ABSTRACT
Staphylococcus aureus is a prominent global nosocomial and community-acquired bacterial pathogen. A strong restriction barrier presents a major hurdle for the introduction of recombinant DNA into clinical isolates of S. aureus. Here, we describe the construction and characterization of the IMXXB series of Escherichia coli strains that mimic the type I adenine methylation profiles of S. aureus clonal complexes 1, 8, 30, and ST93. The IMXXB strains enable direct, high-efficiency transformation and streamlined genetic manipulation of major S. aureus lineages.
IMPORTANCE
The genetic manipulation of clinical S. aureus isolates has been hampered due to the presence of restriction modification barriers that detect and subsequently degrade inappropriately methylated DNA. Current methods allow the introduction of plasmid DNA into a limited subset of S. aureus strains at high efficiency after passage of plasmid DNA through the restriction-negative, modification-proficient strain RN4220. Here, we have constructed and validated a suite of E. coli strains that mimic the adenine methylation profiles of different clonal complexes and show high-efficiency plasmid DNA transfer. The ability to bypass RN4220 will reduce the cost and time involved for plasmid transfer into S. aureus. The IMXXB series of E. coli strains should expedite the process of mutant construction in diverse genetic backgrounds and allow the application of new techniques to the genetic manipulation of S. aureus.
INTRODUCTION
In order to gain an in-depth understanding of bacterial pathogen biology, genetic studies are required. However, a major impediment to genetic manipulation is the presence of restriction modification (RM) barriers, which hinder the uptake of foreign DNA (1–4). In the absence of host-specific methylation profiles, introduced DNA is recognized by the host as foreign and degraded. The RM barrier can help prevent infection by bacteriophage and transfer of conjugative plasmids and can inhibit the natural uptake of DNA via competence mechanisms (1, 5, 6). It can also prevent the transfer of recombinant plasmid DNA constructed in the laboratory workhorse Escherichia coli into organisms such as Staphylococcus aureus. The majority of clinical S. aureus isolates contain an active type IV system which recognizes systems specific to cytosine methylation (7) and clonal complex (CC) type I (8, 9) that can elaborate complex adenine methylation profiles. The type I RM barrier is comprised of three components, hsdM (methylase), hsdS (specificity), and hsdR (restriction). The HsdM2-HsdS1 protein complex recognizes a target recognition motif (TRM) dictated by the HsdS and detects the methylation status. Appropriately hemimethylated DNA (e.g., replicating chromosomal DNA) is then fully methylated, preventing restriction of the DNA by the HsdM2-HsdS1-HsdR2 complex (8). While most strains contain two active type I RM systems (the hsdMS carried on the alpha and beta pathogenicity islands and a single orphan HsdR gene) (10), a third complete HsdRMS can be found in some isolates (11). Additionally, a limited subset of S. aureus strains contain type II RM, which includes the enzyme Sau3AI that recognizes GATC motifs (12). Sau3AI sites on plasmids isolated from E. coli are blocked by Dam methylation. In S. aureus strains of clonal complex 398 (CC398), an active type II RM is encoded that transposes the type IV system. Digestion of CC398 genomic DNA with SmaI is prevented due to cytosine methylation of CCNGG motifs by the cognate type II methylase (13). Plasmids isolated from E. coli strain DC10B lacking cytosine methylation (DH10B Δdcm) can bypass the type IV barrier, but maximal transformation efficiency requires the presence of the CC-specific adenine methylation profile (3). Passage of plasmids through the restriction-deficient, modification-proficient S. aureus strain RN4220 (14) allows transfer into CC8/sequence type 239 (ST239) hosts at high efficiency. Transfer into other CCs is dependent on the TRM sequences on the particular plasmid. The laboratory S. aureus strain RN4220 has been invaluable as an intermediate host for the transfer of plasmid DNA into a restricted set of CCs. However, maintenance of plasmids in S. aureus is not ideal, as it requires additional time for growth at 30°C (temperature-sensitive plasmids) and greater cost (enzymatic lysis with lysostaphin) and provides plasmid DNAs of poor quality/yield compared to those isolated from E. coli.
A novel method to bypass host-encoded RM, termed plasmid artificial modification (PAM), has been described. Plasmid DNA is premethylated in an E. coli strain heterologously expressing methyltransferases of the target host. For Bifidobacterium breve (2) and Bifidobacterium adolescentis (15), the expression of type II methyltransferases in E. coli improved plasmid transfer 1,000 and 10,000-fold, respectively. More recently, a genomic approach was applied to restrictive clones of Bacillus cereus, Bacillus amyloliquefaciens, and Nitrobacter hamburgensis (4). A combination of type II and III RM genes were introduced into E. coli with improved plasmid transfer verified. To apply PAM, first the RM systems encoded by the strain in question need to be evaluated and, second, an appropriate method for the cloning and expression of the modification machinery needs to be devised.
To expand the repertoire of clinically relevant S. aureus strains that can be manipulated routinely in the laboratory, we have developed CC-specific cloning hosts that stably express CC-specific type I modification genes from the chromosome of E. coli DC10B, creating the IMXXB series of E. coli strains (XX denotes CC).
RESULTS
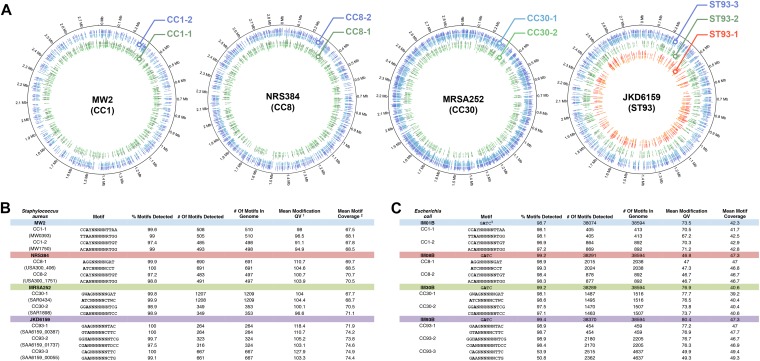
Employing the ability of Pacbio single-molecule, real-time (SMRT) sequencing to detect adenine-methylated DNA (16), we sequenced the genomes of the prototypical methicillin-resistant S. aureus (MRSA) strains MW2 (CC1), NRS384 (CC8), and MRSA252 (CC30) and identified the adenine methylation of two independent TRMs, with each strain harboring two active HsdS alleles (Fig. 1A and B). These results confirmed the TRMs for CC1 and CC8 strains characterized by Roberts et al. (17). We also identified an adenine methylation pattern for MRSA252 that fits with previously uncharacterized HsdS sequences. The sequencing of a fourth strain, JKD6159 (ST93) (11), which is completely refractory to transformation with RN4220 plasmid DNA, revealed the presence of three active, novel HsdS sequences and TRM sites (Fig. 1A and B).
FIG 1 .
SMRT sequencing for the identification of adenine methylation in S. aureus and engineered E. coli. (A) The target recognition motifs (TRMs) for the type I HsdMS systems encoded by each S. aureus strain were identified, and the positions of methylated adenine residues on the chromosome plotted with Circos (34). Each adenine methylation on the chromosome is represented by a line whose length corresponds with the interpulse duration of the read. (B and C) PacBio reads were analyzed by the SMRT suite pipeline version 2.2.0/motif finder 1.3.1 to identify conserved adenine-methylated residues (in boldface) and the TRMs for the HsdS alleles of each S. aureus (B) and IMXXB E. coli (C) strain.1, Mean modification QV is defined as the quality value of the base calls within the motif.2, Mean motif coverage is defined as the average depth of read coverage within a motif.3, GATC methylation encoded by dam in the E. coli strain is not present in S. aureus.
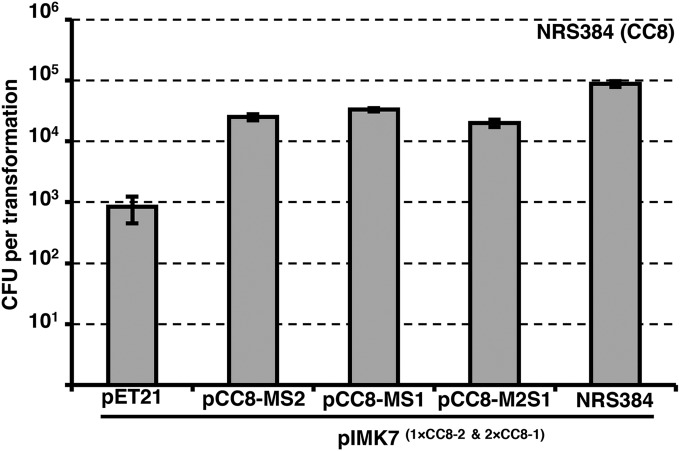
We then attempted to confer on E. coli a CC8 adenine methylation profile. The CC8-1 (nomenclature consistent with Roberts et al. [17]) hsdMS, the CC8-2 hsdMS, and a CC8-2 hsdM/CC8-1 hsdS hybrid were expressed using an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible system (pET21) in E. coli strain BL21 (naturally dcm deficient). The shuttle plasmid pIMK7, which contains two CC8-1 and one CC8-2 TRM, was transformed into the BL21 strains. Methylation of the TRMs was judged to have occurred by the improved transformation of pIMK7 into NRS384 compared to the results for plasmid isolated from the empty pET21 control strain. The frequency of pIMK7 transformation into NRS384 when all CC8-1 and CC8-2 TRM sites were methylated (plasmid isolated from NRS384) was 100-fold greater than for pIMK7 isolated from BL21 containing pET21 (Fig. 2), with the presence of either CC8-1 or CC8-2 methylation alone improving the transformation frequency 30-fold (Fig. 2). Furthermore, we could show that HsdM (CC8-2) could functionally interact with HsdS (CC8-1), because pIMK7 isolated from BL21 containing pCC8-M2S1 improved plasmid transfer ~25-fold, and that maximal transformation efficiency with pIMK7 was dependent on complete methylation of both TRM sites (Fig. 2).
FIG 2 .
Transformation of NRS384 with HsdMS-methylated plasmid. Three different CC8 HsdMS combinations were expressed from an IPTG-inducible plasmid (pET21) in E. coli BL21 (including the hybrid hsdM2-hsdS1). The coextracted shuttle vector pIMK7 (5 µg total plasmid DNA) was transformed into CA-MRSA strain NRS384. pIMK7 isolated from NRS384 was included as the maximal transformation of fully modified plasmid. The number of TRMs on pIMK7 for CC8 strains are indicated next to the plasmid name. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows data representative of the data from three independent experiments.
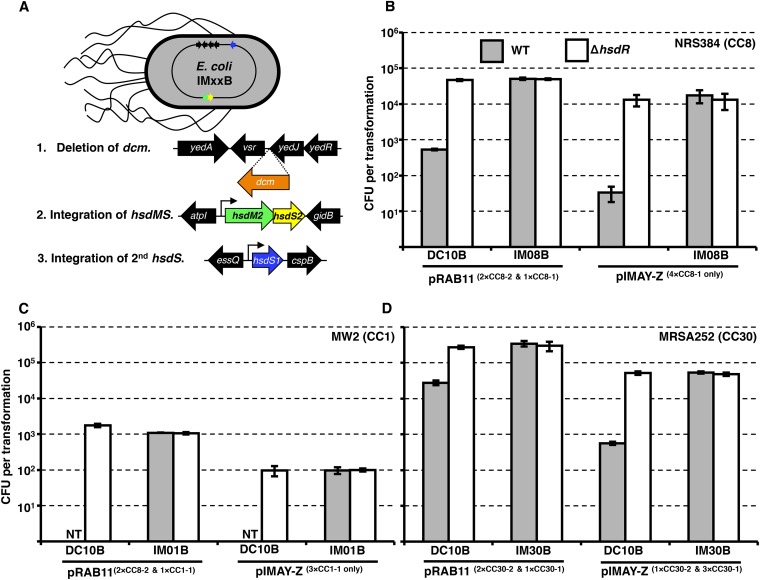
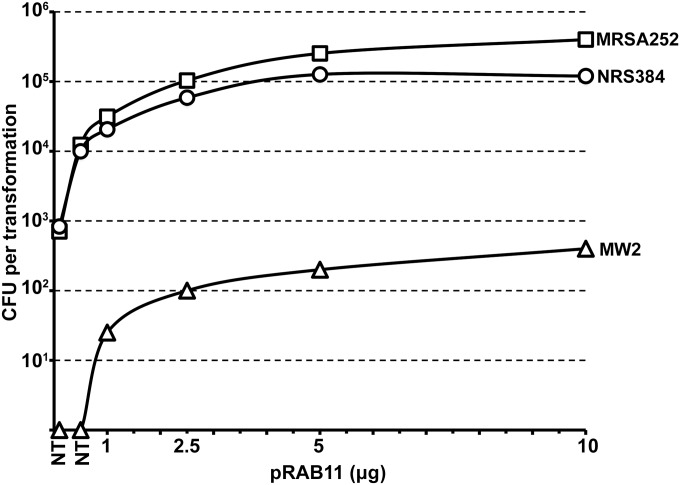
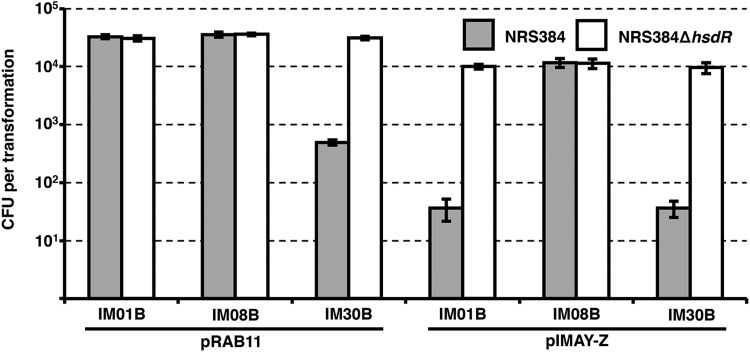
From here, we attempted to express HsdMS CC8-2 from the native or an intermediate or highly expressed promoter on a low-copy-number plasmid (p15A replicon, pIMK series [18]) in the E. coli DC10B background. However, we were unable to obtain positive clones from the intermediate or highly expressed constructs and loss of function was observed upon serial passage of the E. coli cells containing the CC8-2 hsdMS driven by the native promoter (data not shown). These results prompted us to examine the feasibility of integrating the hsdMS genes within neutral locations on the DC10B chromosome to improve stability (19) and to produce unmarked E. coli strains (Fig. 3A). Initially, we attempted to create a CC8 E. coli cloning host to express the equivalent methylation profile of RN4220. By spliced overlap extension (SOE) PCR, we assembled in vitro one set of hsdMS genes (CC8-2), expressed from a strong constitutive promoter. The fragment was targeted by recombineering to a region of the DC10B chromosome between the atpI and gidB genes. This yielded E. coli SA08B after removal of the cat gene. We took advantage of the interaction between HsdM and HsdS to introduce the second hsdS (CC8-1) expressed from the strong coliphage promoter PN25 (20) between the essQ and cspB genes of SA08B. This yielded E. coli strain IM08B after removal of the kanamycin resistance marker. SMRT sequencing of IM08B genomic DNA revealed the methylation of TRM sites identical to those modified in S. aureus strain NRS384 (Fig. 1C) with no detectable off-target modifications by IM08B. Importantly, the efficiency of methylation, as judged by the percentage of motifs methylated in the E. coli background, was equivalent to that observed in NRS384 (Fig. 1B and C). To investigate the effect of plasmid methylation by IM08B, either pRAB11 (two CC8-2 TRMs and one CC8-1 TRM) or pIMAY-Z (four CC8-1 TRMs) was isolated from DC10B (lacks type IV restriction) or IM08B and transformed into NRS384, with NRS384 ΔhsdR used as a control. The presence of a functional methylation profile was confirmed by the improved transformation efficiencies of pRAB11 (~100-fold) and pIMAY-Z (~500-fold) compared to those of the plasmids isolated from DC10B (Fig. 3B). Using the protocol described for making electrocompetent S. aureus cells, we observed a linear increase in the number of transformants recovered with pRAB11 isolated from IM08B for up to 5 µg of plasmid DNA transformed (Fig. 4). Similar results were also observed for pRAB11 isolated from E. coli strains IM01B and IM30B, described below.
FIG 3 .
Construction and characterization of IMXXB E. coli strains. (A) Schematic of the construction of an E. coli strain expressing CC-specific hsdMS alleles from strong promoters at neutral locations in the DC10B (DH10B Δdcm) chromosome. (B to D) Transformation profiles of S. aureus strains (grey bars) and their isogenic hsdR mutants (defective in type I restriction) (white bars) with plasmid pRAB11 (5 µg) or pIMAY-Z (2.5 µg) isolated from DC10B or the respective IMXXB strain of E. coli. The number of TRMs on either plasmid for the CC of the strain is denoted next to the plasmid name. NT, no transformants were detected. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows data representative of the data from three independent experiments.
FIG 4 .
Effect of plasmid concentration for S. aureus electroporation. Different concentrations of plasmid DNA (pRAB11 at 0.1, 0.5, 1, 2.5, 5, and 10 µg) were isolated from E. coli strains IM01B, IM08B, and IM30B for transformation into S. aureus MW2, NRS384, and MRSA252, respectively. A dose-dependent response was observed for up to 5 µg of plasmid with pRAB11. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment.
To extend these results, we sought to introduce the methylation profiles of CC1 (USA400 lineage) and CC30 strains that include common methicillin-susceptible S. aureus (MSSA), MRSA, and community-associated (CA)-MRSA strains (21) into DC10B. As S. aureus strains from CC8 and CC1 share the hsdS allele denoted above as CC8-2 (CC1-2 by Roberts et al. [17]), we introduced the CC1-1 PN25-hsdS into SA08B, creating the CC1 E. coli strain IM01B. Acquisition of the CC1-1 methylation profile in the CC8-2 E. coli background was confirmed by SMRT sequencing (Fig. 1C). We were unable to transform the prototypical CC1 S. aureus strain MW2 (USA400) with pRAB11 (two CC8-2 TRMs and one CC1-1 TRM) or pIMAY-Z (three CC1-1 TRMs) isolated from DC10B. However, plasmid DNA methylated with the CC1 profile exhibited at minimum 100- to 1,000-fold improvements in transformation compared to that of plasmids from DC10B (Fig. 3C). A similar approach was used to integrate the CC30-2 hsdMS and the CC30-1 hsdS genes into the chromosome of DC10B (strain IM30B). The presence of both adenine methylation profiles was confirmed by SMRT sequencing (Fig. 1C). MRSA252 was the most transformable of the S. aureus strains tested with plasmids isolated from DC10B (Fig. 3D), yet IM30B improved pRAB11 (two CC30-2 TRMs and one CC30-1 TRM) transfer 12-fold and pIMAY-Z (one CC30-2 TRM and three CC30-1 TRMs) transfer 100-fold. Plasmids isolated from DC10B or IMXXB strains transformed their respective S. aureus ΔhsdR mutants equivalently, confirming that transformation efficiency is dependent on the plasmid methylation profile and type IV/type I RM barriers.
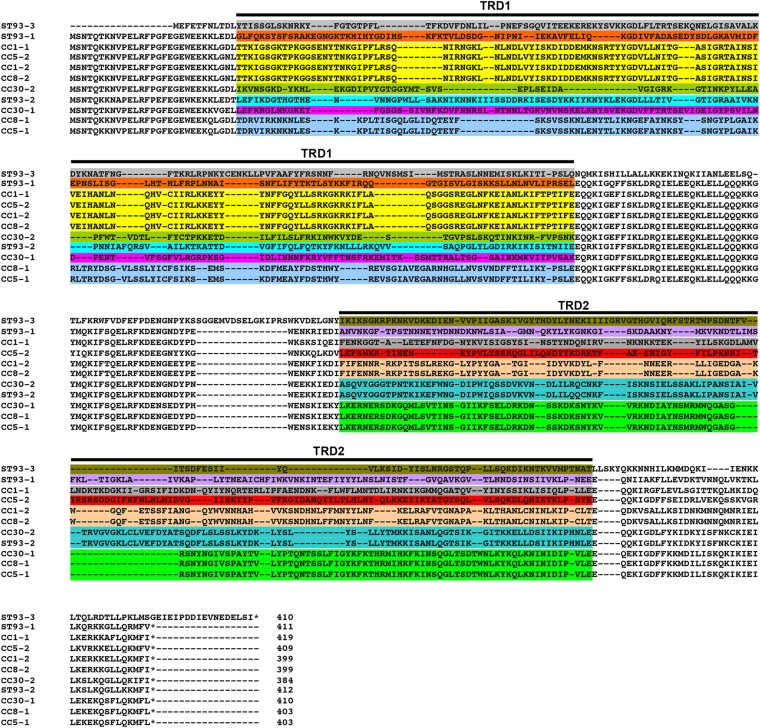
Alignment of the 11 HsdS amino acid sequences (including CC5-1/CC5-2 described previously [17]) allowed us to decipher the TRMs recognized by each of the CC30 HsdS proteins (Table 1). HsdS contains two target recognition domains, designated TRD1 and TRD2 (Fig. 5). Conserved domains were observed between TRD1 of CC1-1, CC1-2, CC5-2, and CC8-2, correlating with the recognition of a CCAY sequence. HsdS from CC8-2/CC1-2 share identical TRD2s that correlate with the recognition of a TGT motif. TRD2s from CC5-1, CC8-1, and CC30-1 are also conserved, with a GAT motif in common between the three alleles. By exclusion, the motif GWAGN5GAT was attributed to CC30-1 and GGAN7TCG to CC30-2 (underlining indicates sites of adenine methylation). Furthermore, the TCG sequence is shared by the CC30-2/CC93-2 HsdS alleles that validated the CC30-2 prediction. In all, recognition sequences for 14/18 of the TRD could be attributed (Table 1) from the SMRT sequencing data.
TABLE 1 .
Assignment of the TRM arms to each TRDa
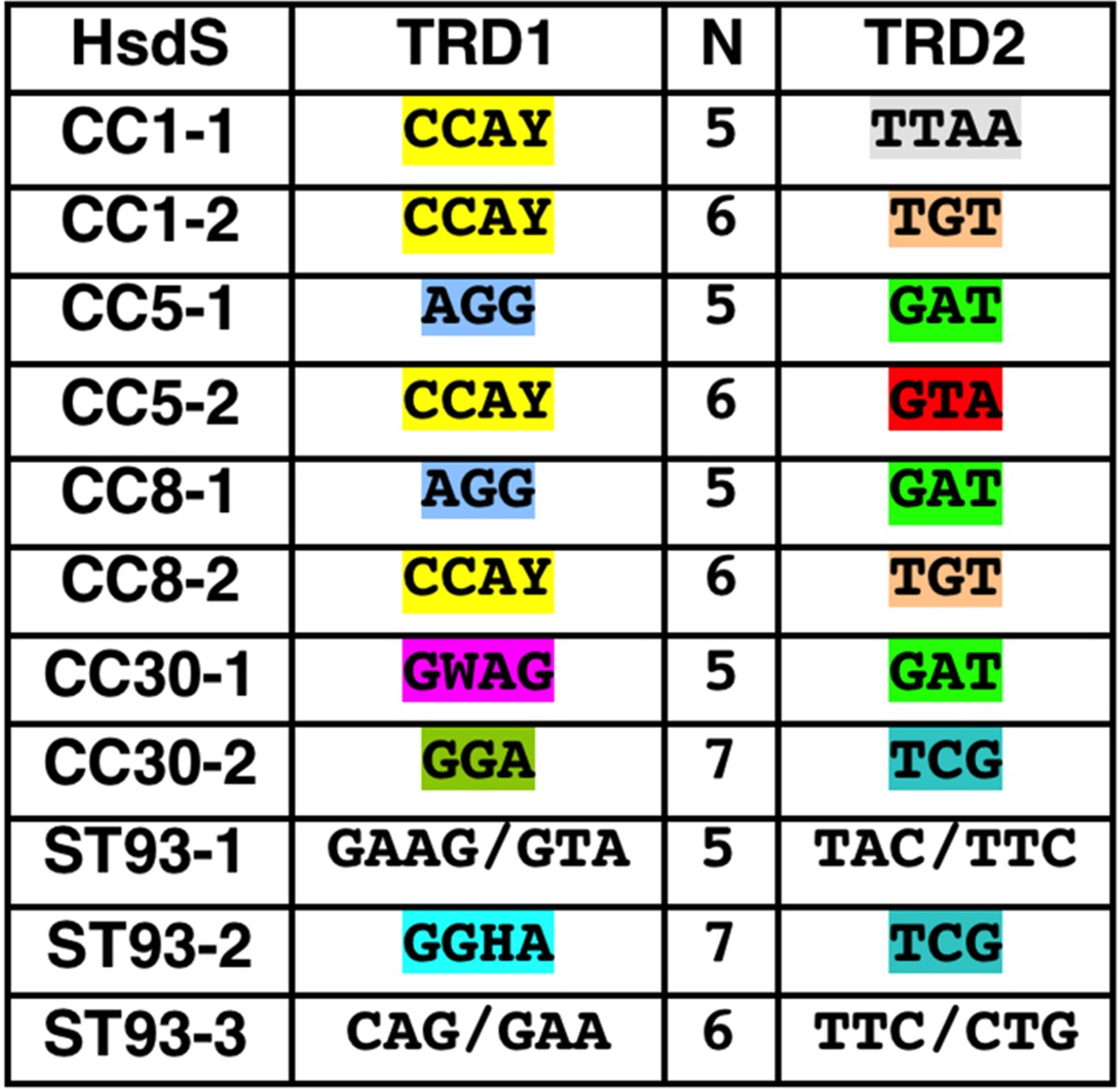
Based on the protein alignment (color coded as in Fig. 5) and TRM (obtained from SMRT sequencing), the DNA motif recognized by the TRD was assigned. For ST93-1/-3, it was not possible to assign the TRM for the TRD.
FIG 5 .
Determination of the TRM for each TRD. Clustal Omega alignment of HsdS proteins from CC1, CC5, CC8, CC30, and ST93. The protein sequences of the HsdS variants were aligned with Clustal Omega, and TRDs that match exactly are color coded in the same color.
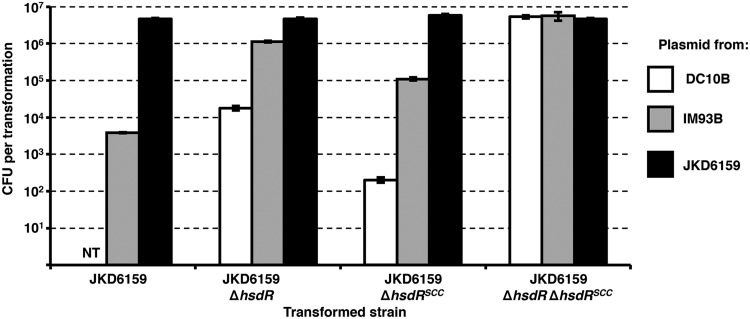
Transformation of S. aureus JKD6159 with plasmid DNA isolated from DC10B, IM93B (ΩhsdMS-2/S-1ΩhsdMS-3; see Materials and Methods for strain construction), and JKD6159 shed light on the motifs recognized by the unassigned HsdS alleles. The creation of IM93B allowed us to routinely transform plasmid into JKD6159. We observed 1,000 times fewer transformants when transforming plasmid from IM93B into JKD6159 compared to the rate of transformation of JKD6159 ΔhsdRSCCmec ΔhsdR, which is deficient in all type I restriction systems (Fig. 6). The plasmid used for the transformation experiments (pRAB11) contained multiple copies of each JKD6159 hsdS TRM. SMRT sequencing of IM93B showed that one of the three alleles was impaired in activity (only ~50% of the motifs were methylated). This was attributed to the HsdS CC93-3 encoded by the gene lying adjacent to the staphylococcal cassette chromosome mec element (SCCmec), as plasmid DNA isolated from IM93B transformed into JKD6159 ΔhsdRSCC 10 times less efficiently than into JKD6159 ΔhsdR. This result is consistent with impaired methylation from CC93-3 in IM93B (Fig. 6). Analysis of the genome sequence of IM93B identified a point mutation at the 3′ end of the CC93-3 hsdS gene, leading to a predicted amino acid change of E to G at position 402 (E402G), which we speculate might reduce the activity of the enzyme. This allowed us to assign the TRMs recognized by the CC93-1 (GAAGN5TAC) and CC93-2 (GGHAN6TCG) HsdS proteins.
FIG 6 .
Transformation of JKD6159 and type I restriction mutants with pRAB11. Plasmid pRAB11 was isolated from E. coli strains DC10B and IM93B and S. aureus strain JKD6159 for transformation into JKD6159 and the respective type I-deficient mutants. pRAB11 contains 4 TRM sites in ST93-1, 2 in ST93-2, and 2 in ST93-3. The transformation efficiencies were expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment. NT, no transformants were isolated.
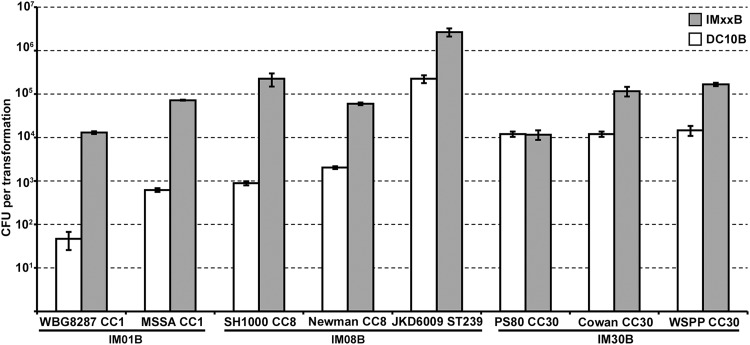
Finally, we tested the transfer of plasmid DNA isolated from IM01B, IM08B, and IM30B into S. aureus strain NRS384. We had earlier observed that pIMK7 required both CC8-1 and CC8-2 methylation for maximal transformation efficiency. Plasmid pRAB11 transferred equally efficiently from IM01B and IM08B, with plasmid isolated from IM30B transferred at a level equivalent to that of the unmethylated plasmid from DC10B (Fig. 7). IM01B and IM08B methylate the two CC8-2 TRM sites present on pRAB11, with one additional different TRM methylated by each strain. IM01B still transferred pRAB11 at high efficiency even though a CC1 methylation profile was present on the plasmid. Therefore, in this case, only one of the two TRMs required modification for maximal plasmid transfer and off-target methylation was tolerated, which was different from the results for pIMK7. Only IM08B allowed the transfer of pIMAY-Z at high efficiency. IM01B and IM30B did not methylate the CC8-1 TRMs (4 sites), and additional off-target methylation patterns led to full type I restriction (plasmid isolated from IM01B/IM30B) (Fig. 7). Analysis of the HsdS repertoire of an S. aureus recipient strain and sequence analysis of the plasmid to be transformed will allow the use of the E. coli strains described here to be applied to S. aureus strains from other CCs. For example, we have found that pIMAY-Z isolated from IM08B had improved transformation efficiency into CC5 S. aureus strains (data not shown). CC5 strains share a CC8-1 hsdS allele (Fig. 5, CC5-1) (17) and encode a novel CC5-2 hsdS allele. Plasmid pIMAY-Z contains both TRMs, but CC8-1 methylation alone appears sufficient to significantly improve transformation efficiency into an S. aureus CC5 strain. We have tested additional S. aureus isolates from CC1, CC8/ST239, and CC30 for their ability to accept plasmid (pRAB11) isolated from either DC10B or the respective IMXXB strain (Fig. 8). Except for PS80 (CC30), we observed improved transformation with plasmid isolated from IMXXB over plasmid isolated from DC10B, showing the general applicability of the IMXXB strains to bypass restriction in these CCs.
FIG 7 .
Transformation of S. aureus NRS384 and NRS384 ΔhsdR with plasmid isolated from IMXXB E. coli. Plasmid pRAB11 or pIMAY-Z was isolated from E. coli strains IM01B, IM08B, and IM30B for transformation into a CC8 host (NRS384) and the respective hsdR mutant. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment.
FIG 8 .
Transformation of additional representative isolates from CC1, CC8/ST239, and CC30 with plasmid isolated from IMXXB strains. Plasmid pRAB11 (5 µg) was isolated from either E. coli strain DC10B or the compatible IMXXB strain for transformation into additional representative S. aureus isolates of CC1, CC8/ST239, and CC30. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment.
DISCUSSION
The method described here is based on engineering E. coli to express active type I hsdMS genes from S. aureus, but this approach can equally be applied to other bacterial species where type I RM is a barrier to genetic manipulation, such as Streptococcus spp., Staphylococcus spp., and E. coli (3, 22–24). Our approach to integrate the hsdMS genes onto the chromosome stabilized the DNA, in contrast to the instability/toxicity observed for hsdMS genes. However, it should be highlighted that functional hsdMS genes have been cloned from multiple bacteria and expressed from plasmids in E. coli (16, 25, 26).
Our research builds on previous studies in this area exploring type II and III RM systems. Zhang et al. (4) conducted an in-depth characterization of the restriction barrier in an untransformable strain of N. hamburgensis and in two bacilli, B. cereus ATCC 10987 and B. amyloliquefacians TA208. Through the expression of an assembled operon of active type II or III and/or orphan methylases (determined by Southwestern blotting and liquid chromatography-mass spectrometry [LC-MS]) from each isolate in E. coli EC135 (TOP10 Δdam Δdcm Δhsd ΔmcrBC ΔmcrA Δmrr), recombinant DNA could be transformed into N. hamburgensis or the two bacilli at high efficiency. An assumption was made that all naturally occurring methylases in the E. coli K-12 TOP10 strain were detrimental to transformation, therefore requiring a “methylase-bald” strain as a starting point for DNA modification (4). No data to support this hypothesis were presented. For all strains of S. aureus that we have tested, we have shown that the presence of dam methylation was not detrimental to DNA transfer, and in one case, its presence was in fact essential (3). Intact dam is important, as deletion of the gene elevates the frequency of transition mutations due to deregulated mismatch repair (27). Furthermore, the deletion of dam in a recA-deficient background is lethal, so the reinstatement of a wild-type recA allele was required prior to dam deletion in EC135 (4). It is widely recognized that the presence of functional recA can destabilize repetitive DNA (28) and, also, lead to plasmid multimer formation through recA-dependent plasmid recombination (29). At least in the context of genetic manipulation of S. aureus, we would contend that a methylase-bald, recA+ E. coli strain is not an ideal cloning host for PAM.
The IMXXB E. coli strains we have developed permit the facile construction of unmarked mutations and transposon libraries in S. aureus lineages that we could not previously examine (e.g., MW2 and JKD6159) (data not shown). The IMXXB strains will open up new avenues for research, such as the assessment of candidate antigen efficacy in vaccine formulations against S. aureus from different lineages that previously could not be assayed (30). The knowledge gained here on the role of type I RM in S. aureus and its application to the E. coli donor strains tailored to specific S. aureus recipients will further improve our ability to conduct sophisticated genetic analysis of this and other important human pathogens.
MATERIALS AND METHODS
Media and reagents.
The bacterial strains and plasmids used in this study are described in Table 2. Oligonucleotides were purchased from Integrated DNA Technologies and are listed in Table 3. Genomic DNA was isolated using the Qiagen 100/G genomic tip (Qiagen). Weakening of the staphylococcal cell wall required the addition of 100 µg of lysostaphin (Ambi) into the lysis buffer and incubation at 37°C for 30 min. Plasmids and PCR products were isolated using the Wizard plus kits (Promega), with T4 DNA ligase also purchased from Promega. Plasmids were isolated from staphylococci as described previously (3). Restriction enzymes, T4 DNA polymerase, and Phusion DNA polymerase were purchased from New England Biolabs. Phire Hotstart DNA polymerase was purchased from Thermofisher. Sanger sequencing was supplied by Eurofins. Routine manipulation of S. aureus and E. coli was performed as described by Monk et al. (3). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Melford) was used at 50 µg/ml in E. coli and 100 µg/ml in S. aureus. Antibiotics were purchased from Sigma Aldrich and used at the following concentrations: carbenicillin (Car), 100 µg/ml; chloramphenicol (Cm), 10 µg/ml; and kanamycin (Kan), 50 µg/ml (E. coli) and 100 µg/ml (S. aureus).
TABLE 2 .
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DC10B | DH10B Δdcm; does not methylate DNA on cytosine | 3 |
| SA08B | DC10BΩPhelp-hsdMS (CC8-2) (SAUSA300_1751) of NRS384 integrated between the atpI and gidB genes | This study |
| SA30B | DC10BΩPhelp-hsdMS (CC30-2) (SAR1898) of MRSA252 integrated between the atpI and gidB genes | This study |
| IM01B | SA08BΩPN25-hsdS (CC1-1) (MW0393) of MW2 integrated between the essQ and cspB genes | This study |
| IM08B | SA08BΩPN25-hsdS (CC8-1) (SAUSA300_0406) of NRS384 integrated between the essQ and cspB genes | This study |
| IM30B | SA30BΩPN25-hsdS (CC30-1) (SAR0434) of MRSA252 integrated between the essQ and cspB genes | This study |
| IM93B | DC10BΩPN25-hsdMS/S (ST93-2/ST93-1) (SAA6159_01737/00387) of JKD6159 integrated between the atpI and gidB genes and ΩPhelp-hsdMS (ST93-3) (SAA6159_00055) of JKD6159 integrated between the essQ and cspB genes | This study |
| BL21(DE3) | F− ompT hsdSB(rB– mB–) gal dcm (DE3), IPTG-inducible T7 RNA polymerase | Novagen |
| Staphylococcus aureus strains | ||
| NRS384 | CC8, USA300, lineage 14, type strain from NARSA collection | BEI resources |
| MW2 | CC1, USA400 lineage, genome sequenced | 35 |
| MRSA252 | ST36 CC30, hospital-acquired MRSA, genome sequenced | 36 |
| JKD6159 | ST93, CA-MRSA, genome sequenced | 11 |
| NRS384 ΔhsdR | Targetron insertion in the hsdR gene of NRS384 | 3 |
| MW2 ΔhsdR | Clean deletion of the hsdR gene in MW2 | This study |
| MRSA252 ΔhsdR | Clean deletion of the hsdR gene in MRSA252 | This study |
| JKD6159 ΔhsdR | Clean deletion of the hsdR gene in JKD6159 | This study |
| JKD6159 ΔhsdRSCC | Clean deletion of the hsdRSCC gene in JKD6159 | This study |
| JKD6159 ΔhsdRSCC ΔhsdR | Clean deletion of the hsdR gene in JKD6159 ΔhsdRSCC | This study |
| WBG8287 | CC1, Western Australian (WA)-MRSA-1, SCCmec IV | 37 |
| MSSA CC1 | CC1, MSSA, pvl+, Rifr, fusidic acid resistant | Laboratory strain |
| SH1000 | CC8, MSSA, 8325-4 rsbU repaired, genome sequenced | 38 |
| Newman | CC8, human clinical MSSA, genome sequenced | 39 |
| JKD6009 | MRSA, ST239, SCCmec III(3A), genome sequenced | 40 |
| PS80 | CC30, MSSA, phage-propagating strain 80, ATCC 27700 | 41 |
| Cowan | CC30, ST30, MSSA, high-level protein A producer, ATCC 12598 | 42 |
| WSSP | CC30, ST30, MRSA, pvl+, SCCmec IV | Laboratory strain |
| Plasmids | ||
| pRAB11 | Anhydrotetracycline-inducible shuttle expression vector, pC194 S. aureus replicon; Carr Cmr | 43 |
| pIMAY | Allelic exchange plasmid for staphylococci; Cmr (IM452/IM453) | 3 |
| pIMAY-Z | Carries Gram-positive ribosome binding site and lacZ cloned downstream from the constitutive cat gene in pIMAY; 8.8 kb, Cmr | This study |
| pIMAY-Z(CC1ΔhsdR) | Carries deletion encompassing the entire hsdR gene (MW_0169 between the ATG and TAA codons); amplified from MW2 (IM243/IM237/IM238/IM244) | This study |
| pIMAY-Z(CC30ΔhsdR) | Carries deletion encompassing the entire hsdR gene (SAR_0196 between the ATG and TAA codons); amplified from MRSA252 (IM236/IM237/IM238/IM239) | This study |
| pIMAY-Z(ST93ΔhsdR) | Carries deletion encompassing the entire hsdR gene (SAA6159_00176 between the GTG and TAA codons); amplified from JKD6159 (IM678/IM679/IM680/IM681) | This study |
| pIMAY-Z(ST93ΔhsdRSCC) | Carries deletion encompassing the entire hsdRSCC gene (SAA6159_00056 between the TTG and TAA codons); amplified from JKD6159 (IM684/IM685/IM686/IM687) | This study |
| pMUTIN4 | Template for E. coli lacZ; Carr (IM454/IM455) | 44 |
| pIMK | E. coli/L. monocytogenes phage integrase vector, does not replicate in S. aureus; Kanr | 18 |
| pIMK2 | Template for Phelp promoter; Kanr | 18 |
| pKD46 | E. coli temperature-sensitive plasmid containing λ red recombinase genes under the control of an arabinose-inducible promoter; Carr | 45 |
| pKD3 | Plasmid for amplification of FRT-cat-FRT for E. coli gene deletion; Carr Cmr | 45 |
| pKD4 | Plasmid for amplification of FRT-kan-FRT for E. coli gene deletion; Carr Kanr | 45 |
| pCP20 | E. coli temperature-sensitive plasmid containing flp required for antibiotic marker excision; Carr Cmr | 45 |
| pIMK7 | Shuttle vector with staphylococcal pC194 replicon and p15a E. coli replicon (IM295/IM296/IM297/IM49) | This study |
| pET21d+ | IPTG-inducible expression vector; Ampr | Novagen |
| pCC8-MS1 | CC8-1 hsdMS genes cloned via NcoI/SacI (IM517/IM521/IM522/IM8) into pET21d+ | This study |
| pCC8-MS2 | CC8-2 hsdMS genes cloned via NcoI/XhoI (IM373/IM374/IM375/IM10) into pET21d+ | This study |
| pCC8-M2S1 | Hybrid CC8-2 hsdM–CC8-1 hsdS cloned via NcoI/SacI (IM373/IM518/IM519/IM8) into pET21d+ | This study |
TABLE 3 .
Oligonucleotides used in the study
| Oligonucleotide purpose, name | Sequence (5′–3′)a | RE siteb |
|---|---|---|
| Creation of pIMAY-Z | ||
| IM452 cat-lacZ F | TCAGATAGGCCTAATGACTGGCTTTTATAAAGGAGGATATCCATGGAAGTTACTGACG | |
| IM453 cat-lacZ R | TGTAAAAAGTACAGTCGGCATTATCTCATATTATTTTTGACACCAGACCAACTGGTAATGG | |
| IM454 pIMAY F | TTATAAAAGCCAGTCATTAGGCCTATCTGAC | |
| IM455 pIMAY R | TATGAGATAATGCCGACTGTACTTTTTACAG | |
| IM427 pIMAY-Z SLIC F | GGTACCCAGCTTTTGTTCCCTTTAGTGAGG | |
| IM428 pIMAY-Z SLIC R | GAGCTCCAATTCGCCCTATAGTGAGTCG | |
| Cloning hsdMS genes | ||
| IM9 ProMS (CC8-2) F | ATATGGATCCGATGCAATTATTCAGCCTGGTAGC | BamHII |
| IM10 hsdMS (CC8-2) R | ATATCTCGAGTTAAATAAACATTTTTTGTAATAGTCC | XhoI |
| IM373 ATG MS (CC8-2) F | ATATATGGTTTTGAAAGCATTTGAAAGCTAC | NcoI |
| IM374 MS (CC8-2) NcoI-R | AGTACCTTCATCGTCTAGGTAATGTACC | |
| IM375 MS (CC8-2) NcoI-F | ACATTACCTAGACGATGAAGGTACTATGGCCGTTGTACTCCCACATGG | |
| IM517 ATG MS (CC8-1) F | ATATCCATGGCTATTACTGAAAAACAACGTCAGC | NcoI |
| IM521 MS (CC8-1) NcoI-R | TGTACCTTCATCGTCTAGGTAATG | |
| IM522 MS (CC8-1) NcoI-R | GGTACATTACCTAGACGATGAAGGTACAATGGCCGTTGTACTCCCACATGGTG | |
| IM8 MS (CC8-1) R | ATATGAGCTCTTATAAGAACATTTTTTGTAAAAAGGATTG | SacI |
| IM518 S (CC8-1) F | CCTGAAAGAACTTGGGGTGTTGAAAGATGAGTAATACACAAAAGAAAAATGTGC | |
| IM519 M (CC8-2) R | CTTTCAACACCCCAAGTTCTTTCAGGTATGC | |
| Recombineering | ||
| IM367 atpI-Phelp F | CAAAAAGCGGTCAAATTATACGGTGCGCCCCCGTGATTTCAAACAATAAGTACGGAGCTCC ATTATGCTTTGGCAG | |
| IM359 MS (CC8-2) −cat F | CAAAAAATGTTTATTTAACTCGAGATATGTGTAGGCTGGAGCTGCTTC | |
| IM360 gidB − cat R | ATAACGTGGCTTTTTTTGGTAAGCAGAAAATAAGTCATTAGTGAAAATATGTCCATATGAA TATCCTCCTTAG | |
| IM434 atpI OUT F | ACTTTCTTTAAGGCTTAGAGTCAAGC | |
| IM435 gidB OUT R | TTTAACGCCACGTTCACTCTTTTGC | |
| IM102 essQ-PN25 F | CCCAAACTGCACCCAAGAGTCAGAACACAGTTTTTCAAGAGTACAAAGGGGTCATAAAAAA TTTATTTGCTTTCAGG | |
| IM103 PN25-S(CC8-1) F | CATAAAAAATTTATTTGCTTTCAGGAAAATTTTTCTGTATAATAGATTCATAAATTTGAGAGAGGAGTTATGAGTAATACACAAAAGAAAAATGTGC | |
| IM104 S (CC8-1) R | TTATAAGAACATTTTTTGTAAAAAGGATTG | |
| IM105 S (CC8-1)-kan F | CAATCCTTTTTACAAAAAATGTTCTTATAAGTGTAGGCTGGAGCTGCTTC | |
| IM106 cspB-kan R | GCTAACCATTGTGGTGAAGTGCAGGTTTGCTGCATGAATAGTTTTACGGTCCATATGAATA TCCTCCTTAG | |
| IM113 essQ OUT F | CGGCCATTTATACAGGAAAAGCCTA | |
| IM114 cspB OUT R | GTTACCTTCTCTATAGAGAGTGGTG | |
| IM132 PN25S (CC30-1) F | CATAAAAAATTTATTTGCTTTCAGGAAAATTTTTCTGTATAATAGATTCATAAATTTGAGAGAGGAGTTATGAGTAATACACAAACGAAAAATGTG | |
| IM133 S (CC1-1) R | TCAAATAAACATTTTCTGTAAAAACG | |
| IM134 S (CC1-1)-kan F | CGTTTTTACAGAAAATGTTTATTTGAGTGTAGGCTGGAGCTGCTTC | |
| pIMK7 | ||
| IM295 pC194 replicon F | ATATGCATGCGCTTTTAAAAAGCAAATATGAGCC | SphI |
| IM296 pC194 replicon R | TTTATCTAAAGTGAATTTAGGAGGC | |
| IM297 pIMC-pC194 F | GCCTCCTAAATTCACTTTAGATAAAATTCTATAATAGAAGGTATGGAGGATG | |
| IM49 pIMC R | AGATCTCCTCTCGCCTGTCCCCTCAGTTCAGTAATTTCC | BglII |
| S. aureus mutants | ||
| IM234 CC30 hsdR OUT F | TGTGCGTTCTAATATAAAGTTAGTTGC | |
| IM235 CC30 hsdR OUT R | TCCGACTGTTGTATCTTTGTATCTAGC | |
| IM236 CC30 hsdR FA | CCTCACTAAAGGGAACAAAAGCTGGGTACCAGTGTGACAAATTAAAGCAAAGTTCAGG | |
| IM237 CC1/CC30 hsdR RB | CATTCATATCCCCTTCCATACACTTTC | |
| IM238 CC1/CC30 hsdR FC | GAAAGTGTATGGAAGGGGATATGAATGTAATGATTCAGCCCCCTCGCTAG | |
| IM239 CC30 hsdR RD | CGACTCACTATAGGGCGAATTGGAGCTCTAATCTCGTAGACAACGCCTTTACC | |
| IM240 CC30 hsdR OUT R | TCCGACTGTTGTATCTTTGTATCTAGC | |
| IM241 CC1 hsdR OUT F | TCAGTTGCTTGATGAAAAATTGTTGC | |
| IM242 CC1 hsdR OUT R | ACTTTGCAAATATCCGCATTCAACC | |
| IM243 CC1 hsdR FA | CCTCACTAAAGGGAACAAAAGCTGGGTACCGACAAATTAAAGCAAAGTTCAGATTTGAGC | |
| IM244 CC1 hsdR RD | CGACTCACTATAGGGCGAATTGGAGCTCAGGACTCTCAGAGACATCATTAGC | |
| IM678 ST93 hsdR FA | CCTCACTAAAGGGAACAAAAGCTGGGTACCAGATTGCATGATTTTTGTGACGAATCAAGG | |
| IM679 ST93 hsdR RB | CATTCATATCCCCTTCCGTACACTTTCTATTGC | |
| IM680 ST93 hsdR FC | GAAAGTGTACGGAAGGGGATATGAATGTGTAATGATTCAGCCCCCTCGCTAGATTAG | |
| IM681 ST93 hsdR RD | CGACTCACTATAGGGCGAATTGGAGCTCCCACGCAAATAAGGATAATACATATTAATCC | |
| IM682 ST93 hsdR OUT F | AAACGCATTTACTTGTGTCAACATTTGC | |
| IM683 ST93 hsdR OUT R | TAGGTTGAATACAATCACCAATCAAACC | |
| IM684 ST93 hsdRSCC FA | CCTCACTAAAGGGAACAAAAGCTGGGTACCACCTAGGAGTTGGAAAGTTGATGAATTAGG | |
| IM685 ST93 hsdRSCC RB | TTCACTAAATTGAAAGCTCATCTTCATTCACC | |
| IM686 ST93 hsdRSCC FC | ATGAAGATGAGCTTTCAATTTAGTGAATAAATACTGTTTATATTGTTGACCTGTTAGATAC | |
| IM687 ST93 hsdRSCC RD | CGACTCACTATAGGGCGAATTGGAGCTCATAAAATTATTCCACTTTTATGCTCTTGCC | |
| IM688 ST93 hsdRSCC OUT F | GGTGGAGAGATGGTTGATAGTGAATTGG | |
| IM689 ST93 hsdRSCC OUT R | GTTTCCTTTATCTCTTTTTCACTCTCACG |
Boldface indicates 5′ primer tails complementary to the E. coli chromosome or pIMAY-Z.
RE, restriction endonuclease.
SMRT sequencing.
Genomic DNA samples were submitted to the Duke IGSP Genome Sequencing and Analysis Core Resource for sequencing on a Pacific Biosciences RS instrument. Libraries of 8 to 11 kb were constructed and sequenced using P5-C2 chemistry. Data were analyzed using the SMRT Portal version 2.2.0 (Pacific Biosciences) and Motif Finder 1.3.1. Reference S. aureus genomes with plasmids omitted were assessed for their type I methylation profiles. DH10B was used as a reference for the IMXXB E. coli strains. Additionally, E. coli strain DC10B was SMRT sequenced, with only dam methylation observed (data not shown). A quality value (QV; prediction of the error probability of a base call) threshold of 30 was set for adenine modification/motif calling.
Electroporation.
E. coli and S. aureus were transformed as described previously (3), except that staphylococci were grown for 40 min rather than 30 min after dilution back to an optical density at 600 nm of 0.5. MW2 competent cells were created as described by Augustin and Götz (31). Routinely, we used 5 µg of pRAB11 and 2.5 µg of pIMAY-Z with the concentration determined by fluorometric assay (Qubit 2.0; Life Technologies) to transform S. aureus. The results presented are from three independent transformation experiments.
SLIC.
To construct pIMAY-Z, we applied sequence- and ligase-independent cloning (SLIC) with the modifications described by Bieniossek et al. (32). SLIC allows rapid, flexible, and efficient cloning into plasmid DNA without restriction enzyme sequence site specificity. To insert lacZ into pIMAY, the entire 5.7 kb pIMAY plasmid was linearized downstream from the cat stop codon and amplified with primers IM452/IM453, and the E. coli lacZ gene was amplified from pMUTIN4 using primers IM454/IM455 including a consensus Gram-positive ribosome binding site. The 5′ tails of the lacZ primers included 30 nucleotides of homology with the ends of the amplified pIMAY. Both fragments were purified to remove unincorporated deoxynucleoside triphosphates (dNTPs) (Wizard SV Gel and PCR clean up system; Promega). A 1 µg aliquot of each PCR product was treated with 3 U of T4 DNA polymerase (to create single-stranded ends) in the NEB 2 restriction enzyme buffer containing, additionally, 200 mM urea (2 M stock), 10 µg/ml bovine serum albumin (1 µg/ml stock), and 5 mM dithiothreitol (1 M stock), final concentration in 40 µl. The reaction mixture was incubated at 23°C for 20 min, and then the enzyme inactivated by the addition of 2 µl of 500 mM EDTA, pH 8, and further incubation at 75°C for 20 min. Both fragments were mixed (125 ng of each product) and incubated on a thermocycler at 65°C for 10 min, followed by decreases of 1°C per minute to 25°C. The annealed vector and insert were ethanol precipitated and electroporated into E. coli strain DC10B, with transformants selected on Luria agar (L agar) plus Cm and X-Gal. To clone into pIMAY-Z, the plasmid was first linearized with KpnI and gel extracted. The vector backbone was amplified (from within the multiple cloning site [MCS] using primers IM427/IM428) with 0.5 U of Phusion and 0.25 U of Phire DNA polymerase (Finnzymes) with annealing at 50°C and extension for 3 min in a 50 µl reaction mixture. Deletion constructs (see below) were amplified by SOE PCR (3) and SLIC cloned into PCR-amplified pIMAY-Z, as described above.
Construction of pIMK7.
A kanamycin-resistant shuttle plasmid was created by amplification of (i) the E. coli p15a replicon, the pBluescript II KS MCS, and the aphA3 kanamycin resistance gene (primers IM297/IM49) from pIMK (18) and (ii) the pC194 S. aureus replicon (primers IM295/IM296) from pRAB11. The two fragments were joined by SOE PCR (primers IM295/IM49) and transformed into DC10B to generate pIMK7 (4.9 kb).
Cloning hsdMS loci into pET21d+.
Type I modification genes from S. aureus NRS384 were cloned into pET21d+, an IPTG-inducible expression plasmid that is active in the BL21 E. coli host (naturally dcm deficient). Modification genes were amplified as follows. (i) The internal NcoI site was removed from CC8-1 hsdMS by SOE PCR (primers IM517, IM521, IM522, and IM8, incorporating flanking NcoI/SacI sites). (ii) The NcoI− variant of the CC8-2 hsdMS gene was amplified from SA08B (see below) genomic DNA (primers IM516/IM10, incorporating NcoI/XhoI sites). (iii) A hybrid of CC8-2 hsdM and CC8-1 hsdS was created by SOE PCR (primers IM516, IM519, IM518, and IM8, incorporating NcoI/SacI sites). All PCR products were ligated into complementarily digested pET21d+. Ligations were cotransformed with pIMK7 into BL21 and transformants selected on L agar plus Kan/Car. Plasmids were isolated from BL21 strains after induction overnight with 0.1 mM IPTG.
Construction of E. coli IM08B for the transformation of CC8 and ST239.
To stably express CC8-2 hsdMS genes in E. coli, we bypassed the cloning step by in vitro PCR assembly of the CC8-2 hsdMS joined to an antibiotic resistance marker. The final construct could then be recombineered into the DC10B chromosome at a neutral location. First, we utilized SOE PCR to remove internal NcoI sites from the CC8-2 hsdMS of NRS384 (using primers IM373, IM374, IM375, and IM10), and then the PCR product was ligated into NcoI/XhoI-cut pIMK2 (allowing high-level expression from the Phelp promoter [18]). PCR products comprising Phelp-hsdMS were then amplified from the ligation mixture with primers IM367/IM10. The cat gene, flanked by Flp recombinase target (FRT) sites, was amplified with primers IM359/IM360 from plasmid pKD3 and joined to the CC8-2 hsdMS amplimer by SOE PCR (IM367/IM360). These primers contained tails with 50 bp of homology for integration into the intergenic region between atpI and gidB in the DC10B chromosome (33). The PCR products were ethanol precipitated and electroporated into DC10B containing pKD46 made competent as described previously (3). Transformants were selected on L agar plus Cm. Colonies were screened by colony PCR for recombination (primers IM434 and IM435), and positive clones were grown overnight at 43°C to promote loss of pKD46. To excise the FRT site-flanked cat gene, the strains were transformed with pCP20 at 28°C, single colony purified at 28°C, and then grown in broth overnight at 43°C to promote loss of the plasmid. Cells were patched onto L agar or L agar plus Cm to confirm the excision of the antibiotic resistance marker, creating strain SA08B. The CC8-1 hsdS gene of NRS384 was amplified with primers IM103/IM104 containing the PN25 promoter within IM103. The kanamycin resistance marker was amplified with IM105/IM106 from pKD4, and SOE PCR performed to join the two products with IM102/IM106, which target the product to integrate upstream from essQ and downstream from cspB (33). The amplimer was transformed into SA08B(pKD46) and processed as described above. The Kans strain was termed IM08B. Shuttle plasmid (pRAB11 or pIMAY-Z) was isolated from either DC10B or IM08B and used to transform NRS384 or NRS384 ΔhsdR to investigate plasmid methylation.
Construction of E. coli IM01B for the transformation of CC1.
The CC1-1 hsdS gene of MW2 was amplified with primers IM103/IM133 containing the PN25 promoter within IM103. The kanamycin resistance marker was amplified with IM134/IM106 from pKD4 and SOE PCR performed to join the two products with primers IM102/IM106. The amplimer was transformed into SA08B(pKD46) and processed as described above. The Kans strain was termed IM01B. Shuttle plasmid (pRAB11 or pIMAY-Z) was isolated from either DC10B or IM01B and used to transform MW2 or MW2 ΔhsdR.
Construction of E. coli IM30B for the transformation of CC30.
The two native NcoI sites of CC30-2 hsdMS of MRSA252 were removed by SOE PCR (using primers IM407, IM408, IM409, IM410, IM411, and IM363, incorporating flanking NcoI/XhoI sites). The MRSA252 CC30-2 hsdMS (NcoI−) genes were amplified and ligated into pIMK2 cut with NcoI and XhoI. The Phelp-hsdMS construct was amplified (primers IM367/IM363) from the pIMK2 ligation, and SOE PCR was performed to join the cat gene flanked with FRT sites (amplified from pKD3 with primers IM364/IM360). PCR products were electroporated into DC10B(pKD46) and screened as described above. Excision of the cat gene yielded E. coli strain SA30B. The CC30-1 hsdS gene of MRSA252 was amplified with primers IM132/IM104 containing the PN25 promoter within IM132. The Kan resistance marker was amplified with IM105/IM106 from pKD4, and SOE PCR with IM102/IM106 performed to join the two products. The amplimer was transformed into SA30B(pKD46) and processed as described above. The Kans strain was termed IM30B. Shuttle plasmid (pRAB11 or pIMAY-Z) was isolated from either DC10B or IM30B and used to transform MRSA252 or MRSA252 ΔhsdR.
Construction of IM93B for the transformation of ST93.
A hybrid hsdMS-hsdS operon from JKD6159 was constructed by SOE PCR. The ST93-2 hsdMS genes were amplified with IM571/IM573 containing the PN25 promoter and joined to the ST93-1 hsdS gene amplified with primers IM574/IM575. The products were gel extracted and used as the template in an SOE PCR reaction (primers IM572/IM575). The resultant product was gel extracted and joined to the cat PCR product (primers IM576/IM548) amplified from pKD3. The essQ-PN25-hsdMS-hsdS-FRT-cat-FRT-cspB PCR product was electroporated into DC10B(pKD46) and screened as described above. Excision of the cat gene yielded E. coli strain SA93B. The hsdMS adjacent to the SCCmec of JKD6159 was amplified with primers IM466/IM467 and cloned into complementarily digested pIMK2 (NcoI/SalI). The cat gene was amplified with primers IM499/IM360 from pKD3, and SOE PCR performed to join the two products with primers IM367/IM360. The amplimer was transformed into SA93B(pKD46) and processed as described above; the Cms strain was termed IM93B. Shuttle plasmid pRAB11 isolated from DC10B, IM93B, and JKD6159 was used to transform JKD6159, JKD6159 ΔhsdR, JKD6159 ΔhsdRSCC, or JKD6159 ΔhsdR ΔhsdRSCC.
Allelic exchange with pIMAY-Z.
The derivatives of pIMAY-Z constructed to delete hsdR from MW2 (primers IM243, IM237, IM238, and IM244), MRSA252 (primers IM236, IM237, IM238, and IM239), JKD6159 (primers IM678, IM679, IM680, and IM681), and JKD6159 hsdRSCC (primers IM684, IM685, IM686, and IM687) were isolated from 25 ml Luria broth cultures of strains with the respective IMXXB backgrounds. The entire plasmid preparation was ethanol precipitated and transformed into electrocompetent S. aureus cells. Transformants were selected on brain heart infusion agar (BHIA) plus Cm at 28°C after 2 days. A single colony containing replicating plasmid was homogenized in 200 µl of tryptic soy broth, and the suspension 10-fold diluted to 10−3. A 100 µl aliquot of each dilution was spread plated on BHIA plus Cm and X-Gal and grown overnight at 37°C. Blue colonies were purified on BHIA plus Cm and X-Gal and grown at 37°C. Potential integrants were screened by colony PCR for loss of the replicating plasmid and the site of integration as described previously (3). Integrants from either side of the gene to be deleted were grown overnight at 28°C without antibiotic selection and then plated on BHIA plus X-Gal at 37°C, screening for white colonies. Plasmid loss was confirmed by the chloramphenicol sensitivity of the white colonies and by colony PCR performed using the chromosomal OUT primers listed in Table 3.
ACKNOWLEDGMENTS
We acknowledge financial support from Science Foundation Ireland (grant no. 08/IN.1/B1845) to TJF and National Health and Medical Research Council, Australia, fellowships to B.P.H. (APP1023526) and T.P.S. (APP1008549).
Footnotes
Citation Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. 2015. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6(3):e00308-15. doi:10.1128/mBio.00308-15.
REFERENCES
- 1.Donahue JP, Israel DA, Peek RM, Blaser MJ, Miller GG. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol Microbiol 37:1066–1074. doi: 10.1046/j.1365-2958.2000.02036.x. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell Motherway M, O’Driscoll J, Fitzgerald GF, Van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol 2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3(2):e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang G, Wang W, Deng A, Sun Z, Zhang Y, Liang Y, Che Y, Wen T. 2012. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet 8:e1002987. doi: 10.1371/journal.pgen.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veiga H, Pinho MG. 2009. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl Environ Microbiol 75:3034–3038. doi: 10.1128/AEM.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roer L, Aarestrup FM, Hasman H. 2015. The EcoKI type I restriction-modification system in Escherichia coli affects but is not an absolute barrier for conjugation. J Bacteriol 197:337–342. doi: 10.1128/JB.02418-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P. 2011. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res 39:5597–5610. doi: 10.1093/nar/gkr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loenen WA, Dryden DT, Raleigh EA, Wilson GG. 2014. Type I restriction enzymes and their relatives. Nucleic Acids Res 42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldron DE, Lindsay JA. 2006. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol 188:5578–5585. doi: 10.1128/JB.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monk IR, Foster TJ. 2012. Genetic manipulation of staphylococci—breaking through the barrier. Front Cell Infect Microbiol 2:49. doi: 10.3389/fcimb.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua KY, Stinear TP, Howden BP. 2013. Functional genomics of Staphylococcus aureus. Brief Funct Genomics 12:305–315. doi: 10.1093/bfgp/elt006. [DOI] [PubMed] [Google Scholar]
- 12.Stobberingh EE, Schiphof R, Sussenbach JS. 1977. Occurrence of a class II restriction endonuclease in Staphylococcus aureus. J Bacteriol 131:645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bens CC, Voss A, Klaassen CH. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J Clin Microbiol 44:1875–1876. doi: 10.1128/JCM.44.5.1875-1876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreiswirth BN, Löfdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 15.Yasui K, Kano Y, Tanaka K, Watanabe K, Shimizu-Kadota M, Yoshikawa H, Suzuki T. 2009. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res 37:e3. doi: 10.1093/nar/gkn884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ. 2012. The methylomes of six bacteria. Nucleic Acids Res 40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts GA, Houston PJ, White JH, Chen K, Stephanou AS, Cooper LP, Dryden DT, Lindsay JA. 2013. Impact of target site distribution for type I restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucleic Acids Res 41:7472–7484. doi: 10.1093/nar/gkt535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friehs K. 2004. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86:47–82. doi: 10.1007/b12440. [DOI] [PubMed] [Google Scholar]
- 20.Brunner M, Bujard H. 1987. Promoter recognition and promoter strength in the Escherichia coli system. EMBO J 6:3139–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGavin MJ, Arsic B, Nickerson NN. 2012. Evolutionary blueprint for host- and niche-adaptation in Staphylococcus aureus clonal complex CC30. Front Cell Infect Microbiol 2:48. doi: 10.3389/fcimb.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada R, Matsumoto M, Zhang Y, Isaka M, Tatsuno I, Hasegawa T. 2014. Emergence of type I restriction modification system-negative emm1 type Streptococcus pyogenes clinical isolates in Japan. APMIS 122:914–921. doi: 10.1111/apm.12230. [DOI] [PubMed] [Google Scholar]
- 23.Hobson N, Price NL, Ward JD, Raivio TL. 2008. Generation of a restriction minus enteropathogenic Escherichia coli E2348/69 strain that is efficiently transformed with large, low copy plasmids. BMC Microbiol 8:134. doi: 10.1186/1471-2180-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krebes J, Morgan RD, Bunk B, Spröer C, Luong K, Parusel R, Anton BP, König C, Josenhans C, Overmann J, Roberts RJ, Korlach J, Suerbaum S. 2014. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res 42:2415–2432. doi: 10.1093/nar/gkt1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Luong K, Murray IA, Davis BM, Keren-Paz A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE. 2012. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wion D, Casadesús J. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bzymek M, Lovett ST. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A 98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedbrook JR, Ausubel FM. 1976. Recombination between bacterial plasmids leading to the formation of plasmid multimers. Cell 9:707–716. doi: 10.1016/0092-8674(76)90134-3. [DOI] [PubMed] [Google Scholar]
- 30.Scully IL, Liberator PA, Jansen KU, Anderson AS. 2014. Covering all the bases: preclinical development of an effective Staphylococcus aureus vaccine. Front Immunol 5:109. doi: 10.3389/fimmu.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augustin J, Götz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett 54:203–207. doi: 10.1016/0378-1097(90)90283-V. [DOI] [PubMed] [Google Scholar]
- 32.Bieniossek C, Nie Y, Frey D, Olieric N, Schaffitzel C, Collinson I, Romier C, Berger P, Richmond TJ, Steinmetz MO, Berger I. 2009. Automated unrestricted multigene recombineering for multiprotein complex production. Nat Methods 6:447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]
- 33.Kuhlman TE, Cox EC. 2010. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res 38:e92. doi: 10.1093/nar/gkp1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 36.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coombs GW, Monecke S, Pearson JC, Tan HL, Chew YK, Wilson L, Ehricht R, O’Brien FG, Christiansen KJ. 2011. Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region. BMC Microbiol 11:215. doi: 10.1186/1471-2180-11-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howden BP, Seemann T, Harrison PF, McEvoy CR, Stanton JA, Rand CJ, Mason CW, Jensen SO, Firth N, Davies JK, Johnson PD, Stinear TP. 2010. Complete genome sequence of Staphylococcus aureus strain JKD6008, an ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate-level vancomycin resistance. J Bacteriol 192:5848–5849. doi: 10.1128/JB.00951-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asheshov EH. 1969. The genetics of penicillinase production in Staphylococcus aureus strain PS80. J Gen Microbiol 59:289–301. doi: 10.1099/00221287-59-3-289. [DOI] [PubMed] [Google Scholar]
- 42.Cowan ST, Shaw C, Williams RE. 1954. Type strain for Staphylococcus aureus Rosenbach. J Gen Microbiol 10:174–176. doi: 10.1099/00221287-10-1-174. [DOI] [PubMed] [Google Scholar]
- 43.Helle L, Kull M, Mayer S, Marincola G, Zelder ME, Goerke C, Wolz C, Bertram R. 2011. Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157:3314–3323. doi: 10.1099/mic.0.052548-0. [DOI] [PubMed] [Google Scholar]
- 44.Vagner V, Dervyn E, Ehrlich SD. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144(Pt 11):3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]