycin resistance and expresses FKBP from the T7 lac promoter. Escherichia coli trxA was amplified from pET32 (Novagen) using the oligonucleotides 5′-TTT GGT ACC ATG GGC GAT AAA ATT ATT CAC CTG ACT GAC G-3′ and 5′-CAT CGT GTC GAC TCA CAG GTT AGC GTC GAG GAA CTC-3′. The gene was ligated into pRSF-1b (Novagen) using the KpnI and SalI linker sites. The resulting plasmid, pRSF-Trx, confers kanamycin resistance and expresses N-terminally His-tagged Trx from the T7 lac promoter. E. coli ADK was amplified from genomic DNA using the oligonucleotides 5′-TTT TTT GGT ACC ATG CGT ATC ATT CTG-3′ and 5′-TTT GGA TCC TTA GCC GAG GAT TTT TTC-3′. The gene was ligated into pRSF-1b using KpnI and BamHI restriction sites. The resulting plasmid, pRSFADK, confers kanamycin resistance and expresses N-terminally His-tagged ADK.
Protein Overexpression
We followed the procedure for preparing REDPRO-labeled NMR samples.29 The three plasmids, pRSF-FKBP, pRSF-Trx, and pRSF-ADK, were separately transformed into E. coli strain BL21(DE3) codon+ (Novagen) for overexpression. Five milliliters of Lurie Broth (LB) medium supplemented with 75 μg/mL kanamycin was inoculated with a single colony and grown overnight at 37 °C while being shaken at 275 rpm. The overnight cultures were added to 50 mL of LB medium containing 75 μg/mL kanamycin at an initial OD600 of 0.07. Cells were grown at 37 °C and 275 rpm until the OD600 reached 0.9–1.0. Cells were pelleted, washed with M9 medium, and resuspended in 50 mL of deuterated M9 medium supplemented with 1.0 g/L 15NH4Cl and 0.2% glucose as the sole nitrogen and carbon sources, respectively. The culture was incubated at 37 °C for 20 min, and expression of REDPRO-labeled protein was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside. Protein expression proceeded for 16–18 h at 37 °C. For in-cell NMR experiments, 50 mL aliquots of the culture were pelleted, washed twice with NMR buffer, 10 mM potassium phosphate buffer (pH 6.5), and resuspended in 450 μL of the same buffer with 60 μL of D2O. Deuterium–hydrogen exchange is very efficient under in-cell conditions.9 Cell viability was checked before and after NMR experiments by using a colony plating method.53 Lysates for NMR samples were prepared by the freeze–thaw method to minimize damage to intracellular components. The freeze–thaw process was repeated five times. The samples were pelleted, and the supernatant was used to perform NMR experiments on lysates.
Protein Purification
REDPRO-labeled proteins, Trx and ADK, were purified using Ni-NTA columns under native conditions. REDPRO-labeled FKBP was purified as described previously.54,55 The proteins were buffer exchanged into NMR buffer, 10 mM potassium phosphate buffer (pH 6.5). Samples containing 100 μM REDPRO-labeled Trx in NMR buffer with 30, 65, 75, and 85% (w/w) d5-glycerol (Sigma-Aldrich, Inc.) were used to calibrate an optimal CRIPT transfer delay,26 Topt.
Total RNA Preparation
Total prokaryotic RNA was prepared as described previously56 from E. coli cells grown on M9 medium supplemented with 1.0 g/L NH4Cl and 0.2% glucose as the sole nitrogen and carbon sources, respectively. Total eukaryotic (yeast) RNA was purchased from Sigma-Aldrich, Inc. To prepare total RNA-REDPRO-labeled Trx and ubiquitin samples, 20 mg of total RNA was added to 20 μM protein samples. For the RNase A-treated total RNA-REDPRO-labeled Trx sample, 10 μL of 7000 units/mL RNase A (Qiagen) was added to 500 μL of a total RNA-REDPRO-labeled Trx sample and incubated at room temperature for 1 h. To prepare total RNA-ADK samples, 0.05 mg of total RNA was added to 20 μM protein.
Cisplatin Assay
A 200 mL cell culture containing REDPRO-labeled Trx was prepared as described above. After overnight expression, the cells were pelleted and washed with NMR buffer. The cells were resuspended in 1 mL of the buffer, and cisplatin was added to a final concentation of 10 μM. The cells were incubated at 37 °C for 1 h, washed twice with NMR buffer, and resuspended in 450 μL of NMR buffer with 60 μL of D2O. Following in-cell NMR spectroscopy, the cells were recovered and cisplatin-modified REDPRO-labeled Trx was purified on a His tag column as described above.
Protein Transfection by Electroporation
HeLa cells (Sigma-Aldrich) were prepared by seeding 2 × 106 cells/flask into six 150 cm2 Corning flasks and culturing the cells for 3 days in complete medium, low-glucose Dulbecco's modified Eagle medium (DMEM, Gibco), supplemented with 10% fetal bovine serum (FBS, Gibco), until 80% confluence was reached (~0.8–1 × 107 cells/flask). Cells were harvested by being exposed to 0.25% trypsin/EDTA (Sigma-Aldrich) for 5 min at 37 °C. Trypsin was neutralized by being diluted 5-fold with complete medium. Cells were pelleted by centrifugation at ~200g for 10 min at 25 °C and washed with 15 mL of prewarmed (37 °C) PBS. The densities of the cell suspensions were adjusted to 2 × 106 cells/mL using PBS. One milliliter of cell suspension was aliquoted into Eppendorf tubes (2 × 106 cells/mL). Cells were pelleted at ~200g for 6 min at 25 °C, and the supernatants were discarded. A protein electroporation mixture was prepared by diluting purified REDPRO-labeled ubiquitin to a final concentration of 800 μM to 1 mM with sterile filtered, freshly prepared electroporation buffer, 50% 100 mM sodium phosphate (pH 7.2), 5 mM KCl, 15 mM MgCl2, 15 mM Hepes, 5 mM ATP, 5 mM reduced glutathione, and 50% Amaxa Nucleofector Solution R (Lonza); 100 μL of the protein electroporation mixture was added to each of the cell pellets and gently mixed. The 100–200 μML cell suspensions were transferred into electroporation cuvettes (Lonza). Protein electroporation, EP, was performed with an Amaxa Nucleofector 2b device (Lonza) by using the Amaxa Nucleofector 2b pulse program, B-28.21 Cells were pulsed two or three times while being gently mixed between pulses. Directly after electroporation, 1 mL of prewarmed (37 °C) and CO2-saturated complete medium was added to each cuvette, and the samples were transferred into prepared cell culture dishes. For in-cell NMR experiments, three or four EP samples were applied to one 14 cm diameter plate. Plates were returned to 5% CO2 incubators. After recovery had proceeded for 2 h, 15 mL of complete medium was added to each plate and incubation continued for an additional 2–3 h. After recovery, nonadherent cells were removed. Cultures were washed three times with PBS (15 mL/plate) and harvested by using 0.25% trypsin/EDTA for 5 min, at 37 °C. Trypsin was neutralized by being diluted 5-fold with complete medium, and cells were pelleted by centrifugation at 200g for 6 min at 25 °C and washed once with 1 mL of PBS and twice with 1 mL of NMR buffer. Cell pellets were resuspended in 250 μL of NMR buffer containing 10% D2O. Cell suspensions were transferred into 5 mm diameter Shigemi tubes (Shigemi) for NMR analysis.
NMR Experiments
All NMR spectra were recorded at 298 K on a 700 MHz Avance II NMR spectrometer (Bruker) equipped with a cryoprobe, except for NMR experiments to calibrate Topt by using glycerol-containing samples, which were acquired at 278 K to increase solvent viscosity.33 For in-cell Topt calibration and total RNA-protein samples, a 1H–15N CRINEPT–HMQC–TROSY26 experiment with Watergate water suppression was used with a recycle delay of 300 ms. The optimal CRIPT transfer delay,26 Topt, was determined for each bacterial in-cell NMR, Topt calibration, and total RNA-protein sample to achieve maximal sensitivity. Because of the instability of REDPRO-labeled ubiquitin-transfected HeLa cells, the CRIPT transfer delay26 was not optimized but set to 1.4 ms.
The number of transients was 512. Spectral widths in the 1H and 15N dimensions were 12 and 30 ppm, respectively, and 1024 and 128 points were collected in the 1H and 15N dimensions, respectively. The total acquisition time for in-cell NMR experiments was 4 h. The interaction of cisplatin and Trx was monitored by using 1H–15N CRINEPT–HMQC–TROSY with a CRIPT transfer delay of 1.4 ms. The lysates of Trx, FKBP, and ADK were run at a Topt of 1.4 ms. For the purified REDPRO-labeled ADK sample, the recycle delay was 1 s and the CRIPT transfer delay was set at 5.2 ms. After each in-cell NMR experiment, the sample was recovered, the cells were sedimented, and a 1H–15N HSQC14 spectrum was collected on the sample supernatant. No protein peaks above noise level were detected, indicating no cell leakage (Figure 1 of the Supporting Information). To take into account small (<0.1 ppm) proton chemical shift changes, chemical shifts of purified Trx,57 FKBP,58 and ADK59 in NMR buffer were reassigned by using standard triple-resonance experiments,14 HNCA, HNCACO, and CBCACONH.
Spectra were processed with Topspin version 2.1 (Bruker) and analyzed by using CARA software.60 The changes in protein peak intensities due to quinary complexation were calculated as relative ΔI = (Ilysate – Iin-cell)/Iin-cell, where Iin-cell and Ilysate are the integrated intensities of the peaks from the in-cell and lysate spectra and were normalized. Peak intensities were calculated by using peak integration with three different integration ranges to estimate a standard error of the mean for ΔI. Residues that exhibited a chemical shift difference of >0.1 ppm between the lysate and the purified protein were discarded. The apparent molecular mass of the in-cell proteins was estimated by measuring Topt.30 Theoretically,30 Topt is a solution of
| (1) |
where JNH is a scalar 15N–1H coupling constant, Rc is the relaxation rate due to cross correlation between amide proton chemical shift anisotropy and 15N–1H dipole–dipole coupling, and RH is the transverse relaxation rate of the amide protons. In the case of large deuterated proteins, both Rc and RH are linearly dependent on the rotational correlation time, τc, and can be approximated as Rc = 1.7τcBo and RH = τc(0.8Bo2 + 1.7), where Rc and RH are in units of seconds, τc is in units of nanoseconds, and Bo is the strength of the magnetic field in proton frequency units of gigahertz. Equation 1 was solved numerically by using MATLAB (Mathwork, Inc.) function fsolve. Using the Debye–Stokes–Einstein relation61 among τc, the protein molecular mass, and viscosity, we obtained a rough estimate for the apparent molecular mass of proteins inside the cells,30 bound to RNA, or in viscous glycerol solutions.
RESULTS
We used four unrelated well-folded proteins with molecular masses from 8 to 24 kDa and pIs from 4.7 to 7.5 (Table 1) to explore quinary interactions inside live prokaryotic and eukaryotic cells by using CRIPT-based in-cell NMR.
Table 1.
Protein Physicochemical Parameters
| protein | molecular mass (kDa) | PI, theoretical | apparent molecular mass (MDa) | apparent molecular mass (MDa), total RNA-protein complex | in-cell protein concentration (μM) |
|---|---|---|---|---|---|
| Trx | 13 | 5.7 | ~1.1 | 0.3 | 300 |
| Trx mutant | 13 | 6.1 | ~1.1 | - | - |
| ADK | 25 | 6.1 | >1.2 | 0.4 | 365 |
| ubiquitin | 8.5 | 6.6 | - | - | - |
| FKBP | 12 | 7.8 | ~1.1 | - | - |
In-Cell NMR of Thioredoxin and FKBP
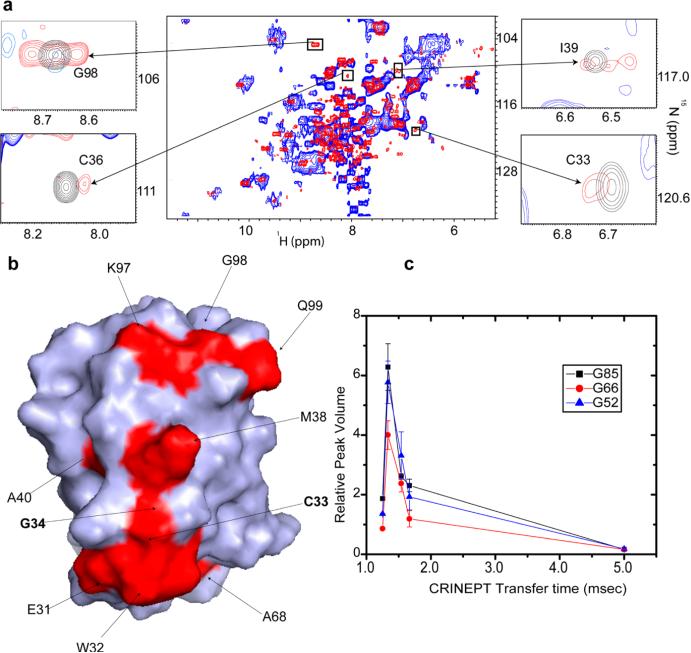
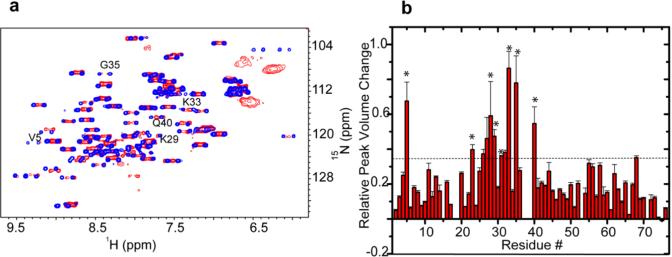
Bacterial thioredoxin (Trx) is a small, highly stable protein that facilitates the reduction of different cellular protein substrates through cysteine thiol–disulfide exchange. The active site of Trx consists of a CGPC motif, of which the cysteines are critical for Trx redox activity. Despite having a small molecular mass, Trx cannot be observed by using conventional solution in-cell NMR; the in-cell 1H–15N HSQC of uniformly labeled [U-15N]Trx contains only sharp peaks from small metabolites and a broad peak from the N-terminal His tag used to simplify protein purification (Figure 2 of the Supporting Information). The solid-state in-cell NMR spectrum is dominated by broad, overlapping signals that likely reflect the heterogeneous environment inside the cells.28 The in-cell 1H–15N CRINEPT–HMQC–TROSY NMR spectrum of E. coli containing REDPRO-labeled Trx exhibits broad peaks at positions close to those observed in cell lysates (Figure 1a). The line shape of the in-cell peaks suggests that Trx binds weakly to a heterogeneous set of cellular components, preventing the in-cell NMR spectrum from being observed by 1H–15N HSQC (Figure 2). The apparent molecular size of the in-cell Trx complex, estimated by measuring the dependence of peak volumes on the CRINEPT transfer time, is ~1.1 MDa30 (Figure 1c and Table 1). The in-cell concentration of Trx was ~300 μM (Figure 3 of the Supporting Information), which is well above the physiological concentration of most intracellular proteins. Some of the in-cell NMR resonances exhibit two multiplets, corresponding to fast and slow transverse relaxing components of the CRINEPT–HMQC peaks30 (Figures 1 and 2), suggesting that free cytosolic Trx is in exchange with a complex inside the cells. Despite the heterogeneity of the Trx complex, the binding between Trx and cellular components is specific; most of the in vitro peaks are present in-cell except for E31, W32, C33, G34, C36, M38, I39 A40, A68, K97, G98, and Q99, which are completely or partially broadened due to in-cell interactions and dynamic exchange processes in the flexible parts of the protein (Figures 1a and 2b). We consider residues both directly and indirectly affected by the interaction with cytosol to form the in-cell protein interaction surface. Mapping the interaction surface residues onto the molecular structure of Trx reveals that they are concentrated in and about the active site (Figure 1b); thus, the Trx active site is involved in quinary interactions, i.e., the transient complexes formed between cellular components and Trx.
Figure 1.
Quinary interactions of Trx in E. coli occlude the active site. (a) Overlay of the in-cell 1H–15N CRINEPT–HMQC–TROSY spectrum of REDPRO-labeled Trx (blue) and that of the cellular lysate (red). The intensities of C33, C36, I39, and G98 peaks, residues involved in quinary interactions, are broadened out. The insets show overlays of the boxed regions of the in-cell spectrum (blue) and the corresponding regions of the 1H–15N CRINEPT–HMQC–TROSY spectrum of lysate (red) and the 1H–15N HSQC spectrum of purified Trx in 10 mM potassium phosphate buffer (pH 6.5) (black). (b) Residues involved in the quinary interactions (red) are mapped onto the molecular surface of Trx (PDB entry 1X0B); active site residues, C33 and G34, are shown in bold. (c) The relative volumes of the G52, G66, and G85 peaks in the in-cell 1H–15N CRINEPT–HMQC–TROSY spectra of Trx are plotted vs CRINEPT transfer delay time. An endogenous tryptophan indole amide peak in the in-cell spectra is used as a reference. The optimal CRINEPT transfer delay for Trx is ~1.3 ms, which corresponds to an apparent molecular mass of ~1.1 MDa.
Figure 2.
Multiple bound states of Trx inside bacterial cells. (a) Overlay of peaks from a 1H–15N CRINEPT–HMQC–TROSY spectrum of REDPRO-labeled Trx in E. coli (blue) with peaks from the 1H–15N HSQC spectrum of purified REDPRO-labeled Trx in 10 mM potassium phosphate buffer (pH 6.5) (black). G52, G66, and G85 exhibit broad in-cell peaks that are characteristic of multiple conformations of Trx in fast exchange on the NMR time scale. This implies that the quinary interactions are inherently transient and dynamic. (b) Bar plot showing the relative changes in in-cell 1H–15N CRINEPT–HMQC–TROSY peak intensities of Trx residues due to quinary interactions. Residues E31, W32, C33, C36, M38, A40, A68, and Q99, annotated with asterisks, are also affected in total RNA-bound Trx. The horizontal line differentiates residues whose NMR peaks undergo significant broadening. (c) Overlays of selected regions of the in-cell spectrum (blue) and 1H–15N HSQC spectrum of purified thioredoxin in 10 mM potassium phosphate buffer (pH 6.5) (black) and 10 mM deuterated sodium acetate buffer (pH 4.7) (red). The chemical shifts of thioredoxin peaks in the in-cell NMR spectrum are shifted downfield by ~0.1 ppm in the 1H dimension compared to the in vitro spectrum of thioredoxin at pH 6.5. Specifically, the chemical shift of A68 resembles that observed at pH 4.7, whereas the indole amide of W29 resonates between pH 4.7 and 6.5.
We used cisplatin31,32 to assess whether covalent modification of cysteines in Trx and its physiological substrates affect the quinary interactions. After incubation with cisplatin, cells overexpressing Trx were resuspended in NMR buffer. The in-cell NMR spectrum revealed further changes in the Trx quinary interactions affecting residues adjacent to the active site: peaks corresponding to F28, W29, W32, G72, L95, and the indole proton and nitrogen of W32 were completely broadened (Figure 3). The in vitro spectrum of Trx purified from cisplatin-treated cells was unchanged from that of untreated purified Trx (Figure 4 of the Supporting Information), confirming that the changes in quinary interactions are due to modified Trx substrates.
Figure 3.
Cisplatin perturbs the quinary interactions of Trx. (a) Overlay of the 1H–15N CRINEPT–HMQC–TROSY spectra of REDPRO-labeled Trx in E. coli treated with (red) and without (blue) 10 μM cisplatin. The insets show peaks from active site residues F28, W32, G72, and L95, which are broadened (arrows) because of changes in the in-cell interaction of Trx induced by cisplatin. In addition, the indole amide of W32 is broadened. (b) The residues affected by quinary interactions (red) and cisplatin (cyan) are mapped onto the molecular surface of Trx (PDB entry 1X0B).
To determine if Trx substrates are the only component of the quinary interactions, we recorded the in-cell NMR spectrum of the active site mutant, CPGC/ILEFGP Trx.34 The in-cell NMR spectrum of CPGC/ILEFGP Trx still exhibits significant broadening, which is characteristic of the quinary interactions (Figure 5a of the Supporting Information). The apparent molecular mass of the mutant was ~1.1 MDa, which is comparable to that of in-cell wild-type Trx (Figure 4 and Figure 5b of the Supporting Information). Thus, the absence of active site residues does not impair the formation of mutant Trx quinary complexes, implying that intracellular components other than substrates contribute to the quinary interactions.
There is an upfield shift of <0.1 ppm in the proton dimension of the Trx in-cell NMR signals compared to that of the lysate, which may be related to the difference in pH between the cytosol and the in-cell NMR buffer35 (Figure 2). To address this issue, we adjusted the lysate pH from 6.5 to 4.7, which is closer to the pH in the proximity of the cell membrane where Trx may be localized,36 and reacquired the spectrum. The positions of the peaks in lysate and in-cell samples could not be completely matched by adjusting the lysate pH from 6.5 to 4.7, although lowering the pH did result in an upfield shift of the lysate sample (Figure 2). The in-cell 1H–15N CRINEPT–HMQC–TROSY NMR spectrum of REDPRO-labeled FKBP, another protein that cannot be observed by using conventional in-cell NMR,28 does not exhibit the upfield shifts observed in the in-cell NMR spectrum of Trx (Figure 6 of the Supporting Information). The effect seems to be specific to the Trx quinary structure and may be related to the membrane proximal localization of Trx.36 Because a decrease in pH inhibits cysteine thiol–disulfide exchange, we suggest that the Trx quinary interactions may create a local decrease in the pH to effectively maintain Trx in the inactive state until it encounters physiological substrates.
The quinary interactions of FKBP overlap the binding site for its small ligand, rapamycin (Figure 6 of the Supporting Information). To test whether these quinary interactions can be mimicked by the interaction of FKBP with any abundant protein, we compared the in vitro NMR spectrum of purified REDPRO-labeled FKBP in a solution of 300 mg/mL bovine serum albumin (BSA) with the in-cell NMR spectrum of REDPRO-labeled FKBP (Figure 7 of the Supporting Information). The in vitro interaction of FKBP with BSA is nonspecific, resulting in near-uniform broadening of all NMR peaks; this is in contrast to the in-cell NMR spectrum of FKBP in which only a specific subset of peaks are broadened. The results suggest that quinary interactions result in unique complexes depending on the individual target protein.
In-Cell NMR of Adenylate Kinase
Adenylate kinase (ADK) catalyzes the phospho transfer from ATP to AMP to create two ADP molecules. The substrate-free form of the enzyme exists in an open conformation when the ATP and AMP binding domains, located on the N- and C-termini, respectively, of the central CORE domain, are apart; ATP and AMP binding moves the domains closer together, resulting in a closed conformation.37 Because of volume exclusion effects, the crowded cytosol may perturb the unligated structure, biasing it toward the closed conformation.38 We used in-cell NMR to probe ADK quinary interactions to examine this possibility.
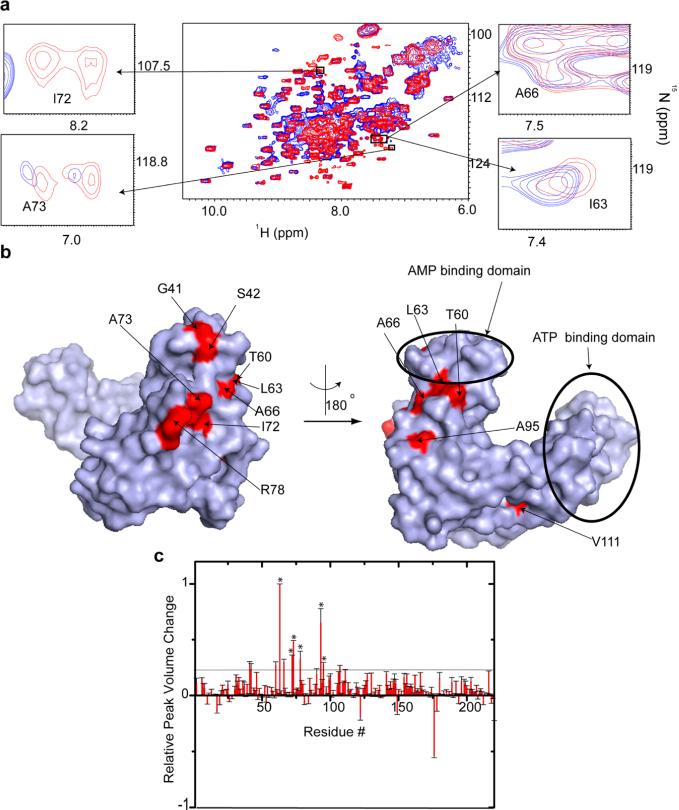
Contrary to computational predictions,38 the in-cell NMR spectrum of ADK closely resembles the open substrate-free conformation observed in vitro (Figure 5 and Figure 8 of the Supporting Information). NMR peaks corresponding to the residues involved in domain closing are unchanged, suggesting that macromolecular crowding does not perturb the tertiary structure of the enzyme. NMR peaks corresponding to ADK residues G42, G56, T60, L63, A66, I72, A73, R78, A93, A95, and V111 are completely or partially broadened in the in-cell NMR spectrum and define the interaction surface involved in the ADK quinary interactions. This surface is located in the CORE domain and lies proximal to the AMP binding domain. Unlike Trx, the active site of ADK is unaffected by quinary interactions and is free to bind both ATP and AMP (Figure 5b).
Figure 5.
Quinary interactions of ADK in E. coli maintain the enzyme in an open conformation. (a) Overlay of the in-cell 1H–15N CRINEPT–HMQC–TROSY spectrum of REDPRO-labeled ADK (blue) and that of the cellular lysate (red). The insets show overlays of the boxed regions of the in-cell spectrum (blue) and the corresponding regions of the 1H–15N CRINEPT–HMQC–TROSY spectrum of lysate (red). The intensities of I63, A66, I72, and A73, which are involved in quinary interactions, are broadened. (b) The residues involved in quinary interactions (red) are mapped onto the molecular surface of ADK (PDB entry 4AKE). The ATP and AMP binding regions are highlighted by circles. The residues involved in quinary interactions mostly lie in the CORE domain of ADK. (c) Bar plot showing the relative changes in in-cell 1H–15N CRINEPT–HMQC–TROSY peak intensities of ADK residues due to quinary interactions. Residues that are affected by the interaction of ADK with total RNA are annotated with asterisks. The horizontal line differentiates residues whose NMR peaks undergo significant broadening.
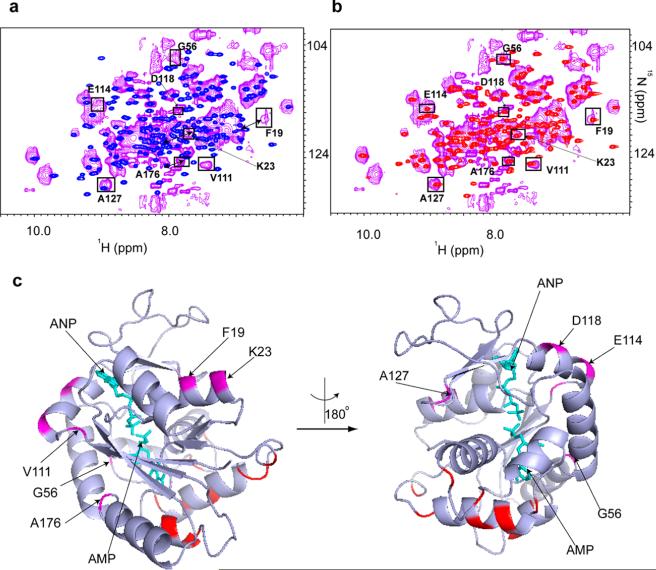
Despite the fact that bacterial cells contain ~3 mM total ATP and the in-cell concentration of ADK is ~300 μM (Figure 3 of the Supporting Information), the observation of an open conformation is consistent with that of Schneider et al.39 and suggests that only a small portion of cytosolic ATP is free to bind ADK. We used chloramphenicol to inhibit the bacterial ribosome and increase the concentration of free ATP in the cell.39 Adding 100 μg/mL chloramphenicol to the cell culture for 45 min prior to the in-cell NMR experiment perturbed the cellular equilibrium among ATP, ADP, and AMP and led to a dramatic change in the in-cell NMR spectrum of ADK. We observed extensive broadening of all ADK peaks (Figure 6 and Figure 9 of the Supporting Information), suggesting that ATP-, ADP-, and AMP-bound ADKs exist inside the cell. The spectrum is similar to that observed in vitro with 3 mM ATP and 200 μM AMP, consistent with a closed conformation of ADK (Figure 6b).
Figure 6.
Chloramphenicol induces binding of the adenine nucleotide to ADK in E. coli. (a) Overlay of the in-cell 1H–15N CRINEPT–HMQC–TROSY spectrum of REDPRO-labeled ADK (purple) in chloramphenicol-treated cells and the 1H–15N HSQC spectrum of purified ADK (blue). Chloramphenicol causes peak shift changes and broadening of the ADK residues due to chemical exchange between the different substrate-bound (AMP, ADP, and ATP) conformations of ADK. (b) Overlay of the in-cell 1H–15N CRINEPT–HMQC–TROSY spectrum of REDPRO-labeled ADK in chloramphenicol-treated cells (magenta) and the 1H–15N HSQC spectrum of purified 50 μM ADK with 3 mM ATP and 200 μM AMP (red). The trajectories of the in-cell peak changes after chloramphenicol treatment correlate with binding of ATP and AMP to ADK, consistent with a closed conformation of ADK. Specifically, F19, K23, G56, V111, E114, D118, A127, and A176 peaks in chloramphenicol-treated cells exhibit chemical shift changes consistent with ADK bound to 3 mM ATP and 200 μM AMP. These peaks are marked in both panels a and b. Double arrows denote F19 and A176 chemical shift changes. (c) The residues (magenta), whose peaks exhibit chemical shift changes and broadening, are mapped onto the cartoon representation of the AMP/ATP analogue, ANP, bound to ADK (PDB entry 1ANK). The ligands are shown as sticks (cyan). All residues indicated in the figure are part of either the ATP or AMP binding domains. The amino acids that form a quinary patch on the crystal structure (red) are located far from the ATP binding domain and are part of the CORE domain of ADK.
In-Cell NMR of Ubiquitin in Human Cells
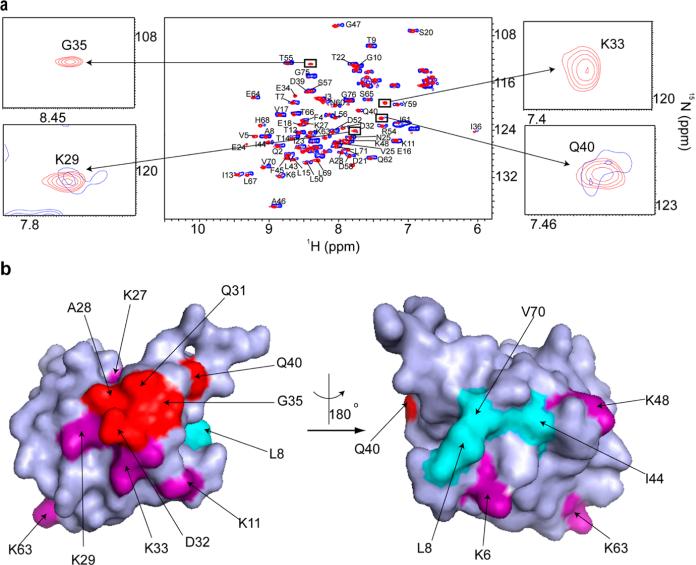
Eukaryotic proteins also engage in quinary interactions.4 Wild-type human ubiquitin labeled with REDPRO29 was electroporated into HeLa cells by using Selenko's electroporation technique.20,21 The quinary interactions of human ubiquitin in HeLa cells affect the Ile36 hydrophobic patch,40 consisting of residues V5, I23, V26, K27, A28, K29, Q31, D32, K33, G35, and Q40, which is involved together with the Ile44 patch40 in interactions between ubiquitin chains (Figure 7a). Importantly, the quinary interaction surface is adjacent to three of the seven surface-exposed lysines that can act as polyubiquitination sites, providing a possible explanation for why K27, K29, and K33 are the least preferred lysines for polyubiquitination41 (Figure 7b).
Figure 7.
Ubiquitin quinary interactions block polyubiquitination sites, K27, K29, and K33. (a) Overlay of the in-cell 1H–15N CRINEPT–HMQC–TROSY spectrum of purified REDPRO-labeled ubiquitin transfected into HeLa cells (blue) and the 1H–15N HSQC spectrum of the cell lysate (red). NMR peaks corresponding to K29, K33, G35, and Q40 (insets) are broadened in the in-cell spectrum, suggesting that ubiquitin is involved in transient interactions with cellular components of the cytosol. (b) Residues (red), whose NMR peaks are broadened out (Figure 10 of the Supporting Information), form a contiguous interaction surface involved in ubiquitin quinary interactions (PDB entry 1D3Z). The seven lysines of ubiquitin, which are used for ubiquitylation, are colored purple. K27, K29, and K33 are a part of the interaction surface and are affected by the quinary interactions. The canonical I44 hydrophobic patch of ubiquitin (cyan) spanning L8, I44, and V70 is unperturbed by quinary interactions. The multiplet structure of the in-cell ubiquitin CRINEPT peaks suggests that there is a large population of free ubiquitin, which is in intermediate exchange on the NMR time scale with bound ubiquitin, in HeLa cells.
RNA Is a Key Component of Quinary Interactions
Besides physiological substrates and nonspecific binding molecules, what are the other molecular components of protein quinary interactions? The in-cell apparent molecular masses of proteins range from 1 to >1.2 MDa, scale linearly with intracellular viscosity, which is ~3–4 times greater than that of water,22,42 and correspond to in vitro apparent molecular masses of 300–400 kDa. Genomic DNA is too large (>10 MDa) and has an abundance that is too low for it to serve as a component of quinary interactions. Despite the abundance of protein in cells, 60% of cell dry weight, it is unlikely that many weakly interacting proteins, with an average molecular mass of ~50 kDa,43 can form such a large complex. Moreover, the observed specificity of quinary interaction surfaces is inconsistent with the heterogeneity of cellular proteins (Figure 7 of the Supporting Information). RNAs, which constitute 20% of a cell's dry weight, have molecular masses of ~20 kDa for tRNAs, 100–500 kDa for mRNAs, and up to 5 MDa for rRNAs. RNA primary structures contain only four bases and are thus vastly less heterogeneous than proteins.
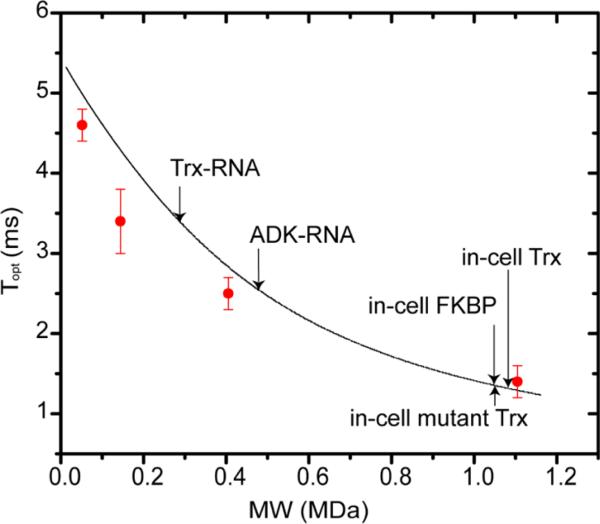
We prepared both bacterial and eukaryotic RNA to test in vitro binding of total RNA to proteins. Total RNA lacks bound protein and can only be considered a crude approximation to cellular RNA. Adding 0.01 mg/mL total RNA to 15 μM REDPRO-labeled FKBP led to the complete disappearance of the NMR spectrum, suggesting that FKBP strongly interacts with very large RNAs, most probably rRNA, which is unlikely to be a physiologically important result. On the other hand, when the concentration of total RNA was increased to 30 mg/ mL, the 1H–15N CRINEPT–HMQC–TROSY NMR spectra of both 15 μM purified REDPRO-labeled Trx and ubiquitin became similar, but not identical, to their in-cell NMR spectra (Figures 2b, 8, and 9), suggesting that total RNA binds weakly to the previously identified quinary interaction sites. The apparent molecular mass of the total RNA–Trx complex was ~250 kDa, which is consistent with the in-cell apparent molecular mass of Trx, 1.1 MDa (Figure 4 and Table 1), and suggests that Trx binds to nonribosomal RNA. At 0.2 mg/mL total RNA, the NMR spectrum of 10 μM REDPRO-labeled ADK becomes similar to its in-cell spectrum (Figure 5c). The apparent molecular mass of the total RNA–ADK complex is
Figure 8.
Total RNA–ubiquitin interactions and quinary interactions affect similar residues. (a) Overlay of in-cell 1H–15N CRINEPT–HMQC–TROSY spectra of REDPRO-labeled ubiquitin treated with yeast total RNA (red) and untreated (blue). V5, K29, K33, G35, and Q40 peaks, indicated in the figure, are differentially broadened because of the interaction of ubiquitin with total RNA. (b) Bar plot showing ubiquitin residues whose peaks are differentially broadened because of quinary interactions in HeLa cells. Residues V5, I23, A28, K29, Q31, K33, G35, and Q40, annotated with asterisks, are also affected in total RNA-bound ubiquitin. The horizontal line differentiates residues whose peaks undergo significant broadening. The similarity between residues affected by both RNA binding and quinary interactions suggests that RNA is an important component of quinary interactions.
Figure 9.
RNA is a key component of Trx quinary structure. (a) Overlay of in-cell 1H–15N CRINEPT–HMQC–TROSY spectra of 15 μM REDPRO-labeled purified wild-type Trx with (red) and without (blue) 30 mg/mL total E. coli RNA. The indole NH of W29 exhibits a downfield shift in the RNA-bound and in-cell NMR spectra (Figures 1 and 2). The indole NH of W32, along with backbone amide peaks of E31, C33, C36, K37, I39, and A40 (boxed), is broadened due to the protein–RNA interaction that is similar to the quinary interaction observed in-cell. (b) The relative volumes of the G52, G66, and G85 peaks in the in vitro 1H–15N CRINEPT–HMQC–TROSY spectrum of total RNA-bound wild-type Trx are plotted vs the CRINEPT transfer delay times. An endogenous tryptophan indole amide peak in the in vitro spectra is used as a reference. The optimal CRINEPT transfer delay is 3.5 ms, which corresponds to a molecular mass of ~0.3 MDa. (c) Overlay of in-cell 1H–15N CRINEPT–HMQC–TROSY spectra of 15 μM REDPRO-labeled purified wild-type Trx treated with 15 mg/mL total E. coli RNA in the presence (black) and absence (red) of RNase A. The indole NH of W29 exhibits a downfield shift in the RNA-bound and RNase A-treated NMR spectra. The indole NH of W32, along with backbone amide peaks of E31, C33, C36, K37, I39, A40, and I42 (boxed), is broadened due to the protein–RNA interaction that is similar to the quinary interaction observed in-cell. (d) The relative volumes of the G52, G66, and G85 peaks in the in vitro 1H–15N CRINEPT–HMQC–TROSY spectrum of RNase A-treated total RNA-bound wild-type Trx are plotted vs the CRINEPT transfer delay times. The optimal CRINEPT transfer delay is ~5.4 ms, which corresponds to a molecular mass of ~12 kDa.
Figure 4.
In-cell proteins exhibit megadalton apparent molecular masses. The curve, calculated using eq 1, shows the dependence of the apparent molecular mass at 700 MHz on the CRINEPT transfer delay that provides the optimal transfer efficiency, Topt. To calibrate Topt, 100 μM REDPRO-labeled Trx was dissolved in NMR buffer with 30, 65, 75, and 85% (w/w) d5-glycerol and corresponding viscosities of 4, 34, 92, and 343 cP, respectively.33 Topt was experimentally determined for each sample at 5 °C (red symbols). The in-cell apparent molecular masses of proteins used in this study and the in vitro apparent molecular masses of protein–RNA complexes are indicated. The in-cell apparent molecular mass of ADK (not shown) is greater than 1.2 MDa (Figure 10 of the Supporting Information).
~400 kDa (Figure 4 and Table 1 and Figure 10 of the Supporting Information), which is consistent with the apparent in-cell molecular mass of >1.2 MDa.
As a control experiment to show that RNA is an important component of protein quinary interactions, a mixture of purified Trx and total E. coli RNA was treated with RNase A. RNase A endonuclease activity cuts RNA after C and U residues to yield a pool of small RNA fragments and nucleotides. NMR spectra of the Trx and total RNA mixture were acquired before and after the addition of RNase A. If short RNA oligonucleotides act as ligands, the digestion procedure will increase the concentration of RNA-bound Trx because of the increase in the molar concentration of total RNA. Indeed, following RNase A digestion, further changes in the Trx chemical shifts were observed relative to those of the total RNA–Trx complex consistent with a greater population of bound Trx (Figure 9). In addition, the apparent molecular mass of thioredoxin decreased from 200 kDa when it was bound with total RNA to ~20 kDa for the RNase A-treated sample (Figure 9 and Table 1). The reduced increase in the apparent molecular mass corresponds well with Trx bound to small RNA oligonucleotides. The same set of Trx peaks are affected by both total RNA and RNase A-treated total RNA, implying that the quinary interaction surface is specific.
DISCUSSION
Our observation that RNA is an important component of protein quinary interactions is unlikely to be the result of protein overexpression; recent proteomics studies of the mRNA interactome, which were conducted at physiological concentrations, uncovered nearly 800 proteins bound to eukaryotic transcripts.44–46 The eukaryotic homologues of Trx and ADK, as well as FKBP and ubiquitin, were among the RNA binding proteins identified in these studies.45 Interestingly, RNA was key to driving a protein, ΔTAT-GB1, into supercrowded “clusters” in vitro.47 For some proteins, the contribution of RNA to quinary interactions may be an example of “fossil” molecular interactions left from the time of the “RNA world”.48 For other proteins, such as post-transcription or translation factors, the functional role of protein–RNA interactions is well-established.49 For the rest of the proteins associated with the mRNA interactome,44–46 the functional role of protein–RNA binding and quinary interactions has yet to be determined.
The amino acids involved in the quinary interactions of Trx, FKBP, and ubiquitin are located in or about the protein active sites and are highly conserved across species, as originally proposed by McConkey.6 ADK residues involved in quinary interactions are highly conserved within the Enterobacteriaceae family of Gram-negative bacteria (Figure 11 of the Supporting Information). The interaction surfaces that form between target proteins and cytosol are specific. We identified both hydro-phobic and hydrophilic patches on the surface of Trx, ADK, FKBP, and ubiquitin that are involved in quinary interactions. Mutating residues in ubiquitin9 near the identified patches or in the functional analogue of FKBP, Pin1,24 resulted in weaker target protein–cytosol interactions, consistent with a disruption of quinary interactions.
Protein quinary interactions are weak but highly specific, mediated by RNA–protein interactions. Quinary interactions may alter the physicochemical properties of proteins, i.e., Trx, or overlap protein active sites, i.e., Trx, ubiquitin, and FKBP. Such functional ramifications may provide a molecular basis for recent observations showing that cytosol affects protein stability7,50 and short-lived conformations important for protein activity.51,52 These results may also explain why drugs such as cisplatin, and ligands such as AMP, ADP, and ATP, bind differently to proteins in-cell from what is observed in vitro. Nonetheless, the question of whether quinary interactions affect protein function will need to be assessed on a case by case basis, because despite their pervasive nature, they appear to be unique for each protein.
CONCLUSIONS
Our observations provide compelling evidence that quinary interactions are linked to RNA biology and thus are important for regulating protein function and activity.4 The atomic resolution study of protein quinary interactions afforded by in-cell NMR adds another unique layer of complexity to physiological processes inside a living cell.
Supplementary Material
Leakage control of proteins during in-cell NMR experiments (Figure 1), Trx His tag that does not interact with cytosol (Figure 2), quantification of proteins overexpressed in E. coli (Figure 3), in-cell thioredoxin that is not modified by cisplatin (Figure 4), active site mutant of thioredoxin that engages in quinary interactions (Figure 5), quinary interactions of FKBP in E. coli (Figure 6), quinary interactions of FKBP that cannot be reconstituted in vitro with a high concentration of BSA (Figure 7), in-cell ADK that is not bound to ATP (Figure 8), chloramphenicol that alters the physiological state of ADK in E. coli (Figure 9), in-cell apparent molecular mass of ADK that is consistent with that of a total RNA–ADK complex (Figure 10), and quinary interactions of ADK that affect residues that are conserved in Enterobacteriaceae (Figure 11).
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant 5R01GM085006.
ABBREVIATIONS
- CRIPT
cross-correlated relaxation-induced polarization transfer
- HMQC
heteronuclear multiple-quantum coherence
- TROSY
transverse relaxation-optimized spectroscopy
- Trx
thioredoxin
- FKBP
FK506 binding protein
- ADK
adenylate kinase
- REDPRO
reduced proton density
- EDTA
ethylenediaminetetraacetic acid
- PDB
Protein Data Bank
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00036.
REFERENCES
- 1.Goodsell D. The machinery of life. Vol. 167. Springer; New York: 2009. [Google Scholar]
- 2.Persson E, Halle B. Cell water dynamics on multiple time scales. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6266–6271. doi: 10.1073/pnas.0709585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedek GB, Villars FMH. Physics: With Illustrative Examples from Medicine and Biology: Electricity and Magnetism. Springer-Verlag; New York: 2000. [Google Scholar]
- 4.Wirth AJ, Gruebele M. Quinary protein structure and the consequences of crowding in living cells: Leaving the test-tube behind. BioEssays. 2013;35:984–993. doi: 10.1002/bies.201300080. [DOI] [PubMed] [Google Scholar]
- 5.Chien P, Gierasch LM. Challenges and dreams: Physics of weak interactions essential to life. Mol. Biol. Cell. 2014;25:3474–3477. doi: 10.1091/mbc.E14-06-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConkey EH. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc. Natl. Acad. Sci. U.S.A. 1982;79:3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serber Z, Dotsch V. In-cell NMR spectroscopy. Biochemistry. 2001;40:14317–14323. doi: 10.1021/bi011751w. [DOI] [PubMed] [Google Scholar]
- 9.Inomata K, et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 10.Sakakibara D, et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 2009;458:102–105. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- 11.Bonetta L. Protein-protein interactions: Interactome under construction. Nature. 2010;468:851–854. doi: 10.1038/468851a. [DOI] [PubMed] [Google Scholar]
- 12.Burz DS, Dutta K, Cowburn D, Shekhtman A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods. 2006;3:91–93. doi: 10.1038/nmeth851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedberg DI, Selenko P. Live cell NMR. Annu. Rev. Biophys. 2014;43:171–192. doi: 10.1146/annurev-biophys-051013-023136. [DOI] [PubMed] [Google Scholar]
- 14.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR spectroscopy. Academic Press; New York: 2007. [Google Scholar]
- 15.Banci L, et al. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand K, Reverdatto S, Burz DS, Zitomer R, Shekhtman A. Structure of proteins in eukaryotic compartments. J. Am. Chem. Soc. 2012;134:12798–12806. doi: 10.1021/ja304809s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamatsu J, et al. High-resolution heteronuclear multidimensional NMR of proteins in living insect cells using a baculovirus protein expression system. J. Am. Chem. Soc. 2013;135:1688–1691. doi: 10.1021/ja310928u. [DOI] [PubMed] [Google Scholar]
- 18.Ogino S, et al. Observation of NMR Signals from Proteins Introduced into Living Mammalian Cells by Reversible Membrane Permeabilization Using a Pore-Forming Toxin, Streptolysin O. J. Am. Chem. Soc. 2009;131:10834–10835. doi: 10.1021/ja904407w. [DOI] [PubMed] [Google Scholar]
- 19.Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selenko P. In: In-cell NMR in Mammalian Cells. In The 25th ICMRBS. Delepierre A. L. a. M., editor. Vol. 26. Lyon; France: 2012. [Google Scholar]
- 21.Bekei B. Ph.D. Dissertation. Freie Universität Berlin; Berlin: 2013. In-cell NMR Spectroscopy in Mammalian Cells. [Google Scholar]
- 22.Ye Y, et al. 19F NMR spectroscopy as a probe of cytoplasmic viscosity and weak protein interactions in living cells. Chemistry. 2013;19:12705–12710. doi: 10.1002/chem.201301657. [DOI] [PubMed] [Google Scholar]
- 23.Harada R, Tochio N, Kigawa T, Sugita Y, Feig M. Reduced native state stability in crowded cellular environment due to protein-protein interactions. J. Am. Chem. Soc. 2013;135:3696–3701. doi: 10.1021/ja3126992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luh LM, et al. Molecular crowding drives active pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition. J. Am. Chem. Soc. 2013;135:13796–13803. doi: 10.1021/ja405244v. [DOI] [PubMed] [Google Scholar]
- 25.Li C, et al. Protein 19F NMR in Escherichia coli. J. Am. Chem. Soc. 2010;132:321–327. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riek R, Wider G, Pervushin K, Wuthrich K. Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4918–4923. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wider G. NMR techniques used with very large biological macromolecules in solution. Methods Enzymol. 2005;394:382–398. doi: 10.1016/S0076-6879(05)94015-9. [DOI] [PubMed] [Google Scholar]
- 28.Reckel S, Lopez JJ, Lohr F, Glaubitz C, Dotsch V. In-cell solid-state NMR as a tool to study proteins in large complexes. ChemBioChem. 2012;13:534–537. doi: 10.1002/cbic.201100721. [DOI] [PubMed] [Google Scholar]
- 29.Shekhtman A, Ghose R, Goger M, Cowburn D. NMR structure determination and investigation using a reduced proton (REDPRO) labeling strategy for proteins. FEBS Lett. 2002;524:177–182. doi: 10.1016/s0014-5793(02)03051-x. [DOI] [PubMed] [Google Scholar]
- 30.Riek R, Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. Solution NMR techniques for large molecular and supramolecular structures. J. Am. Chem. Soc. 2002;124:12144–12153. doi: 10.1021/ja026763z. [DOI] [PubMed] [Google Scholar]
- 31.Yokomizo A, et al. Cellular levels of thioredoxin associated with drug sensitivity to cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res. 1995;55:4293–4296. [PubMed] [Google Scholar]
- 32.Arnesano F, et al. Probing the Interaction of Cisplatin with the Human Copper Chaperone Atox1 by Solution and In-Cell NMR Spectroscopy. J. Am. Chem. Soc. 2011;133:18361–18369. doi: 10.1021/ja207346p. [DOI] [PubMed] [Google Scholar]
- 33.Segur J, Oberstar H. Viscosity of Glycerol and its Aqueous Solutions. Ind. Eng. Chem. 1951;43:2117–2120. [Google Scholar]
- 34.Reverdatto S, et al. Combinatorial Library of Improved Peptide Aptamers (CLIPs) to inhibit RAGE signal transduction in mammalian cells. PLoS One. 2013;8:e65180. doi: 10.1371/journal.pone.0065180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waudby CA, et al. In-cell NMR characterization of the secondary structure populations of a disordered conformation of α-synuclein within E. coli cells. PLoS One. 2013;8:e72286. doi: 10.1371/journal.pone.0072286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar JK, Tabor S, Richardson CC. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3759–3764. doi: 10.1073/pnas.0308701101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 38.Dong H, Qin S, Zhou HX. Effects of macromolecular crowding on protein conformational changes. PLoS Comput. Biol. 2010;6:e1000833. doi: 10.1371/journal.pcbi.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider DA, Gourse RL. Relationship between growth rate and ATP concentration in Escherichia coli: A bioassay for available cellular ATP. J. Biol. Chem. 2004;279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- 40.Pickart CM, Fushman D. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verkman AS. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem. Sci. 2002;27:27–33. doi: 10.1016/s0968-0004(01)02003-5. [DOI] [PubMed] [Google Scholar]
- 43.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 44.Freeberg MA, et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013;14:R13. doi: 10.1186/gb-2013-14-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 46.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 47.Kyne C, Ruhle B, Gautier VW, Crowley PB. Specific ion effects on macromolecular interactions in Escherichia coli extracts. Protein Sci. 2015;24:310–318. doi: 10.1002/pro.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffares DC, Poole AM, Penny D. Relics from the RNA world. J. Mol. Evol. 1998;46:18–36. doi: 10.1007/pl00006280. [DOI] [PubMed] [Google Scholar]
- 49.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 50.Monteith WB, Cohen RD, Smith AE, Guzman-Cisneros E, Pielak GJ. Quinary structure modulates protein stability in cells. Proc. Natl. Acad. Sci. U.S.A. 2015;112:1739–1742. doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Latham MP, Kay LE. A Similar In Vitro and In Cell Lysate Folding Intermediate for the FF Domain. J. Mol. Biol. 2014;426:3214–3220. doi: 10.1016/j.jmb.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 52.Latham MP, Kay LE. Probing non-specific interactions of Ca2+-calmodulin in E. coli lysate. J. Biomol. NMR. 2013;55:239–247. doi: 10.1007/s10858-013-9705-2. [DOI] [PubMed] [Google Scholar]
- 53.Burz DS, Shekhtman A. Current Protocols in Protein Science. Unit. Vol. 17. Wiley; New York: 2010. The STINT-NMR method for studying in-cell protein-protein interactions. p. 11. Chapter 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park ST, Aldape RA, Futer O, DeCenzo MT, Livingston DJ. PPIase catalysis by human FK506-binding protein proceeds through a conformational twist mechanism. J. Biol. Chem. 1992;267:3316–3324. [PubMed] [Google Scholar]
- 55.Xie J, Thapa R, Reverdatto S, Burz DS, Shekhtman A. Screening of small molecule interactor library by using in-cell NMR spectroscopy (SMILI-NMR). J. Med. Chem. 2009;52:3516–3522. doi: 10.1021/jm9000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15:532–534. [PubMed] [Google Scholar]
- 57.Dyson HJ, Holmgren A, Wright PE. Assignment of the proton NMR spectrum of reduced and oxidized thioredoxin: Sequence-specific assignments, secondary structure, and global fold. Biochemistry. 1989;28:7074–7087. doi: 10.1021/bi00443a044. [DOI] [PubMed] [Google Scholar]
- 58.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 59.Aden J, Verma A, Schug A, Wolf-Watz M. Modulation of a pre-existing conformational equilibrium tunes adenylate kinase activity. J. Am. Chem. Soc. 2012;134:16562–16570. doi: 10.1021/ja3032482. [DOI] [PubMed] [Google Scholar]
- 60.Masse JE, Keller R. AutoLink: Automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J. Magn. Reson. 2005;174:133–151. doi: 10.1016/j.jmr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 61.Schimmel PR, Cantor CR. Biophysical Chemistry: Part II; Techniques for the Study of Biological Structure and Function. W. H. Freeman; New York: 1980. p. 650. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Leakage control of proteins during in-cell NMR experiments (Figure 1), Trx His tag that does not interact with cytosol (Figure 2), quantification of proteins overexpressed in E. coli (Figure 3), in-cell thioredoxin that is not modified by cisplatin (Figure 4), active site mutant of thioredoxin that engages in quinary interactions (Figure 5), quinary interactions of FKBP in E. coli (Figure 6), quinary interactions of FKBP that cannot be reconstituted in vitro with a high concentration of BSA (Figure 7), in-cell ADK that is not bound to ATP (Figure 8), chloramphenicol that alters the physiological state of ADK in E. coli (Figure 9), in-cell apparent molecular mass of ADK that is consistent with that of a total RNA–ADK complex (Figure 10), and quinary interactions of ADK that affect residues that are conserved in Enterobacteriaceae (Figure 11).