ABSTRACT
The endocarditis and biofilm-associated pili (Ebp) are important in Enterococcus faecalis pathogenesis, and the pilus tip, EbpA, has been shown to play a major role in pilus biogenesis, biofilm formation, and experimental infections. Based on in silico analyses, we previously predicted that ATT is the EbpA translational start codon, not the ATG codon, 120 bp downstream of ATT, which is annotated as the translational start. ATT is rarely used to initiate protein synthesis, leading to our hypothesis that this codon participates in translational regulation of Ebp production. To investigate this possibility, site-directed mutagenesis was used to introduce consecutive stop codons in place of two lysines at positions 5 and 6 from the ATT, to replace the ATT codon in situ with ATG, and then to revert this ATG to ATT; translational fusions of ebpA to lacZ were also constructed to investigate the effect of these start codons on translation. Our results showed that the annotated ATG does not start translation of EbpA, implicating ATT as the start codon; moreover, the presence of ATT, compared to the engineered ATG, resulted in significantly decreased EbpA surface display, attenuated biofilm, and reduced adherence to fibrinogen. Corroborating these findings, the translational fusion with the native ATT as the initiation codon showed significantly decreased expression of β-galactosidase compared to the construct with ATG in place of ATT. Thus, these results demonstrate that the rare initiation codon of EbpA negatively regulates EbpA surface display and negatively affects Ebp-associated functions, including biofilm and adherence to fibrinogen.
IMPORTANCE
Enterococcus faecalis is among the leading causes of serious infections in the hospital setting, and the endocarditis and biofilm-associated pili (Ebp) have been shown to play significant roles in E. faecalis pathogenesis. Understanding the regulation of virulence is important for the development of new approaches to counteract multidrug-resistant pathogens. We previously predicted that ATT, which has been reported to start protein synthesis only in rare instances, is the most likely translational start codon of EbpA in E. faecalis. Here, we demonstrate that ATT is the initiation codon of EbpA and, relative to a constructed ATG start codon, results in smaller amounts of EbpA on the surface of the cells, attenuating biofilm formation and fibrinogen adherence, phenotypes associated with the ability of E. faecalis to cause infections. This provides the first example of pilus regulation through the use of an ATT initiation codon.
INTRODUCTION
Enterococcus faecalis is a Gram-positive commensal of the gastrointestinal tract of humans (1) but also ranks as one of the leading causes of hospital-acquired infections (HAIs) (2), including urinary tract infections (UTIs), bacteremia, and infective endocarditis (1, 3), among others. The endocarditis and biofilm-associated pili (Ebp) are considered important contributors to the pathogenicity of E. faecalis (4–7). We have previously shown that Ebp are important for adherence of E. faecalis OG1RF to host extracellular matrix (ECM) proteins, including fibrinogen and collagen, a process that is considered crucial in the initial steps of infection (8, 9). Furthermore, we have demonstrated that Ebp play a role in biofilm formation (6, 7) and in experimental animal models of endocarditis (7) and ascending urinary tract infections (UTIs) (4). The role of Ebp has also been established in a model of catheter-associated urinary tract infection (CAUTI) (5). The ebp locus consists of an operon of three genes, ebpA, ebpB, and ebpC, which encode the pilus subunits, or pilins, and bps, encoding a class C sortase (6, 7). The three pilin genes and the downstream sortase are cotranscribed, but a second promoter controls the independent expression of bps (6). Regulation of pilus expression has been shown to occur at the transcriptional level (10) through the action of two positive regulators, EbpR and rnjB (11, 12), while the fsr system was shown to be a weak repressor (13). Furthermore, environmental conditions, including bicarbonate (13) and the presence of serum (7), positively affect Ebp production.
Several reports have shown that EbpC forms the backbone polymer, while EbpA and EbpB are present at the tip and at the base of the pilus fiber, respectively (5–7, 14). In addition, our electron microscopy studies indicated that the majority of the EbpA protein is found on the surface of E. faecalis OG1RF cells (6), and analyses of the contribution of each structural subunit of the Ebp revealed the importance of EbpA to pilus biogenesis (5, 6, 14). We previously reported that deletion of ebpA resulted in the formation of fewer but extremely long pili compared to wild-type (WT) OG1RF, which suggested a role for EbpA in the initiation as well as termination of pilus polymerization (6). We corroborated the role of EbpA as a factor that influences the length of pili by controlled overexpression of EbpA from a nisin-inducible promoter in an ΔebpA mutant, which led to a gradual decrease in pilus length as the concentration of nisin was increased (6). Deletion of ebpA also affected the overall levels of the other Ebp pilin subunits, without altering the transcription levels of the downstream genes (6). EbpA has been demonstrated to play a crucial role in biofilm formation and in a model of urinary tract infection, with the ebpA deletion mutant attenuated approximately to the same level as that observed with the ebpABC operon deletion (6). In addition, Nielsen et al. showed the contribution of EbpA in the colonization of bladders and intrabladder implants in CAUTIs and revealed that the metal ion-dependent adhesion site (MIDAS) motif present in EbpA’s von Willebrand factor A (VWA) domain is important for pilus function (5). More recently, this group demonstrated that immunization with EbpA inhibits binding of E. faecalis to fibrinogen and provides protection in catheter-associated bladder infection in mice (15). Additionally, we showed that a monoclonal antibody targeting the shaft of the pili, EbpC, prevents endocarditis in rats (16).
We previously predicted that the most likely translational start codon of EbpA in E. faecalis OG1RF is the very rarely used triplet ATT (AUU in the corresponding mRNA), which is located 9 bp downstream of a suitable ribosomal binding site (RBS) and 120 bp upstream of the current genome database annotation of an ATG start codon (Fig. 1) (7). In the majority of mRNAs, translation initiates from the codon AUG that in prokaryotes codes for formylmethionine; nevertheless, other NUG codons are occasionally found as translational starts (17, 18). In only very few instances has the AUU codon been reported as the translational start for protein synthesis in bacteria (19, 20). The rarity of AUU as a start codon is attributed to the fact that initiation factor 3 (IF3) discriminates against noncanonical start codons (18). As observed in other prokaryotic species, in E. faecalis OG1RF, the use of ATT as the initiation of protein synthesis is very rare, and there is no evidence indicating that other E. faecalis proteins, besides EbpA, start translation with this codon. In addition, when our search of several other sequenced E. faecalis strains found conservation of this codon, we postulated that this codon participates in regulation of EbpA expression. To test this hypothesis and our prediction of ATT as the start codon of EbpA protein synthesis, we first used site-directed mutagenesis to introduce, in E. faecalis OG1RF, two successive stop codons between the ATT predicted initiation codon and the currently annotated ATG start codon, to experimentally show that the ATT indeed determines the start of EbpA protein synthesis. Then, we constructed a derivative of OG1RF in which the ATT codon of ebpA was replaced with ATG and investigated the effect of this change on translation, EbpA and EbpC surface display, biofilm formation, and adherence to fibrinogen.
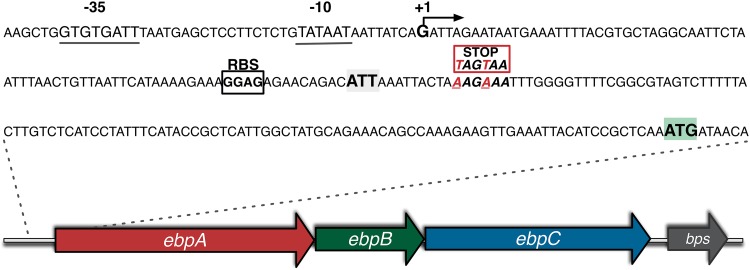
FIG 1 .
Schematic representation of the ebpABC-bps locus of E. faecalis OG1RF. The locus consists of three genes, ebpA, ebpB, and ebpC, encoding the pilin subunits, and bps, encoding a class C sortase. The putative promoter region (with the −35 and −10 promoter boxes), the predicted transcriptional start (+1), the predicted ribosomal binding site (RBS), the ATT postulated start codon, and the ATG annotated start codon are shown. The positions of the two successive stop codons introduced to generate strain TX5751 are also shown.
RESULTS
Introduction of stop codons indicates that ATT (AUU) is the start codon of EbpA protein synthesis.
The ebpA gene is the first gene of the ebpABC operon that encodes the E. faecalis Ebp (Fig. 1). The current genome annotation of the E. faecalis strain V583 indicates that EbpA is an 1,103-residue protein with a VWA domain; however, closer examination of the operon sequence revealed that no recognizable RBS is present upstream of the ebpA annotated start codon, ATG (7). In addition, no recognizable signal peptide or cleavage site was found downstream of the ATG annotated as the start codon of the predicted 1,103-residue EbpA protein (EbpA-1103) (see Fig. S1A in the supplemental material). We predicted that the most likely, but very unusual, start codon of ebpA is the triplet ATT, located 9 bp downstream of a recognizable RBS and 120 bp upstream of the currently annotated ATG start codon (Fig. 1). Using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/), we found a potential cleavage site at position 30 after the ATT codon (EbpA-1143) (see Fig. S1B). In addition, the other two proteins encoded by the ebpABC operon, EbpB and EbpC, have well-defined signal sequences as well as cleavage sites (see Fig. S1C and D). Furthermore, protein alignments of EbpA-1103 and EbpA-1143 with EmpA (EbpA homolog in Enterococcus faecium) support the ATT start for EbpA protein synthesis.
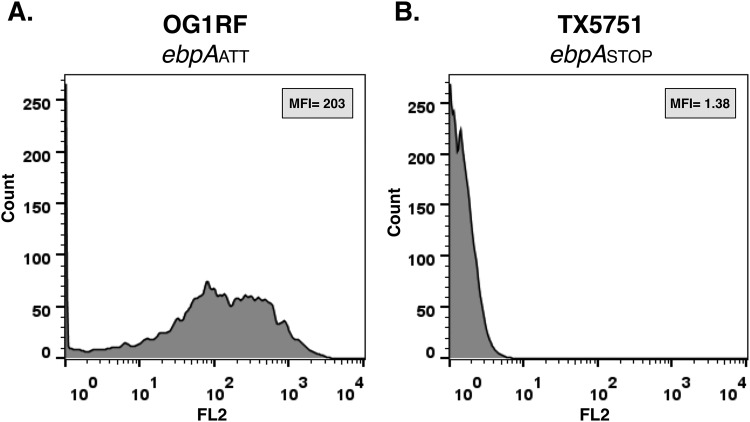
To experimentally confirm that the designated ATG (located 120 bp downstream of our predicted ATT start) is not the start codon for EbpA protein synthesis, we used site-directed oligonucleotide mutagenesis to generate the strain TX5751, in which two lysine residues, AAG and AAA at positions 5 and 6 after the ATT, respectively, were changed to two stop codons, TAG (amber) and TAA (ocher) (Fig. 1; Table 1). Our rationale was that, if translation starts from ATT, the introduction of a stop codon would signal termination of translation and no EbpA protein would be produced; for this purpose, we introduced two stop codons in order to avoid readthrough, as was reported when only one stop codon was inserted at the 5′ end of a chromosomal copy of a gfp reporter gene in Bacillus subtilis (21). In contrast, if the currently annotated ATG is the start of EbpA synthesis, the introduction of the stop codons before this ATG would not affect EbpA translation. As we predicted, flow cytometry showed that E. faecalis OG1RF displayed a strong EbpA signal (Fig. 2A), while strain TX5751 was negative for EbpA surface display (Fig. 2B). Next, we investigated the conservation of this unusual codon as the start of EbpA protein synthesis and found 100% conservation of the ATT codon in the 347 E. faecalis strains with available genome sequences in NCBI (data not shown). In addition, we interrogated the genome annotation for EbpA homologs in other enterococcal species, including E. faecium, Enterococcus hirae, Enterococcus casseliflavus, Enterococcus mundtii, and Enterococcus gallinarum and found that, in all but E. gallinarum, ATG was annotated as the start codon of these EbpA homologs (see Fig. S2 in the supplemental material). Furthermore, a recognizable RBS was present appropriately upstream of these ebpA-like genes’ ATG annotated start codons. Interestingly, no recognizable RBS was observed upstream of the annotated ebpA ATC start codon in E. gallinarum (see Fig. S2), raising the possibility of an alternative start for EbpA protein synthesis in this enterococcal species.
TABLE 1 .
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| OG1RF | Laboratory strain; Rifr Fusr | 35 |
| TX5751 | OG1RF with two point mutations at nucleotides 13 and 16 after the putative ebpA ATT initiation codon that changed two lysine residues (AAG and AAA) to two stop codons (TAG and TAA, respectively) (ebpASTOP) | This study |
| TX5731 | OG1RF with a point mutation that changed the ebpA putative ATT start codon ATT to ATG (ebpAATG) | This study |
| TX5732 | Restored OG1RF ebpAATT; the mutated ebpAATG codon was restored to wild-type ebpAATT | This study |
| TX5608 | OG1RFΔebpABC; ebpABC operon deletion mutant | 12 |
| TX5620 | OG1RFΔebpA; ebpA deletion mutant | 6 |
| E. coli | ||
| TG1 | E. coli host strain used for routine cloning | |
| EC1000 | E. coli host strain for cloning of RepA-dependent plasmids | 36 |
| Plasmids | ||
| pGEM-T Easy | Plasmid used for initial cloning of PCR fragments; Ampr | Promega |
| pHOU1 | Conjugative donor plasmid used for the introduction of point mutations into E. faecalis; confers Genr and carries the counterselectable pheS* gene | 31 |
| pSD2 | Plasmid used to construct the translational lacZ fusions; the lacZ gene lacks a promoter, an RBS, and a start codon; confers Ampr and Ermr | 32 |
| pTEX5749 | pSD2 plasmid containing a fragment from −261 bp upstream to 31 bp downstream of the ATT start codon of the ebpA gene of E. faecalis OG1RF(pSD2-ebpAATT::lacZ) | This study |
| pTEX5750 | pSD2 plasmid containing a fragment from −261 bp upstream to 31 bp downstream of the mutated ATG start codon of the ebpA gene of E. faecalis TX5731(pSD2-ebpAATG::lacZ) | This study |
Amp, ampicillin; Erm, erythromycin; Gen, gentamicin; Fus, fusidic acid; Rif, rifampin.
FIG 2 .
Flow cytometry analysis of EbpA surface display by wild-type E. faecalis OG1RF and TX5751. Flow cytometry profile of E. faecalis OG1RF (ebpAATT) (A) and TX5751 (ebpASTOP) (B), each grown in TSBG to exponential phase and labeled with anti-rEbpA antibody. The mean fluorescence intensity (MFI) is indicated.
The start codon ATT affects EbpA surface display.
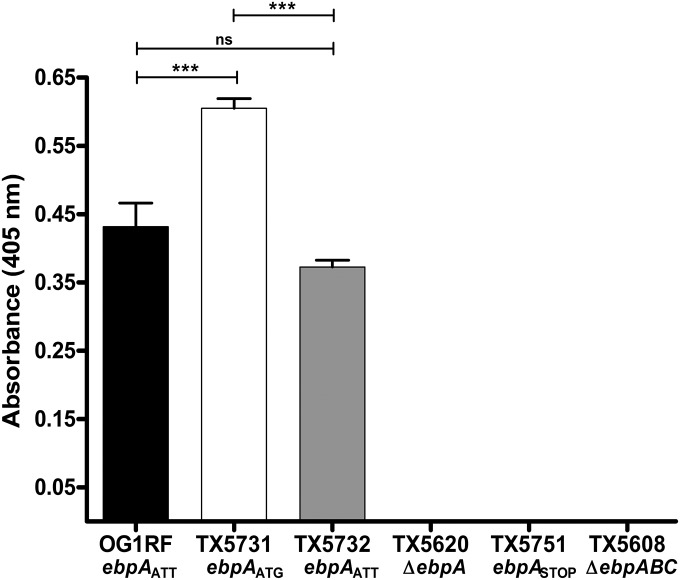
The use of a rare start codon as the translational start of EbpA, along with its conservation in all published E. faecalis genomes, led us to the hypothesis that this codon may play a role in the regulation of Ebp expression. To investigate this, we constructed a single-nucleotide variant, named TX5731, in which the ebpA ATT triplet of E. faecalis OG1RF was replaced with ATG (ebpAATG), and then explored the effect of this initiation codon change on the levels of EbpA surface display. Whole-cell enzyme-linked immunosorbent assay (WC-ELISA) using anti-recombinant EbpA (anti-rEbpA), performed after growing the cells to exponential phase in TSBG medium (tryptic soy broth supplemented with 0.25% [vol/vol] glucose), revealed that strain TX5731 carrying ATG as the initiation codon of ebpA had significantly increased amounts of EbpA on the surface compared to OG1RF (P < 0.001) (Fig. 3). To confirm that the differences observed between WT OG1RF and its ebpAATG mutant (TX5731) were due to the mutation in the ebpA start codon, we generated a revertant strain (TX5732), by replacing the ATG of TX5731 with the original ebpA start codon ATT (ebpAATT), in the native location. TX5732 displayed EbpA levels on the surface similar to those observed on WT OG1RF, demonstrating the role of the initiation codon in the regulation of EbpA protein levels (Fig. 3). In addition, WC-ELISA did not show any EbpA on the surface of the strain carrying the two consecutive stop codons after the ATT initiation codon, TX5751, nor on the ebpA deletion mutant, TX5620 (Fig. 3). EbpC surface display was also investigated under the same growth conditions, and the results revealed a small but nonsignificant increase in EbpC on the surface of TX5731 compared to OG1RF and TX5732 (see Fig. S3 in the supplemental material). As expected, no surface display of either EbpA or EbpC was observed on the surface of TX5608, the ebpABC operon deletion mutant (Fig. 3; see also Fig. S2) (6, 7). Similarly, when the cells were grown in BHI-S (brain heart infusion broth supplemented with 40% [vol/vol] horse serum), the ebpAATG mutant, TX5731, also showed a significant increase in surface display of EbpA compared to the ebpAATT counterparts, OG1RF and TX5732 (P < 0.001) (data not shown). Quantitation of surface-localized EbpA protein was also performed by flow cytometry. The mutant strain TX5731 with ATG showed increased levels of EbpA on the cell surface, compared to OG1RF and TX5732, corroborating the results obtained with WC-ELISA (data not shown).
FIG 3 .
Effect of the ebpA initiation codon on EbpA surface display measured by WC-ELISA. EbpA surface display was detected using anti-rEbpA. Bars represent the means of absorbance measured at 405 nm ± standard deviations (SD) from two independent experiments representing six wells per strain. The mean absorbance values were compared using ANOVA with Bonferroni’s post-test (***, P ≤ 0.001; ns, P > 0.05).
β-Galactosidase production is reduced by the presence of ATT as the start codon of an ebpA-lacZ fusion.
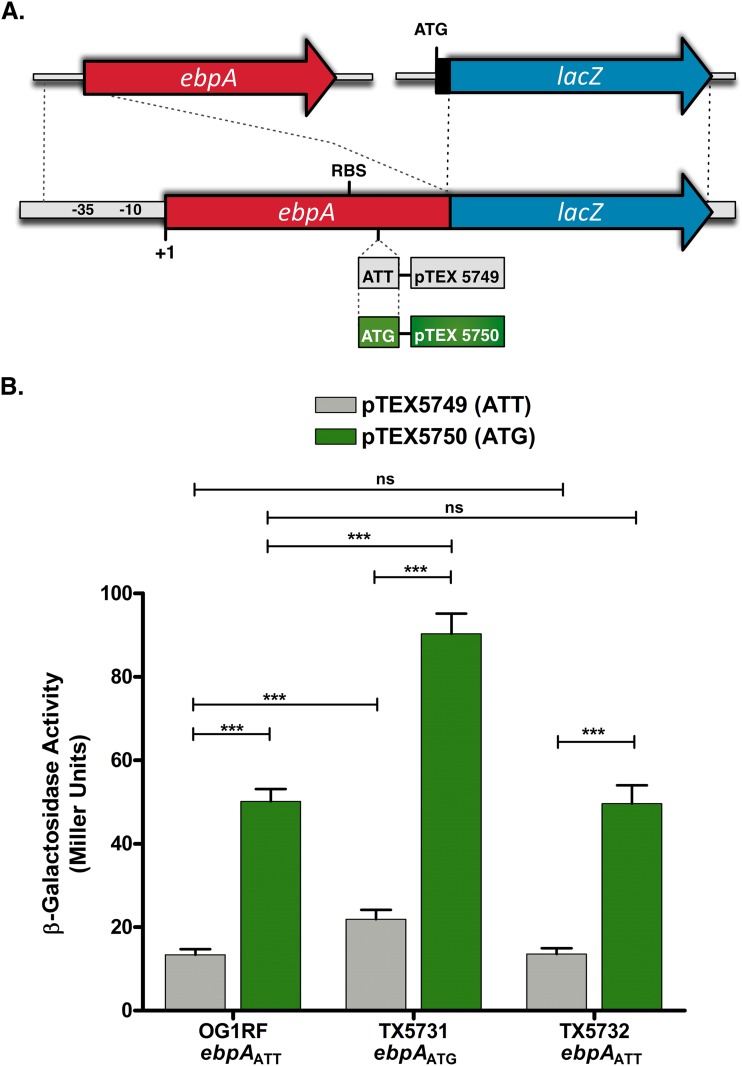
To explore the direct contribution of the EbpA initiation codon to translational efficiency, two translational reporter fusions, pTEX5749 and pTEX5750, were generated by amplifying a 292-bp fragment from −261, including the promoter region of ebpA, to 31 bp downstream of the translational start codon from E. faecalis OG1RF and TX5731, respectively (Fig. 4A). These fragments were fused to the reporter gene lacZ (Fig. 4A), and after electroporation of the fusion constructs into E. faecalis OG1RF, TX5731, and TX5732, β-galactosidase activity was assayed following growth in TSBG (Fig. 4B) and BHI-S (see Fig. S4 in the supplemental material). Under both growth conditions and in each of the strain backgrounds, β-galactosidase activity from cells carrying the reporter fusion pTEX5750 with ATG as the start codon was significantly greater than that from cells carrying the reporter fusion pTEX5749 with the triplet ATT as the initiation codon of translation (P < 0.001) (Fig. 4B; see also Fig. S4). It is interesting that strain TX5731 carrying either pTEX5749 or pTEX5750 expressed significantly higher levels of the reporter protein than did OG1RF and TX5732 carrying the corresponding reporter fusions (P < 0.001), which could suggest a positive-feedback loop controlling EbpA expression. In contrast, no differences in β-galactosidase activity were observed between OG1RF and TX5732 (Fig. 4B; see also Fig. S4). In addition, we observed that in BHI-S, β-galactosidase activity was increased approximately 3-fold over that in TSBG-grown cells in both the ATT (pTEX5749) and ATG (pTEX5750) constructs, but the relationship of β-galactosidase from the ATT and the ATG start codon was still maintained (see Fig. S4 versus Fig. 4B). Importantly, the differences in β-galactosidase activity are not a consequence of differences in growth rate, as OG1RF and its derivatives carrying the two plasmids, pTEX5749 and pTEX5750, exhibited equivalent growth kinetics (data not shown).
FIG 4 .
β-Galactosidase expression from ebpA::lacZ translational fusions is dependent on the identity of the start codon. (A) Schematic representation of the ebpA::lacZ fusions pTEX5749 (pSD2-ebpAATT::lacZ) and pTEX5750 (pSD2-ebpAATG::lacZ) carrying ATT and ATG as the start of EbpA::LacZ fusion protein synthesis, respectively. (B) β-Galactosidase activity in E. faecalis OG1RF and its ebpA start codon derivatives, TX5731 and TX5732, containing either pTEX5749 (gray bars) or pTEX5750 (green bars) after growth to mid-log phase in TSBG. Bars represent the means ± standard deviations of results from four independent assays, each with two duplicates. The mean values were compared using ANOVA with Bonferroni’s posttest (***, P ≤ 0.001; ns, P > 0.05).
ATT as the initiation codon of ebpA translation correlates with less biofilm formation.
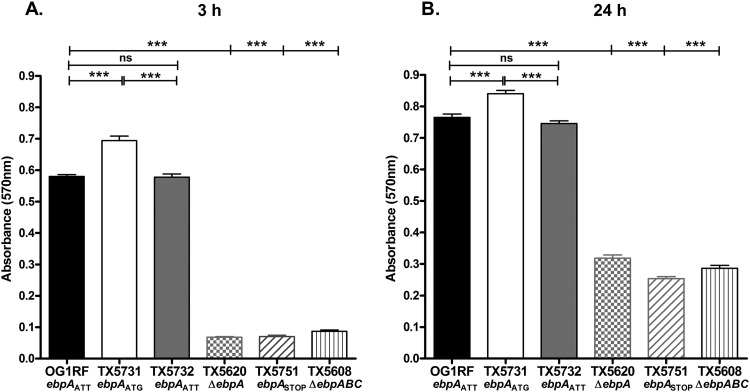
Previous studies showed that deletion of ebpA had a marked effect on the ability of E. faecalis OG1RF to form biofilm (6). We therefore investigated the impact of the ebpA initiation codon on early biofilm development (3 h) and on mature biofilm (24 h). When we scored biofilm after 3 h of static incubation, the strain carrying ATG as the initiation codon of EbpA, TX5731, showed a significant increase in biofilm density compared to the strains carrying ATT, WT OG1RF, and the revertant TX5732 (median for TX5731, 0.68, versus 0.58 and 0.57 for WT and TX5732, respectively; P < 0.001) (Fig. 5A). A smaller but still significant increase was observed in biofilm density after 24 h of static incubation of the ebpAATG mutant, TX5731, compared to OG1RF and TX5732 (P < 0.001). Consistent with previous findings (6, 7, 11, 12), a marked reduction in biofilm formation was observed when the ebpABC operon (TX5608) or ebpA (TX5620) had been deleted (P < 0.001) (Fig. 5A and B). In addition, TX5751, the strain in which two successive stop codons were introduced after the ATT EbpA start codon, showed reduction in biofilm density comparable to these two deletion mutants, corroborating the role played by EbpA in biofilm formation. The greater difference in biofilm density observed at the earlier time point than at 24 h between the strain carrying ATG as the start codon of EbpA (TX5731) and the strains carrying ATT (OG1RF and TX5732) may be related to the data of Bourgogne et al., who demonstrated that ebpA expression peaked at log phase, followed by a decline during stationary phase (13). Hence, our results corroborate the importance of EbpA in biofilm production and demonstrate that the levels of EbpA protein on the surface of the cells are important for E. faecalis biofilm formation, in particular during its initial stages.
FIG 5 .
Effect of the ebpA initiation codon on biofilm formation. Bacterial cells grown for 3 h (A) or 24 h (B) in TSBG were analyzed for biofilm formation using a crystal violet-based assay. Bars represent the means of absorbance at 570 nm ± standard deviations from four independent assays (40 wells per strain). ANOVA with Bonferroni’s posttest was used to compare biofilm density values (***, P ≤ 0.001; ns, P > 0.05).
ATT as the initiation codon of ebpA translation correlates with less binding to fibrinogen.
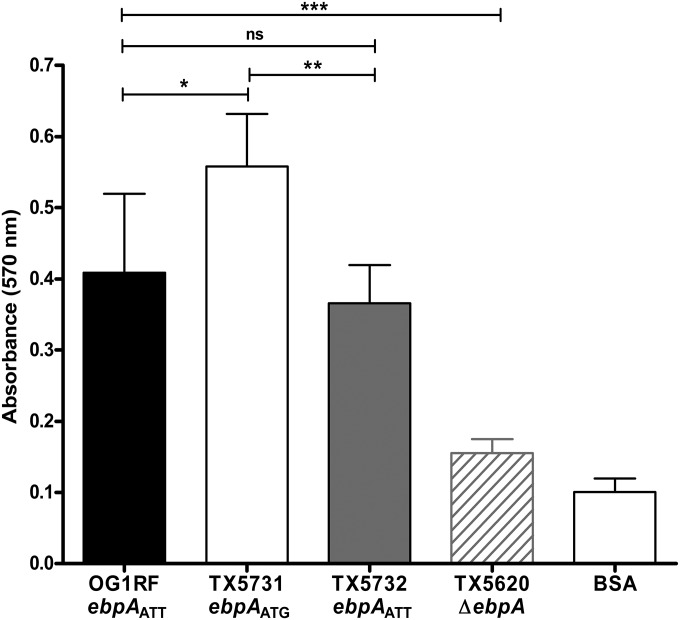
Nallapareddy et al. demonstrated that Ebp-deficient mutants of E. faecalis OG1RF showed reduced binding to fibrinogen and to collagen type I (8); in addition, Flores-Mireles et al. confirmed that EbpA mediates attachment of E. faecalis to host fibrinogen (15). Therefore, we investigated the abilities of OG1RF, its ebpAATG initiation codon mutant (TX5731), and the revertant strain ebpAATT (TX5732) to bind to fibrinogen. A slight but significant increase in binding to fibrinogen was observed when the initiation codon of ebpA was changed to ATG (TX5731) compared to OG1RF and TX5732 with the ATT initiation codon (P ≤ 0.05 and P ≤ 0.01, respectively) (Fig. 6). This result therefore indicates that the ATT initiation codon of EbpA also affects E. faecalis adherence to fibrinogen.
FIG 6 .
Effect of the ebpA initiation codon on the adherence of E. faecalis to immobilized fibrinogen. Bars represent means ± standard deviations of absorbance measured at 570 nm (for 4 wells per strain). The mean values between E. faecalis OG1RF and its derivatives were analyzed using ANOVA with Bonferroni’s posttest (***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05).
DISCUSSION
In bacteria, the most frequently used translational start codon is AUG (90% of the Escherichia coli mRNAs); however, alternative initiation codons, including GUG (8%) and UUG (1%), are occasionally found (17, 18). In contrast, the triplet AUU has been reported in only two instances in E. coli, namely, the pcnB gene encoding poly(A) polymerase (PAPI) (19) and the infC gene encoding translation initiation factor IF3 (20). In both instances, expression of the corresponding proteins was limited by the presence of AUU as the translational initiation codon (19, 20, 22). We analyzed the genome of E. faecalis OG1RF and determined that, in 81.5% of the open reading frames (ORFs), AUG is annotated to be the initiation codon while, in 10% and 8.5% of the instances, the codons GUG and UUG are predicted to initiate protein synthesis, respectively. However, we observed that the most likely translational start codon of EbpA in E. faecalis OG1RF is the rare triplet AUU (corresponding to ATT in the DNA) (Fig. 1), while the E. faecalis infC gene, encoding IF3, is annotated to start with the canonical ATG. The presence of a rare start codon as the most likely start of EbpA protein synthesis and its conservation in all sequenced E. faecalis strains led us to the hypothesis that this codon plays a role in the translational regulation of EbpA expression. First, we experimentally confirmed that ATG is not the start of EbpA translation by introducing two successive stop codons in between the ATT that we predicted is the initiation codon and the currently annotated ATG start codon of ebpA; as we expected, these stop codons abolished EbpA surface display (Fig. 2 and 3). Then, we constructed a derivative of OG1RF in which we replaced, in its native location, the ATT start codon of ebpA with ATG (TX5731) and then reverted this ATG to ATT (TX5732) and demonstrated that E. faecalis OG1RF and TX5732, carrying ATT as the start of EbpA protein synthesis, had reduced levels of EbpA on their surfaces compared to the strain TX5731, carrying ATG as the translational start (Fig. 3). We previously demonstrated that EbpA levels influence the length and number of pilus fibers, which led us to propose that EbpA is important for initiation as well as termination of pilus polymerization (6). It could be suggested that increased levels of EbpA protein observed on TX5731 with the ATG change (Fig. 3) may result in a small increase in the number of pilus fibers, albeit with decreased pilus length, consistent with a small (but nonsignificant) increase in EbpC surface display (see Fig. S3 in the supplemental material); however, we postulate that a major consequence of the ATT start codon is on EbpA levels exposed on E. faecalis cells, as we previously described that there are considerable amounts of EbpA monomers on the cell surface (6).
We inferred that the reduced levels of EbpA on the surface of E. faecalis OG1RF and TX5732 are a consequence of reduced rates of translation when ATT is present as the start of EbpA protein synthesis. Our results using translational reporter fusions to lacZ also indicate that, in E. faecalis, ATG is a more efficient start codon for the initiation of EbpA translation than ATT (Fig. 4; also see Fig. S4 in the supplemental material), which is in accordance with the hierarchy of start codon efficiencies proposed in E. coli (23). Although other signals and factors play a role in the rate of translation initiation (23), including the Shine-Dalgarno sequence and the initiation factors IF1, IF2, and IF3 (24), evidence in E. coli indicates that, in the presence of an ATT start codon, IF3 increases the dissociation of the initiation complex, which includes the 30S ribosomal subunit, the specific initiator tRNA, the mRNA, and the three initiation factors (18). It is interesting that β-galactosidase activity, which is a reflection of the levels of the EbpA-LacZ fusion protein inside the cell, was increased approximately 3.5-fold when the reporter fusion start codon was ATG versus ATT (Fig. 4), but a more modest increase (approximately 1.5-fold) was observed in EbpA surface display on TX5731 carrying ATG as the ebpA start codon than on the strains carrying ATT (OG1RF and TX5732) (Fig. 3). One possibility that could explain this difference would be the existence of additional regulatory mechanisms participating in the regulation of EbpA levels on the surface of the cells. In light of the current model of Ebp assembly in E. faecalis (6, 14), one could infer that modulating the ratios of the major backbone subunit, EbpC, to the minor subunits, EbpA and EbpB, could be important for pilus biogenesis, since an individual pilus fiber is composed of multiple EbpC subunits while in theory only one EbpA and one EbpB subunit are required for the tip and base of one pilus fiber, respectively (6, 14). Therefore, it seems plausible that the ATT start codon of ebpA is a way to regulate the ratio of EbpA to EbpC in E. faecalis.
Ebp are considered one of the major virulence factors of E. faecalis, playing a role in biofilm formation, adherence to fibrinogen, and the ability of E. faecalis to cause endocarditis and infection in mouse models of ascending UTI and CAUTI (4–8, 14, 15). EbpA has been demonstrated to be the most important pilin in biofilm formation (6, 15), while deletion of ebpC, encoding the major pilin, had a minor effect (6) despite abrogating pilus formation; this suggests that EbpA, in monomeric or dimeric form, on the surface of E. faecalis cells is capable of sustaining biofilm formation even when it is not part of a pilus polymer (6, 15). In addition, it has been shown that the MIDAS motif present in EbpA’s VWA domain is crucial for EbpA-mediated biofilm formation and fibrinogen binding (16). Proteins containing VWA domains, which are widely distributed among the three domains of life, Eukarya, Archaea, and Bacteria, often participate in cell adhesion and protein-protein interactions (25). Furthermore, other VWA-containing tip pilin proteins, including PilA of Streptococcus agalactiae and RrgA of Streptococcus pneumoniae, have also been implicated in binding to ECM proteins (26, 27). Considering the demonstrated role of EbpA in biofilm formation (6, 15) and fibrinogen adherence (16), we believed that identity of the EbpA start codon would impact these processes, which have been associated with the ability of E. faecalis to cause infection. Indeed, we found that the presence of ATT, compared to ATG, as the start codon of EbpA protein synthesis resulted in less biofilm formation (Fig. 5) and decreased adherence to fibrinogen (Fig. 6). Although the reasons behind the advantages or disadvantages of negatively regulating the levels of pilation are unknown, it has been suggested that high expression of pilin surface proteins could involve a fitness cost to the bacteria due to the selective pressure exerted by the immune system (28). Indeed, Danne et al. demonstrated in Streptococcus gallolyticus that a mutant overexpressing pili showed reduced survival in human blood compared to a nonpiliated mutant (28). In addition, they demonstrated that THP-1 human macrophages showed better opsonophagocytosis of highly piliated bacterial cells than did their nonpiliated counterparts. Although we cannot discard the possibility that ATT as the start codon of EbpA protein synthesis has a role in E. faecalis human infections, the conservation of this rare codon in all sequenced E. faecalis strains implies that this change appeared long before enterococci became common human colonizers and pathogens, and we speculate that its presence aided E. faecalis in some way in the environment or an early host, perhaps due to the decreased adherence observed in weakly piliated cells, thus favoring dispersal. Regardless of why this change occurred, our results, taken together, provide the first example of pilus regulation through the use of a very rare initiation codon and support our hypothesis that “ATTenuation starts with ATT.”
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine growth conditions.
Bacterial strains and plasmids used in this study and their relevant characteristics are listed in Table 1. E. faecalis strains were routinely grown at 37°C using brain heart infusion (BHI) agar (Becton, Dickinson [BD], France). Tryptic soy broth (BD) supplemented with 0.25% (vol/vol) glucose (TSBG) and BHI broth supplemented with 40% (vol/vol) horse serum (Sigma-Aldrich, Saint Louis, MO) (BHI-S) were used for some experiments. Escherichia coli strains used for cloning experiments were grown at 37°C in/on Luria-Bertani (LB) medium (BD). Growth characteristics of OG1RF and its derivatives were assessed in BHI broth based on the optical density at 600 nm (OD600). In addition, samples were taken at 0, 3, 6, 8, and 24 h for CFU determination on BHI agar, as previously described (29).
Construction of mutants.
Specific point mutations were generated by modifying a previously described methodology (30), based on the pHOU1 vector (31) that carries the pheS* allele that confers susceptibility to p-chloro-phenylalanine. Strains TX5751 and TX5731 were constructed using E. faecalis OG1RF as the parental strain, while a revertant strain, named TX5732, was generated by placing back the ebpAATT start codon into the TX5731 strain background (Table 1). In brief, two external primers, ebpA-Ext-F-BamHI and ebpA-Ext-R-PstI, containing the BamHI and PstI restriction sites, respectively, and two internal complementary primers containing the desired change were designed (see Table S1 in the supplemental material). First, two independent PCRs using the respective external and internal primers were carried out. Then, the two PCR amplicons were joined together by a crossover PCR and the generated fragments, now containing the desired mutations (confirmed by sequencing), were cloned into the pGEM vector and then subcloned into pHOU1 (31) using the BamHI and PstI sites. The recombinant pHOU1 plasmids were propagated in E. coli EC1000 and then electroporated into E. faecalis CK111 (30). Subsequently, the recombinant pHOU1 plasmids were transferred into E. faecalis OG1RF (or TX5731) by filter mating with CK111, followed by culturing the gentamicin-resistant colonies that integrated the plasmid on MM9–yeast extract-glucose (MM9YEG) medium supplemented with 10 mM p-chloro-phenylalanine as described in references 30 and 31, to select for the colonies from which the plasmid had excised. Sequencing and pulsed-field gel electrophoresis (PFGE) were performed to detect the mutations in the correct background.
Construction of translational lacZ fusion vectors and β-galactosidase assay.
The region extending from 261 bp upstream to 31 bp downstream ofhe postulated ebpA ATT translational start codon was PCR amplified from E. faecalis OG1RF and its ebpAATG mutant, TX5731, digested with SalI and BamHI, and cloned into the pSD2 vector (32) (Table 1). The reporter gene of pSD2, lacZ, lacks a promoter, an RBS, and a start codon. Constructs, harboring either the native (pTEX5749) or the mutated (pTEX5750) form of the ebpA start codon, were propagated in E. coli TG1 before electroporation into E. faecalis OG1RF, TX5731, and TX5732. β-Galactosidase activity was assayed in TSBG or BHI-S, as previously described (33).
Flow cytometry.
Flow cytometry analysis of E. faecalis strains was performed as previously described (12) with minor modifications. Cells grown in TSBG to mid-logarithmic phase were collected by centrifugation and washed twice with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS; pH 7.4). The bacterial cells were resuspended in 100 µl of the previously described affinity-purified polyclonal antibodies against EbpA (7) (1 µg/ml) and incubated for 30 min at room temperature (RT). After a washing step, secondary labeling was performed with a 1:100 dilution of phycoerythrin-conjugated goat anti-rabbit IgG for 30 min, and cells were washed and fixed with 1% paraformaldehyde for analysis using a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA).
WC-ELISA.
EbpA and EbpC surface display by OG1RF and the panel of mutants was evaluated as described above, with minor modifications (6). Briefly, bacteria grown overnight in TSBG were diluted in the same medium to an OD600 of 0.1 and incubated until they reached mid-logarithmic phase. Cells were collected by centrifugation and washed twice with PBS before they were resuspended in 50 mM carbonate-bicarbonate buffer, pH 9.6, to an OD600 of 1.0. Immulon 1B plate wells (Thermo Scientific, Woburn, MA) were coated for 1 h with E. faecalis cells, followed by two washes with PBS containing 0.05% Tween 20 (PBS-T). The wells were then blocked for 1 h with 2% bovine serum albumin (BSA), followed by a 1 h incubation with affinity-purified polyclonal antibodies against EbpA and EbpC, respectively (1:5,000 dilution of 1 mg/ml) (7). After three washes with PBS-T to remove the unbound antibodies, goat anti-rabbit F(ab′)2 fragment conjugated to alkaline phosphatase (AP) (Jackson ImmunoResearch Laboratories, West Grove, PA) (1:5,000 dilution) was added and incubated for 1 h. Next, the wells were washed twice with PBS-T and once with PBS, followed by the addition of AP substrate solution. The absorbance at 405 nm was measured with a microplate reader (Thermo Scientific, Waltham, MA).
Biofilm formation assay.
Biofilm density was measured as previously described (34), with some modifications. In brief, E. faecalis strains from overnight cultures in TSBG broth were diluted in the same medium to an OD600 of 0.1 and grown statically for 3 h or 24 h at 37°C in 96-well polystyrene plates (BD, Franklin Lakes, NJ). The plates were gently washed with PBS, and then the cells were fixed with Bouin’s solution (Sigma-Aldrich Co., St. Louis, MO) for 30 min. After two washes with PBS, bacterial cells were stained with a 1% crystal violet solution (Sigma-Aldrich Co., St. Louis, MO) for 30 min. Excess crystal violet was removed by rinsing thoroughly with distilled water followed by the addition of ethanol-acetone (80:20) to solubilize the dye and dissolve the biofilms. The absorbance at 570 nm was measured with a microplate reader (Thermo Scientific, Waltham, MA). Two independent experiments were performed in duplicate (8 wells per strain each in duplicate).
Fibrinogen binding assay.
E. faecalis adherence to immobilized fibrinogen was assayed using the CytoSelect cell adhesion assay kit (Cell Biolabs, San Diego, CA). First, E. faecalis OG1RF and its derivatives from overnight cultures grown at 37°C in BHI-S were normalized to an OD600 of 0.05 and cultured to mid-log phase. Bacterial cells were collected by centrifugation, washed three times with PBS, and resuspended in 0.5% BSA to an OD600 of 1.0. A volume of 150 µl of the cell suspension was added to the fibrinogen-precoated wells and incubated for 1 h at 37°C. The unbound bacteria were removed by gently washing each well two times with PBS. Next, 200 µl of the cell stain solution was added to each well and incubated for 10 min at RT, followed by two washes with deionized water. After air-drying the wells, 200 µl of extraction solution was added per well. The plate was incubated for 10 min on a shaker, and then 150 µl from each extracted sample was transferred to a 96-well microtiter plate (Becton, Dickinson, Franklin Lakes, NJ). The absorbance at 570 nm was measured with a microplate reader (Thermo Scientific, Waltham, MA).
Statistical analyses.
Analysis of variance (ANOVA) with Bonferroni’s multiple comparison posttest was used to compare the results. GraphPad Prism version 4.00 (GraphPad Software, San Diego CA, USA) was used for the statistical analyses.
SUPPLEMENTAL MATERIAL
Prediction of signal peptides and cleavage sites of the proteins encoded by the ebpABC operon. The presence and location of signal peptides and cleavage sites were predicted using the SignalP 4.1 server from the amino acid sequence of the annotated 1,103-residue EbpA protein (EbpA-1103) (A), the 1,143-residue EbpA protein (EbpA-1143) (B), EbpB (C), and EbpC (D). The C score (raw cleavage site score), S score (signal peptide score), and Y score (combined cleavage site score) are shown for each sequence. In addition, the average S score (mean S) of the possible signal peptide (from position 1 to the position immediately before the maximal Y score [combined cleavage site score]) and the D score (discrimination score), used to discriminate signal peptides from nonsignal peptides, are indicated. The D cutoff value used was 0.420. Download
DNA sequence alignment of 18 nucleotides upstream of the annotated initiation codon of E. faecalis ebpA and its homologs in five enterococcal species. The annotated start codons and RBSs are indicated for E. faecalis ebpA and its homologs in E. faecium (TX16), E. hirae (ATCC 9790), E. casseliflavus (EC20), E. mundtii (QU25), and E. gallinarum (A6981). Download
EbpC surface display measured by WC-ELISA. EbpC surface display was detected using affinity-purified polyclonal antibodies against EbpC. Bars represent the means of absorbance measured at 405 nm ± standard deviations from two independent experiments representing six wells per strain. The mean absorbance values were compared using ANOVA with Bonferroni’s posttest (ns, P > 0.05). Download
β-Galactosidase expression of an ebpA::lacZ translational fusion in BHI-S. β-Galactosidase activity in E. faecalis OG1RF and its ebpA start codon derivatives, TX5731 and TX5732, containing either pTEX5749 (gray bars) or pTEX5750 (green bars) after growth to mid-log phase in BHI-S. Bars represent the means ± standard deviations from four independent assays each with two duplicates. The mean values were compared using ANOVA with Bonferroni’s posttest (***, P ≤ 0.001; ns, P > 0.05). Download
Oligonucleotides used in this study.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 AI047923 from the Division of Microbiology and Infectious Diseases, NIAID, to B.E.M.
We thank Kavindra V. Singh for his scientific advice and Karen Jacques-Palaz, Emily Stinemetz, and Ken Pinkston for their technical assistance.
Footnotes
Citation Montealegre MC, La Rosa SL, Roh JH, Harvey BR, Murray BE. 2015. The Enterococcus faecalis EbpA pilus protein: attenuation of expression, biofilm formation, and adherence to fibrinogen start with the rare initiation codon ATT. mBio 6(3):e00467-15. doi:10.1128/mBio.00467-15.
REFERENCES
- 1.Murray BE. 1990. The life and times of the enterococcus. Clin Microbiol Rev 3:46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities . 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 3.Arias CA, Murray BE. 2012. The rise of the enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh KV, Nallapareddy SR, Murray BE. 2007. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis 195:1671–1677. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen HV, Guiton PS, Kline KA, Port GC, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. 2012. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. mBio 3(4):e00177-12. doi: 10.1128/mBio.00177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sillanpää J, Chang C, Singh KV, Montealegre MC, Nallapareddy SR, Harvey BR, Ton-That H, Murray BE. 2013. Contribution of individual Ebp pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS One 8:e68813. doi: 10.1371/journal.pone.0068813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, Erlandsen SL, Murray BE. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nallapareddy SR, Singh KV, Sillanpää J, Zhao M, Murray BE. 2011. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun 79:2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nallapareddy SR, Murray BE. 2008. Role played by serum, a biological cue, in the adherence of Enterococcus faecalis to extracellular matrix proteins, collagen, fibrinogen, and fibronectin. J Infect Dis 197:1728–1736. doi: 10.1086/588143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danne C, Dramsi S. 2012. Pili of gram-positive bacteria: roles in host colonization. Res Microbiol 163:645–658. doi: 10.1016/j.resmic.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Bourgogne A, Singh KV, Fox KA, Pflughoeft KJ, Murray BE, Garsin DA. 2007. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol 189:6490–6493. doi: 10.1128/JB.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. 2010. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol 192:5489–5498. doi: 10.1128/JB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourgogne A, Thomson LC, Murray BE. 2010. Bicarbonate enhances expression of the endocarditis and biofilm associated pilus locus, ebpR-ebpABC, in Enterococcus faecalis. BMC Microbiol 10:17. doi: 10.1186/1471-2180-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen HV, Flores-Mireles AL, Kau AL, Kline KA, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. 2013. Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J Bacteriol 195:4484–4495. doi: 10.1128/JB.00451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores-Mireles AL, Pinkner JS, Caparon MG, Hultgren SJ. 2014. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci Transl Med 6:254ra127. doi: 10.1126/scitranslmed.3009384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinkston KL, Singh KV, Gao P, Wilganowski N, Robinson H, Ghosh S, Azhdarinia A, Sevick-Muraca EM, Murray BE, Harvey BR. 2014. Targeting pili in enterococcal pathogenesis. Infect Immun 82:1540–1547. doi: 10.1128/IAI.01403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M. 1983. Comparison of initiation of protein synthesis in prokaryotes, eukaryotes, and organelles. Microbiol Rev 47:1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. 2005. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binns N, Masters M. 2002. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol Microbiol 44:1287–1298. doi: 10.1046/j.1365-2958.2002.02945.x. [DOI] [PubMed] [Google Scholar]
- 20.Sacerdot C, Fayat G, Dessen P, Springer M, Plumbridge JA, Grunberg-Manago M, Blanquet S. 1982. Sequence of a 1.26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J 1:311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyerovich M, Mamou G, Ben-Yehuda S. 2010. Visualizing high error levels during gene expression in living bacterial cells. Proc Natl Acad Sci U S A 107:11543–11548. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. 1996. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol Microbiol 21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- 23.O’Donnell SM, Janssen GR. 2001. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J Bacteriol 183:1277–1283. doi: 10.1128/JB.183.4.1277-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. 2009. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol 19:300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konto-Ghiorghi Y, Mairey E, Mallet A, Duménil G, Caliot E, Trieu-Cuot P, Dramsi S. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog 5:e1000422. doi: 10.1371/journal.ppat.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izoré T, Contreras-Martel C, El Mortaji L, Manzano C, Terrasse R, Vernet T, Di Guilmi AM, Dessen A. 2010. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18:106–115. doi: 10.1016/j.str.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Danne C, Dubrac S, Trieu-Cuot P, Dramsi S. 2014. Single cell stochastic regulation of pilus phase variation by an attenuation-like mechanism. PLoS Pathog 10:e1003860. doi: 10.1371/journal.ppat.1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sillanpää J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, Murray BE. 2010. Characterization of the ebpfm pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1:236–246. doi: 10.4161/viru.1.4.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristich CJ, Chandler JR, Dunny GM. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131–144. doi: 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panesso D, Montealegre MC, Rincón S, Mojica MF, Rice LB, Singh KV, Murray BE, Arias CA. 2011. The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BMC Microbiol 11:20. doi: 10.1186/1471-2180-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. 2012. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet 8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. 2011. Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO2-signalling. Mol Microbiol 82:634–647. doi: 10.1111/j.1365-2958.2011.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun 72:3658–3663. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Weinstock GM. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol 175:5216–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction of signal peptides and cleavage sites of the proteins encoded by the ebpABC operon. The presence and location of signal peptides and cleavage sites were predicted using the SignalP 4.1 server from the amino acid sequence of the annotated 1,103-residue EbpA protein (EbpA-1103) (A), the 1,143-residue EbpA protein (EbpA-1143) (B), EbpB (C), and EbpC (D). The C score (raw cleavage site score), S score (signal peptide score), and Y score (combined cleavage site score) are shown for each sequence. In addition, the average S score (mean S) of the possible signal peptide (from position 1 to the position immediately before the maximal Y score [combined cleavage site score]) and the D score (discrimination score), used to discriminate signal peptides from nonsignal peptides, are indicated. The D cutoff value used was 0.420. Download
DNA sequence alignment of 18 nucleotides upstream of the annotated initiation codon of E. faecalis ebpA and its homologs in five enterococcal species. The annotated start codons and RBSs are indicated for E. faecalis ebpA and its homologs in E. faecium (TX16), E. hirae (ATCC 9790), E. casseliflavus (EC20), E. mundtii (QU25), and E. gallinarum (A6981). Download
EbpC surface display measured by WC-ELISA. EbpC surface display was detected using affinity-purified polyclonal antibodies against EbpC. Bars represent the means of absorbance measured at 405 nm ± standard deviations from two independent experiments representing six wells per strain. The mean absorbance values were compared using ANOVA with Bonferroni’s posttest (ns, P > 0.05). Download
β-Galactosidase expression of an ebpA::lacZ translational fusion in BHI-S. β-Galactosidase activity in E. faecalis OG1RF and its ebpA start codon derivatives, TX5731 and TX5732, containing either pTEX5749 (gray bars) or pTEX5750 (green bars) after growth to mid-log phase in BHI-S. Bars represent the means ± standard deviations from four independent assays each with two duplicates. The mean values were compared using ANOVA with Bonferroni’s posttest (***, P ≤ 0.001; ns, P > 0.05). Download
Oligonucleotides used in this study.