Abstract
Invasion of the malaria vector Anopheles gambiae midgut by Plasmodium parasites triggers transcriptional changes of immune genes that mediate the antiparasitic defense. This response is largely regulated by the Toll and Immune deficiency (IMD) pathways. To determine whether An. gambiae microRNAs (miRNAs) are involved in regulating the anti-Plasmodium defense, we showed that suppression of miRNA biogenesis results in increased resistance to Plasmodium falciparum infection. In silico analysis of An. gambiae immune effector genes identified multiple transcripts with miRNA binding sites. A comparative miRNA microarray abundance analysis of P. falciparum infected and naïve mosquito midgut tissues showed elevated abundance of miRNAs aga-miR-989 and aga-miR-305 in infected midguts. Antagomir inhibition of aga-miR-305 increased resistance to P. falciparum infection and suppressed the midgut microbiota. Conversely, treatment of mosquitoes with an artificial aga-miR-305 mimic increased susceptibility to P. falciparum infection and resulted in expansion of midgut microbiota, suggesting that aga-miR-305 acts as a P. falciparum and gut microbiota agonist by negatively regulating the mosquito immune response. In silico prediction of aga-miR-305 target genes identified several anti-Plasmodium effectors. Our study shows that An. gambiae aga-miR-305 regulates the anti-Plasmodium response and midgut microbiota, likely through post-transcriptional modification of immune effector genes.
Keywords: microRNAs, Anopheles gambiae, immunity, Plasmodium, microbiota
1. Introduction
Female Anopheles gambiae mosquitoes are the principal vector of the malaria parasite Plasmodium falciparum, and the parasite has to complete a complex infection cycle in the mosquito vector to accomplish transmission. Invasion of mosquito tissues by P. falciparum ookinete-stage parasites results in extensive transcriptional changes of immune genes that mediate the host defense response, along with genes playing roles in other infection-responsive physiological systems (Dong et al., 2006). Mosquitoes lack an adaptive immune response and rely solely upon an innate immune system that is triggered through the recognition of pathogen associated molecular patterns (PAMPS) by pattern recognition receptors (PRRs). Plasmodium infection of the mosquito midgut epithelium triggers the activation of the highly conserved NF-κB TOLL and IMD signaling cascades, with the TOLL pathway primarily suppressing infection with the rodent parasite P. berghei and the IMD pathway limiting human P. falciparum infection. Activation of the IMD pathway induces expression of key anti-Plasmodium effectors such as APL1, TEP1, and LRRD7, through the nuclear translocation of the NF-κB transcription factor REL2. The immune response can be tempered by the negative regulators Caspar and Caudal, which inhibit IMD pathway signal transduction and prevent REL2-mediated transcription of immune effectors, respectively (reviewed in (Clayton et al., 2014)).
Over-activation of the immune response could exert a negative effect on the individual mosquito's fitness, and therefore mechanisms must be in place to either tolerate or limit the response. Post-transcriptional gene regulation has been proposed as a mechanism to fine-tune immune responses and other physiological processes and to prevent any negative effects of over-activation (reviewed in (Chen et al., 2013)). Because transcriptional changes are central to the A. gambiae anti-Plasmodium defense, it is plausible to hypothesize that post-transcriptional regulation also plays a role in the host's defense response. MicroRNAs (miRNA) are small regulatory non-coding RNAs responsible for sequence-specific post-transcriptional regulation (Lau et al., 2001). miRNAs are transcribed by RNA polymerase II to form long pri-miRNAs, cleaved by the RNase III enzyme Drosha within the nucleus to form pre-miRNAs (~ 70 nt), and then cleaved into their mature forms (21-25 nt) by a second RNase III, Dicer-1, following their export to the cytoplasm (Hutvagner et al., 2001; Lee et al., 2003; Lee et al., 2004). Argonaute-1 (Ago-1), which is part of the RNA-induced silencing complex (RISC) then guides the mature miRNAs to target mRNA 3’-untranslated regions, according to the classic pathway (Forstemann et al., 2007; Tomari et al., 2007). Sequence complementarity of the miRNA seed region, a heptamer spanning nucleotides 2–8 at the 5’ end of the mature miRNA, to its target mRNA is critical for post-transcriptional regulation (Brennecke et al., 2005). Binding of the RISC complex to target mRNAs results in either mRNA transcript degradation or repression of translation (reviewed in (Filipowicz et al., 2008)).
The biological function of insect miRNAs has predominantly been studied in Drosophila melanogaster, and shown to comprise many processes including development and differentiation (reviewed in (Lucas and Raikhel, 2013)). Recently, a role for miRNAs in insect immunity has been shown in D. melanogaster; where dme-miR-8 controls basal immune homeostasis (Choi and Hyun, 2012; Lee and Hyun, 2014). This sequence is conserved in the diamondback moth Plutella xylostella and up-regulates the expression of the TOLL pathway negative regulator serpin 27 (Etebari and Asgari, 2013). Dengue virus infection of the vector mosquito Aedes aegypti modulates the expression of 35 mosquito miRNAs (Campbell et al., 2014). A specific miRNA regulates the expression of two TOLL pathway-related immune genes, specifically up-regulating the negative regulator cactus and down-regulating the transcription factor Rel1 (Hussain et al., 2013). The direct interaction of this miRNA with target genes renders mosquitoes more susceptible to dengue virus infection (Hussain et al., 2013). The A. gambiae miRNA biogenesis pathway is involved in the host response to Plasmodium infection. P. falciparum infection causes transcripts of the miRNA biogenesis components Dicer1 and Drosha to exhibit increased polysome loading (Mead et al., 2012). The rodent malaria parasite P. berghei affects the expression of A. gambiae miRNAs, and RNA interference (RNAi) targeting of Ago-1 and Dicer-1 renders mosquitoes more susceptible to P. berghei infection (Winter et al., 2007). In addition, P. vinckei petteri and P. berghei infection of A. stephensi and A. gambiae respectively causes differential expression of multiple miRNAs (Biryukova et al., 2014; Jain et al., 2014).
Two lines of evidence suggest that specific miRNAs may modulate A. gambiae susceptibility to P. falciparum infection. First, IMD pathway-controlled transcriptional changes are key to regulating permissiveness to this parasite. Second, there is evidence that the mosquito miRNA pathway modulates susceptibility to infection. Through microarray-based profiling of P. falciparum-infected and -naïve tissues, we show that A. gambiae miRNAs are regulated by parasitic infection of the midgut tissue. Through functional assays, we demonstrate that miRNA aga-miR-305 negatively regulates the A. gambiae immune response to P. falciparum infection, and our in silico analysis predicts several potential aga-miR-305 target immune genes. In summary, we show that specific A. gambiae miRNAs can regulate the innate immune response and anti-Plasmodium defense. Identification of Plasmodium-responsive miRNAs offers the possibility of generating transgenic miRNA sponge-expressing mosquitoes that are more resistant to P. falciparum infection as part of the development of a novel malaria control strategy.
2. Materials and Methods
2.1 Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Mice were only used for mosquito rearing as a blood source according to approved protocol. The protocol was approved by the Animal Care and Use Committee of the Johns Hopkins University (Permit Number: M006H300). Commercial anonymous human blood from Interstate Bloodbank was used for Plasmodium infection assays in mosquitoes, and informed consent was therefore not applicable. The Johns Hopkins School of Public Health Ethics Committee has approved this protocol.
2.2 Mosquito Rearing
A. gambiae Keele strain mosquitoes (Hurd et al., 2005) were reared in the Insectary Core Facility of the Johns Hopkins School of Public Health. Mosquito cages were maintained in controlled chambers with 12-hr light/dark cycles at 27°C and 70% humidity. Adult mosquitoes were maintained on 10% sucrose solution.
2.3 RNA isolation and miRNA abundance determination
All samples for RNA extraction were homogenized in TRIzol® (Life Technologies, Carlsbad, CA.) using RNAse-free plastic pestles and stored at −80°C prior to extraction, according to the manufacturer's protocol. Isolated RNA was treated with Turbo DNase I (5U, Life Technologies) to remove contaminating DNA. For quantification of miRNA abundance, the miScript PCR system (Qiagen, Valencia, CA, USA) was used according to the manufacturer's guidelines. In brief, the kit contains an enzyme mix containing both poly(A) polymerase and reverse transcriptase. Mature miRNAs within total RNA samples were selectively polyadenylated and reverse-transcribed to cDNA using the miScript HiSpec Buffer and oligo(dT) primers that contain a universal tag sequence. miRNA abundance was then quantified using the miScript Sybr Green PCR kit (Qiagen) and a universal primer complementary to the tag sequence and miRNA-specific primers (Table S2). For each target gene, three biological replicates were performed and normalized to the abundance levels of miRNAs showing equal levels across all samples, as determined by the NormFinder software as previously described (Andersen et al., 2004). To determine silencing efficiencies of selected miRNAs, the HiFlex buffer, which allows reverse transcription of both miRNAs and mRNAs, was used to prepare cDNA to enable normalization to the ribosomal S7 gene, as miRNAs with invariant expression in these samples was not known. All reactions were performed in triplicate. Fold changes between samples were calculated using the ΔΔct method (Pfaffl, 2001). The following PCR conditions were used for all reactions: 95°C for 15 min, 40 cycles of 94°C for 15 s, 56°C for 30 s and 70°C for 30 s. Melting curve analysis was performed to confirm the amplification of a unique product.
2.4 RNAi gene silencing and real-time PCR
To silence the miRNA pathway genes Ago1 (AGAP011717) and Dicer1 (AGAP002836), we used an RNAi approach as previously described (Dong et al., 2006). In brief, specific dsRNA was synthesized using the HiScribe T7 in vitro Transcription Kit (New England Biolabs, Ipswich, MA) from purified PCR-amplified products, and 69 nl of each dsRNA (3 μg/μl) in sterile water was injected into the thorax of 3- to 4-day-old cold-anesthetized female mosquitoes using a nanoinjector (Nanoject, Drummond). A dsRNA corresponding to GFP was used as a control. To determine efficiency of gene silencing, total midgut RNA from 10 mosquitoes was isolated three days post injection and cDNA prepared using the M-MLV Reverse Transcriptase 1st-Strand cDNA Synthesis Kit (Promega, Madison, WI, USA). Knockdown efficiency was determined by quantitative real-time PCR (qRT-PCR) as previously described (Dong et al., 2006) using the SYBR Green PCR Mastermix (Applied Biosystems, Life Technologies, Grand Island, NY, USA) . cDNA templates were normalized to the ribosomal S7 gene and fold change in gene expression levels following silencing was determined using the standard ΔΔct method. Three independent biological replicates were performed for each candidate gene. The following PCR conditions were used for all reactions: 95°C for 10 min, 40 cycles of 94°C for 15 s, 56°C for 30 s and 72°C for 30 s. Melting curve analysis was performed to confirm the amplification of a unique product. Primer sequences used for amplification of dsRNA PCR templates and verification of silencing efficiency are listed in Table S2.
2.5 miRNA microarray analysis
A total of 194 mosquito miRNA sequences were accessed from miRBase (www.mirbase.org) and integrated into a custom Agilent miRNA microarray design (Agilent Technologies, CA, USA) (Wang et al., 2007). For microarray-based miRNA abundance analysis, midguts were dissected from mosquitoes following either a P. falciparum-infected blood meal or a non-infected human blood meal at 18 and 24 hr post-feeding. Three independent replicates of 15 midguts were dissected for each condition. A separate cohort of mosquito midguts, from the same experiment, were dissected 7 days after feeding and stained with 0.1% mercurochrome, and oocyst numbers were enumerated using a light-contrast microscope to establish infection of the P. falciparum-fed group. Total RNA (100 ng) from dissected midguts from each group was labeled with Cyanine3-pCp using the Agilent miRNA Complete Labeling and Hyb Kit and added to the miRNA microarray. Arrays were hybridized for 20 hr at 55°C and scanned with an Agilent scanner. Scanned images were analyzed using Feature Extraction v11.5 and GeneSpring GX 12.6 (Agilent Technologies), and the data were normalized to the 75th percentile (Koufaris et al., 2012). Only miRNAs detected above background level in at least two of the three biological replicates were considered. miRNAs were selected if they showed a log2-fold change >1, using a p-value cutoff of <0.05 following a moderated t-test.
2.6 Antagomir-mediated inhibition of miRNAs
To silence selected miRNAs, we used antagomirs, chemically synthesized short RNA oligonucleotides with complementary miRNA sequences that bind and sequester their sequence-specific miRNAs. Antagomirs were designed for the target miRNAs and synthesized by Dharmacon, Thermo Fisher Scientific. miRBase-annotated sequences (http://www.mirbase.org/) were used to design the complementary antagomirs. Missense controls containing a scrambled sequence were also designed. Antagomirs contained the following sequences: ant.305, 5’ CAGAGCACCUGAUGAAGUACAAU 3’; mis.305, 5’ CAGAGGGCCGCAUGCCUAGUGCU 3’; ant.989, 5’ GUACCACUACGUCACAUCACA 3’; and mis989, 5’ GUCAUACCACACCACGCAGUA 3’. All antagomirs contained the following modifications as previously described (Krutzfeldt et al., 2005): 1) a 3’ cholesterol group at the antagomir 3’ end, 2) a phosphorothioate backbone in place of a phosphodiester at the first two and the last four nucleotides, and 3) a 2’ O-methyl modification of all nucleotide ribose molecules. Antagomirs were injected into the thoraces of 3- to 4-day-old cold-anesthetized female mosquitoes using a nanoinjector (Nanoject, Drummond), in a volume of 69 nl at the concentrations indicated within the figures.
2.7 Treatment of mosquitoes with miRNA mimics
We designed and synthesized artificial miRNAs that mimic Dicer-processed mature miRNAs, as previously described (Boutz et al., 2007). In brief, DNA oligonucleotides were designed as template sequences for transcription of miRNA sense and antisense strands. Oligonucleotides contained an eight-nucleotide tag to allow annealing with a universal T7 promoter containing oligonucleotide (5’ TAATACGACTCACTATAGGGAGACAGG 3’) and were then filled in using DNA polymerase I Klenow fragment (New England BioLabs, NEB) to create dsDNA sense and antisense templates. ssRNA strands were then synthesized using HiScribe T7 RNA polymerase (NEB), and complementary RNA strands were annealed. The duplex RNA was digested with Turbo DNase 1 (5 U; Life Technologies) to remove the DNA template and with RNase T1 (200 U; Thermoscientific) to generate the characteristic miRNA 3’ two-nucleotide overhang (Lee et al., 2003). Synthetic miRNAs were then extracted with phenol-chloroform and precipitated with ethanol, then resuspended in sterile H20 to a concentration of 250 μM. The aga-miR-305 specific primers were: sense, 5’ CAGAGCACCTGATGAAGTACAATCCTGTCTC 3’ and antisense, 5’ TTAGTGTACTTCATCAGGTGCTCCCTGTCTC 3’. The control mimic primers used were: sense, 5’ CTTAGTGCAGGTCGATACTAGCTCCTGTCTC 3’ and antisense, 5’ TTATCTAGTATCGACCTGCACTACCTGTCTC 3’. Mimics and control mimics were injected with a nanoinjector into the thoraces of cold-anesthetized 3- to 4-day-old female mosquitoes in a volume of 69 nl.
2.8 Plasmodium infection assays
Mosquitoes were fed on NF54W P. falciparum gametocytes (0.01% gametocytemia) (provided by the Johns Hopkins Malaria Institute Parasitology Core Facility) in human blood through an artificial membrane feeder at 37°C for 30 min. The adult mosquitoes were starved for 8–12 hr prior to feeding to ensure engorgement, and unfed mosquitoes were removed from the cohort within 2 hr. Mosquitoes were incubated for a further 8 days at 27°C, and midguts were dissected in PBS, stained with 0.1% mercurochrome, and examined using a light-contrast microscope (Olympus) to determine oocyst numbers. At least three biological replicates were performed for each P. falciparum infection assay.
2.9 Quantification of culturable mosquito midgut bacteria
The culturable bacteria of mosquito midguts were quantified by colony forming unit (CFU) assay as previously described (Blumberg et al., 2013). In brief, individual female mosquitoes were surface-sterilized by washing them in 100% ethanol for 2 min and then rinsing them for 1 min in sterile 1x PBS. This process was repeated twice, and sterilized mosquitoes were streaked onto LB agar plates to confirm sterilization of the external surface. Individual mosquitoes with greater than 10 CFU on the external surface were excluded from the analysis. Midguts were then dissected from each individual mosquito over a sterile glass slide containing a drop of 1x PBS, transferred to a microcentrifuge tube containing 200 μl of sterile 1x PBS, and homogenized for 1 min. Ten-fold serial dilutions were plated onto LB agar plates and incubated at room temperature for 72 hr, and CFUs per plate (minimum 25) were counted and used to determine CFU/midgut. If the CFU number fell below the limit of detection, the CFU number was determined to be that of the lowest possible value from the dilutions plated. Eight to ten midguts were used per replicate, and three replicates were performed for each experiment. Fold change in number of culturable bacteria was determined from the geometric mean of CFU number across all three replicates for each experimental group.
2.10 In silico prediction of miRNA targets
Potential immune target genes of A. gambiae miRNAs were identified using three different miRNA prediction algorithms, RNAHybrid, miRanda and PITA. A. gambiae miRNAs (a total of 66, accessed from miRBase http://www.mirbase.org/) were used to search for potential targets within A. gambiae 3’ UTR sequences (Vectorbase AgamP3.7). RNAHybrid was used with the following parameters: energy value ≤23 kcal/mol and >6-nucleotide match within the seed region (Rehmsmeier et al., 2004). Miranda was utilized, and candidates with a minimum free energy value ≤23 kcal/mol and a score >80 were selected (Enright et al., 2003). PITA was implemented, using the following parameters: a ΔΔG energy value ≤-10 and a minimum six-nucleotide similarity within the seed region (Kertesz et al., 2007). The identities of predicted miRNA targets were determined using Kobas default parameters, with A. gambiae as the reference species (Xie et al., 2011). Potential targets of differentially regulated miRNAs were predicted using the same software and parameters.
3. Results and Discussion
3.1 RNAi depletion of miRNA biogenesis factors renders mosquitoes more resistant to P. falciparum infection
To study the influence of miRNA biogenesis on P. falciapium infection, we coupled gene silencing with P. falciparum infection experiments. We inhibited the biogenesis of miRNAs by targeting two key genes within the pathway, ago1 and dicer1, prior to infecting mosquitoes with the parasite. Ago1 is required for recruitment of the RISC complex to target mRNAs, and Dicer1 is an RNase III enzyme that processes miRNAs into their mature forms (Hutvagner et al., 2001; Tomari et al., 2007). It has previously been shown that genes of the A. gambiae miRNA biogenesis pathway exhibit increased polysome loading following P. falciparum infection (Mead et al., 2012). In addition, RNAi-mediated silencing of dicer1 and ago1 results in an increase in P. berghei infection (Winter et al., 2007). Silencing of both genes independently resulted in a decreased median number of oocysts (infection intensity) when compared to infected GFP dsRNA-injected control mosquitoes (Fig.1A). This effect was not significant (Kruskal-Wallis p = 0.0812), despite a three-fold reduction in the median number of oocysts following knockdown of ago1 transcripts. However, the reduction in infection intensity was significant for Ago1-silenced mosquitoes when the data concerning the silencing-mediated infection phenotype were separately compared to those for the GFP dsRNA control cohort using the Mann-Whitney test (ago1: p = 0.0363, dicer1: p = 0.1026). A significant reduction in prevalence, from 71.8% to 52.4% (p = 0.0278), was observed for dsAgo1 but not for dsDicer1 (reduced to 59.7% [p = 0.1578]) (Fig.1B) when compared to the GFP dsRNA-treated control cohort. Transcripts of both ago1 and dicer1 were efficiently silenced, reducing transcript abundance by 49.6 ± 10.7 % and 58.9 ± 5.5 % respectively.
Figure 1. RNAi silencing of miRNA biogenesis pathway genes.
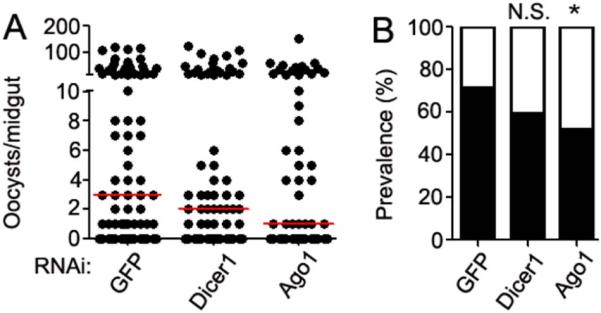
(A) P. falciparum oocyst loads in the midguts of A. gambiae following RNAi-mediated depletion of dicer1 and ago1 transcripts or injection with a dsGFP control. Oocysts were counted at 8 days post-infection, and data shown are pooled from three biological replicates. Dots represent the number of oocysts per individual mosquito, and the red horizontal line is the median number. Statistical significance for differences between groups was determined using a Kruskal-Wallis test (p=0.0812). Additional statistical analysis is described within the main text. (B) Prevalence of P. falciparum infection following RNAi treatment. The filled portion of bars represent the percentage of all mosquitoes harboring at least one oocyst; the open portion represents those in the group that were uninfected. Significance was determined by chi-square with Yates' correction. * p<0.5.
The differential effects of gene silencing on P. falciparum infection intensity and prevalence have been previously documented (Mitri et al., 2009). For example, targeted gene silencing of APL1 anti-Plasmodium family members results in a significant decrease in P. falciparum prevalence but not intensity (Mitri et al., 2009). Hence, a significant impact upon infection prevalence following RNAi silencing of ago1 demonstrates that the miRNA biogenesis pathway affects Plasmodium development. A reduction in infection prevalence is relevant, since even a single oocyst can produce over a thousand sporozoites that can result in disease transmission (Stone et al., 2013). We speculate that the non-significant effect of dicer1 knockdown may be as a result of some miRNAs undergoing maturation in a Dicer-independent pathway, which would limit the impact of dsDicer1 treatment, as previously described (Cheloufi et al., 2010). Our data indicate that interrupting the miRNA biogenesis pathway interferes with P. falciparum development, but in the opposite direction from its influence on P. berghei infection (Winter et al., 2007). The previously shown opposite effect of Ago-1 silencing on P. berghei infection likely relates to differences in regulation of the immune responses to the rodent and human malaria parasites (Dong et al., 2006). As noted previously, P. berghei infection is known to be controlled by the TOLL pathway whereas P. falciparum infection is inhibited by the IMD pathway (Clayton et al., 2014).
3.2 In silico prediction of A. gambiae miRNA immune targets
miRNAs post-transcriptionally regulate mRNAs in a sequence-specific manner, with the 5’ seed region (positions 2 to 8) being highly important for regulation (Brennecke et al., 2005). This sequence similarity has been used to design algorithms to aid in the prediction of miRNA target genes (Enright et al., 2003). To identify putative miRNAs involved in immune regulation and response to Plasmodium infection, we took an in silico screening approach, as performed previously in D. melanogaster (Fullaondo and Lee, 2012). Since the IMD pathway has been linked extensively to the mosquito anti-Plasmodium defense (Dong et al., 2011), we screened for potential miRNAs that target IMD pathway-associated genes. We used three different prediction softwares to minimize the number of false-positives. RNAhybrid, miRanda, and PITA were used to search 3’UTR sequences (Vectorbase AgamP3.7) for potential targets of 66 A. gambiae miRNAs accessed from miRBase (http://www.mirbase.org). RNAhybrid determines the most energetically favorable hybridization between the miRNA and target mRNA while ensuring strict sequence similarity within the seed region (Rehmsmeier et al., 2004). miRanda determines interactions through free energy dynamics across the entire miRNA:mRNA interaction, with increased weighting given to the 5’ miRNA seed region (Enright et al., 2003). PITA requires sequence similarity within the seed region but also determines the accessibility of the mRNA 3’ untranslated region based upon free energy (Kertesz et al., 2007). We identified a total of 23 miRNAs that potentially regulate the expression of eight IMD genes, including those with known anti-Plasmodium roles (Table 1). Of particular interest were aga-miR-2-1 and aga-miR-11, which were predicted by all three programs to contain binding sites within the 3’-UTR of the P. falciparum antagonists IMD and PGRP-LC, respectively (Garver et al., 2012; Meister et al., 2009). In addition, both RNAhybrid and miRanda predicted binding sites for aga-miR-305 within the 3’-UTR of the anti-Plasmodium effector gene APL1C (Garver et al., 2012). The short length of the APL1C 3’-UTR sequence prevented PITA from predicting aga-miR-305 binding sites, since it is below the minimum required length. It should be noted that not all IMD pathway genes have annotated 3’-UTRs; therefore, this analysis may underestimate the number of potential immune regulatory miRNAs.
Table 1.
Predicted targets of the IMD pathway.

|
Potential IMD target genes were identified using in silico analysis of 66 A. gambiae miRNAs (accessed from miRBase http://www.mirbase.org/) using RNAhybrid, miRanda, and PITA software. miRNAs predicted by two and three of the programs are highlighted in red and blue, respectively. The software parameters used are detailed in Materials and Methods. A. gambiae immune gene names, accession numbers, and miRbase miRNA names are given in the table.
We extended our analysis to include members of the TOLL pathway and selected antimicrobial peptides. There are 23, 19, and 4 miRNAs with potential binding sites within the transcripts of IMD pathway genes, antimicrobial peptide (AMP) genes, and putative TOLL genes, respectively (Table S1). A total of 8, 6, and 3 transcripts of IMD, AMP, and TOLL genes, respectively, were targeted by at least one miRNA. Although some miRNAs were specific to an individual group, three miRNAs were identified as potentially regulating genes related to both the IMD and TOLL pathways, and two miRNAs were identified that may regulate genes within both pathways and an antimicrobial peptide. These findings suggest that a single miRNA can influence multiple immune factors, and the ability of a single miRNA to regulate multiple transcripts has previously been demonstrated (Hussain et al., 2013). For example, aga-miR-34 has predicted binding sites within the 3’-UTR of the TOLL pathway transcription factor REL1, the IMD pathway negative regulator Caspar, and an antimicrobial peptide gene encoding Cecropin3. Thus, using an in silico approach, we have shown here that several A. gambiae miRNAs potentially regulate the expression of known immune genes, including those involved in the anti-Plasmodium defense.
3.3 A. gambiae miRNAs are differentially expressed following an infectious blood meal
Since our in silico analysis identified A. gambiae miRNAs that potentially regulate anti-Plasmodium genes (Table 1), we next used a microarray approach to investigate whether any of these miRNAs or other miRNAs are differentially expressed upon P. falciparum infection of the midgut tissue. A. gambiae miRNA abundances were compared between P. falciparum-infected (median oocyst number = 13) and -naive midguts from three independent biological experiments. Fold changes in miRNA abundance levels between infected and non-infected mosquitoes were determined below a significance threshold (p<0.05), with a log2-fold change (FC) >1. Differential profiling of 194 mosquito miRNAs, including 66 A. gambiae miRNAs (available from miRBase), was conducted at two time points (18 and 24 hr) post-feeding on a P. falciparum gametocyte culture, the time at which the ookinetes invade the midgut epithelium (Fig. S1). At 18 hr, 65 miRNAs (31 A. gambiae) were detected in both naïve and infected mosquitoes, and 8 (2 A. gambiae) of these showed differential abundance between the compared samples. At 24 hr, 62 miRNAs (31 A. gambiae) were detected in both naïve and infected mosquitoes, and 8 of these (5 A. gambiae) showed differential abundance. Of the differentially expressed miRNAs (Table 2), the abundance of aga-miR-989 (log2 FC = 1.74, −log10p value = 3.31) and aga-miR-305 (log2 FC = 7.24, −log10p value = 7.13) was significantly elevated at 18 and 24 hr, respectively. Interestingly, aga-miR-989 has previously been shown to exhibit a four-fold increased abundance in the midgut at 24 to 48 hr after a P. berghei-infected blood meal (Winter et al., 2007). This miRNA contains predicted binding sites within the 3’UTR of the PRR gene PGRP-LD (Table 1). Expression of aga-miR-305 has also previously been observed within the midgut tissue (Winter et al., 2007), and it has in silico-predicted binding sites within the 3’-UTR of the anti-Plasmodium genes PGRP-LC and APL1C (Table 1). We confirmed the elevated abundance of aga-miR-305 and aga-miR-989 using Sybr Green qPCR assays and three reference miRNAs that displayed equal abundance across infected and control samples, as determined by NormFinder (Andersen et al., 2004) (Fig. S2). Thus, two miRNAs, aga-miR-989 and aga-miR-305, are up-regulated following a P.falciparum infected blood meal at 18 and 24 hr, respectively, when ookinete invasion of the midgut epithelium occurs.
Table 2.
Differential expression of A. gambiae miRNAs in response to P. falciparum at 18 and 24 hr post-infection.
| 18 hr | Log2 FC | −Log10 p value |
|---|---|---|
| aae-miR-276 | 4.55 | 1.20 |
| aae-miR-285 | 2.38 | 0.38 |
| aae-miR-2946 | −2.41 | 0.48 |
| aga-miR-11 | −2.75 | 0.70 |
| aga-miR-989 | 1.74 | 3.31 |
| cqu-miR-210 | 2.54 | 0.38 |
| cqu-miR-2951 | −3.10 | 0.69 |
| cqu-miR-92 | 2.06 | 0.32 |
| 24 hr | Log2 FC | −Log10 p value |
|---|---|---|
| aae-miR-2946 | 2.85 | 0.68 |
| aae-miR-34* | −0.14 | 0.35 |
| aga-bantam | 2.42 | 0.63 |
| aga-miR-11 | 2.23 | 0.61 |
| aga-miR-305 | 7.24 | 7.13 |
| aga-miR-317 | 2.06 | 0.31 |
| aga-miR-989 | 2.42 | 0.40 |
| cqu-miR-2951 | 2.75 | 0.66 |
A custom Agilent miRNA microarray was used to compare the miRNA expression profile of A. gambiae midguts at 18 and 24 hr after P. falciparum infection to that of mosquitoes fed on non-infected human blood. Log2FC values were determined from the mean expression value across three biological replicates, following background subtraction and normalization to the 75th percentile. P-values were determined using a moderated t-test. Only miRNAs detected above background level in at least two of the three biological replicates are shown.
3.4 Antagomir-mediated depletion of aga-miR-305, but not aga-miR-989, increases mosquito resistance to P. falciparum infection
To determine whether aga-miR-989 and aga-miR-305 influence A. gambiae's susceptibility to P. falciparum infection, we used an miRNA inhibition approach based on antagomirs, chemically synthesized short oligonucleotides with the reverse-complementary sequence to a specific miRNA. Antagomirs bind and sequester their complementary miRNAs, preventing regulation of target transcripts. Depletion of miRNAs in vivo by antagomir delivery has been demonstrated in D. melanogaster (Gehrke et al., 2010) and Ae. aegypti (Bryant et al., 2010). We designed two pairs of antagomirs, ant.305 and ant.989, with reverse-complementary sequence to that of the mature aga-miR-305 and aga-miR-989 sequences, respectively. Control antagomirs (mis.305 and mis.989) were designed containing scrambled sequences of the mature miRNA. Treatment of mosquitoes with ant.305 prior to feeding on a P. falciparum gametocyte culture resulted in a significant reduction in oocyst intensity when compared to the missense control antagomir-treated infected mosquitoes (Fig. 2A). For aga-miR-305, we conducted antagomir-mediated depletion at two different concentrations (100 μM and 250 μM) to determine which would be optimal for inhibition. Both concentrations resulted in a significant decrease in infection intensity (100 μM, p=0.0041; 250 μM, p=0.0181) across three biological replicates. Delivery of the antagomir resulted in a significant decrease in parasite prevalence from 89.7% to 66.7% (p= 0.0049) at 100 μM and from 80.5% to 67.8% that was marginally non-significant (p= 0.0834) at 250 μM. Microarray analysis revealed that aga-miR-305 is expressed at low levels when compared to other miRNAs (data not shown). This low expression of aga-miR-305 in P. falciparum-infected tissues likely results in complete inhibition of the miRNA, even when treated with the lower concentrations of antagomir. We note that in two additional single replicate experiments, we observed no significant difference (100 μM, p=0.5108 and 250 μM, p=0.1697) between the intensity of infection in antagomir- and control-injected mosquitoes (data not shown). We therefore confirmed the efficiency of ant.305-mediated silencing of aga-miR-305 using miRNA qRT-PCR, observing a 98.7 ± 1.1 % and 97.1 ± 3.1 % reduction in aga-miR-305 abundance after 100 μM and 250 μM ant.305 treatment, respectively. Highly efficient silencing of individual miRNAs in mosquito whole bodies using antagomirs has previously been observed (Bryant et al., 2010). As aga-miR-305 was effectively silenced, we hypothesize that this variability is a result of the low level expression of aga-miR-305 and that silencing of a limited number of miRNAs does not always significantly influence susceptibility. One experiment that showed no significant impact of aga-miR-305 silencing had a high infection intensity (median oocysts >25) in the control cohort, suggesting that aga-miR-305 may have a limited impact when the infection levels are unnaturally high. It is also possible that some variation in the abundance and composition of the mosquito microbiota that is known to modualte P. falciparum susceptibility (Dong et al., 2009), between cohorts, may have contributed to the observed variation. Despite this variability, our data show that inhibiting aga-miR-305 can increase the resistance to P. falciparum infection. Targeted inhibition of aga-miR-305 resulted in a significant decrease in both infection intensity and prevalence (Fig. 2A), whereas silencing of the miRNA biogenesis pathway only affected prevalence (Fig.1B). RNAi-mediated knockdown of ago1 and dicer1 has a lesser effect upon individual miRNA expression than does the delivery of specific antagomirs (Bryant et al., 2010). We speculate that the depletion of miRNA biogenesis pathway components will not completely silence individual miRNAs, and that this result may explain the difference in infection phenotype seen when targeting ago1 and aga-miR-305 individually.
Figure 2. P. falciparum infection intensity following antagomir inhibition of aga-miR-305 and aga-miR-989.
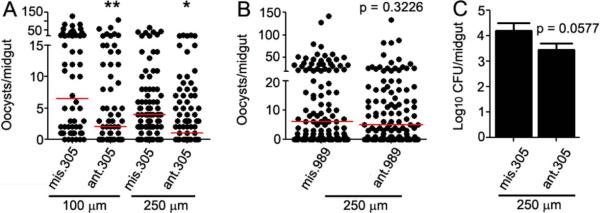
P. falciparum oocyst loads in the midguts of A. gambiae mosquitoes following inhibition of (A) aga-miR-305 and (B) aga-miR-989 using antagomirs at the concentrations indicated in the figure. Oocysts were counted at 8 days post-infection, and data shown are pooled from three biological replicates. Circles represent the number of oocysts per individual mosquito, and the red horizontal line is the median number. The indicated statistical significance was determined by Mann-Whitney tests. **, p<0.01, *, p<0.05. (C) Delivery of ant.305 results in a marginally non-significant decrease in the colony forming units (CFU) of cultivable bacteria when compared to the mis.305 control. Data were pooled from three independent biological replicates, and statistical significance was determined by an unpaired t-test. ant = antagomir, mis = missense control.
Treatment of mosquitoes with ant.989 at the higher (250 μM) concentration did not affect the intensity (p>0.05) or prevalence (p>0.05) of infection when compared to its missense control (Fig. 2B). This non-significant effect of aga-miR-989 inhibition may be a result of its incomplete inhibition of the endogenous miRNAs. Alternatively, aga-miR-989 may be regulated by infection but not target transcripts involved in defense-related processes. A previous study has shown that miRNA antagomir treatment of Ae. albopictus can result in different degrees of miRNA inhibition (Puthiyakunnon et al., 2013), and antagomir treatment of Ae. aegypti has been shown to result in a low-penetrance phenotype despite miRNA inhibition (Bryant et al., 2010). Therefore, we cannot currently conclude whether the lack of impact of aga-miR-989 on P. falciparum infection is the result of a lack of biological impact or of technical limitations.
Anti-Plasmodium and antibacterial defenses are largely regulated by the same immune factors in A. gambiae, and the IMD pathway is involved in controlling the mosquito midgut microbiota (Dong et al., 2006). Since aga-miR-305 can regulate P. falciparum infection, we hypothesized that it is also likely to regulate the mosquito midgut microbiota. To test this possibility, we used CFU assays to assess the midgut microbial load of aga-miR-305-depleted and control mosquitoes. Depletion of aga-miR-305 resulted in a ~5-fold decrease in the number of cultivable bacteria per mosquito midgut that was nearly statistically significant (p = 0.0577) (Fig. 2C). This non-significant effect may have been a result of the very low-level expression of aga-miR-305 in non-Plasmodium-infected mosquitoes (data not shown). In the absence of P. falciparum infection-dependent induction of aga-miR-305 expression, inhibition of the basal levels of aga-miR-305 miRNAs may simply be insufficient to result in a significant phenotype. In summary, depletion of aga-miR-305 influences both the anti-Plasmodium and antibacterial defense of the mosquito, resulting in an increased resistance.
3.5 Injection of artificial aga-miR-305 miRNAs decreases mosquito resistance to P. falciparum infection
To further confirm the involvement of aga-miR-305 in the post-transcriptional regulation of the anti-Plasmodium defense, we synthesized artificial miRNAs that mimic Dicer-processed mature miRNAs, as previously described (Boutz et al., 2007). We designed an aga-miR-305 mimic (mimic305) in such a way that the mature miRNA sense strand of aga-miR-305 would be incorporated into the RISC and function as an endogenous miRNA (Materials and Methods). We also designed a missense control mimic (ctrl305) with a scrambled sequence to prevent binding to target mRNAs. Mosquitoes injected with the mimic prior to feeding on a P. falciparum gametocyte culture displayed a significant increase (p=0.0068) in infection intensity when compared to the missense control mimic-treated, infected cohort (Fig. 3A). Infection prevalence increased from 81.4% in ctrl.305-injected individuals to 92.1% in mimic.305-injected individuals, although this result was marginally non-significant (P=0.0603). Injection of the aga-miR-305 mimic resulted in the opposite phenotype to that observed following antagomir inhibition of aga-miR-305, thereby demonstrating that this miRNA is involved in regulating A. gambiae resistance to Plasmodium infection. In addition, we also observed a ~8-fold increase (p = 0.0541) in the midgut microbial load of mimic-injected mosquitoes when compared to the missense control (Fig. 3B). This increase is directly correlated with the decreased microbial load observed when aga-miR-305 is depleted through antagomir inhibition. These data confirm that aga-miR-305 regulates both the anti-Plasmodium and antibacterial host defense.
Figure 3. P. falciparum infection intensity following injection of an aga-miR-305 mimic.
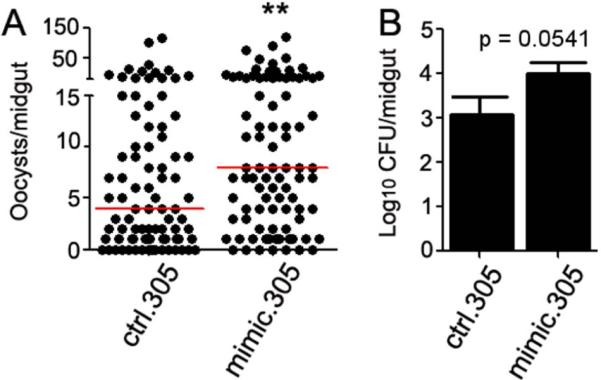
(A) P. falciparum oocysts laods in the midguts of A. gambiae following injection of an aga-miR-305 mimic (mimic.305) or a scrambled missense control (ctrl.305). Oocysts were counted at 8 days post-infection, and data shown are pooled from three biological replicates. Circles represent the number of oocysts per individual mosquito, and the red horizontal line is the median number. The indicated statistical significance was determined by Mann-Whitney tests. **, p<0.01. (B) Delivery of mimic.305 results in a marginally non-significant increase in the colony forming units (CFU) of cultivable bacteria when compared to the ctrl.305 control. Data were pooled from three independent biological replicates, and statistical significance was determined by an unpaired t-test.
3.6 aga-miR-305 potentially targets multiple immune genes
The degenerate nature of the regulatory mechanism of miRNA action allows a single miRNA to regulate a network of multiple genes. In addition, ookinete invasion of the midgut epithelia results in changes in the transcription of many genes (Dong et al., 2006). Therefore, it is likely that the infection phenotypes observed following manipulation of aga-miR-305 abundance were the result of the targeting of multiple genes by aga-miR-305, rather than a single miRNA-gene relationship. miRNA target genes are commonly predicted through sequence similarity between the miRNA seed region, at nucleotide positions 2-8, and the 3’-UTR of target transcripts (Brennecke et al., 2005). More recently, miRNA binding sites have been identified within the 5’-UTR as well as the coding regions of transcripts (Hussain et al., 2013). Therefore, we undertook a comprehensive in silico approach to identify all potential targets of aga-miR-305 that may have contributed to the anti-P.falciparum and antibacterial phenotypes. We used RNAHybrid, miRanda, and PITA software to predict aga-miR-305 binding sites within the annotated 5’-UTR, the coding sequence, and the 3’-UTR sequences of all A. gambiae genes (Vectorbase AgamP3.7). Using the software-predicted transcripts, we pursued three approaches to isolate candidate aga-miR-305 targets that may account for the observed infection phenotypes: 1) We first focused on candidate genes that were predicted by at least two of the software programs; 2) we next compared these targets to transcripts that have previously been shown to be differentially regulated upon P. falciparum infection; and, finally, 3) we identified known anti-Plasmodium genes with binding sites for aga-miR-305.
In total 35, 1330, and 44 targets were predicted for the 5’-UTR, coding sequence, and 3’-UTR, respectively, by at least two of the software programs (Fig. 4). RNAhybrid consistently predicted a higher number of targets than PITA and miRanda did (Fig. 4). Intriguingly, RNAHybrid and miRanda predicted aga-miR-305 binding sites within the 3’-UTR of APL1C (Table 1). APL1C is part of a group of three paralogous leucine-rich repeat (LRR) proteins found within the Plasmodium-Resistance Island (PRI) locus that is linked to natural resistance to P. falciparum infection (Riehle et al., 2006). APL1 forms a complex with a second LRR protein (LRIM1) that stabilizes the mature form of the complement-like factor TEP1, which then binds to the parasite surface and targets the parasite for destruction (Fraiture et al., 2009; Povelones et al., 2009). Gene-silencing experiments have also demonstrated that APL1C protects A. gambiae mosquitoes against P. falciparum (Garver et al., 2012). Because injection of the aga-miR-305 mimic resulted in an increase in susceptibility to P. falciparum, we hypothesize that the miRNA negatively regulates the P. falciparum antagonist APL1C to prevent an over-activation of the immune system. P. falciparum-induced expression of aga-miR-305 (Table 2) may therefore act as a negative regulator to fine-tune the immune response and prevent an over-allocation of mosquito resources to the defense against invading parasites.
Figure 4. In silico prediction of A. gambiae aga-miR-305 targets using RNAHybrid, miRanda, and PITA.
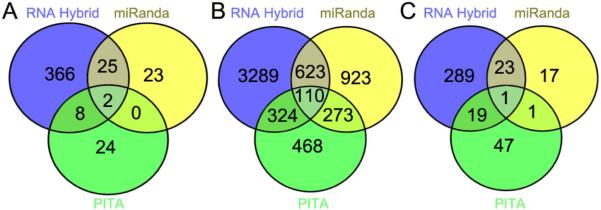
Gene targets of aga-miR-305 were predicted within the (A) 5’ untranslated region, (B) coding sequence, and (C) 3’ untranslated region of A. gambiae genes (Vectorbase: AgamP.37) using the software programs RNAHybrid, miRanda, and PITA. Details of the parameters used for each program are provided in Materials and Methods. Venn diagrams were created using the online software Venny (Oliveros, (2007, accessed June 2014)).
In addition to APL1C, aga-miR-305 binding sites were predicted within transcripts of other immune genes; binding sites were detected by two software programs within the 5’-UTR of PGRP-LB, a PRR that is up-regulated in response to both bacterial and Plasmodium challenge (Dimopoulos et al., 2002). Binding sites within the 3’-UTR of a second PRR, PGRP-LC, were also predicted by the PITA software. PGRP-LC activates the IMD pathway following recognition of bacterial peptidoglycan and mediates bacteria-dependent anti-P. falciparum activity (Meister et al., 2009). SCRBQ2, part of a group of scavenger receptors that recognize polyanionic ligands, contains aga-miR-305 binding sites within the coding region. Expression of scavenger receptor proteins are up-regulated following P. falciparum and P. berghei infection, and SCRBQ2 acts as a Plasmodium agonist, aiding the establishment of parasite infection (Blumberg et al., 2013; Dong et al., 2006; Gonzalez-Lazaro et al., 2009). Binding sites were also predicted within the coding region of five different clip-domain serine protease (CLIP) genes. CLIPs mediate serine protease cascades that activate pro-phenoloxidases (PPO), resulting in the killing of Plasmodium parasites (Volz et al., 2006). Of particular note is CLIPB17, which negatively regulates P. berghei infection by promoting melanization of the invading parasite (Volz et al., 2006). Serpin2 (SRPN2), an inhibitor of PPO that promotes the survival of invading Plasmodium parasites, contains aga-miR-305 binding sites within the coding region (Michel et al., 2005). It is interesting to note that SRPN2 and SCRBQ2 are agonists of P. falciparum development; therefore, we hypothesize that aga-miR-305 may stabilize SRPN2 and SCRBQ2 transcripts to temper the anti-Plasmodium PPO response. Similarly, miRNAs have previously been shown to increase the relative transcript levels of negative immune regulators in Ae. aegypti (Hussain et al., 2013).
We also compared the aga-miR-305 targets of midgut transcripts (predicted by at least two of our software programs) that are known to be differentially regulated 24 hr after the mosquito feeds on P. falciparum-infected blood, when the ookinetes invade the midgut epithelium (Blumberg et al., 2013). Of the 1,397 predicted transcript targets of aga-miR-305, 196 (96 up-regulated and 100 down-regulated) were also differentially expressed upon P. falciparum infection (Blumberg et al., 2013). Of particular interest are Spz2, which has been suggested to activate the TOLL pathway (Akhouayri et al., 2011); the serine proteases CLIPB17 and CLIPB36; the pattern recognition protein PGRP-LB; and a family member of the gal-lectin PRRs that are up-regulated in response to P. falciparum infection (Dong et al., 2006). Our in silico analysis suggests that aga-miR-305 regulation of the anti-P. falciparum response is likely to be complex and not mediated by a single miRNA target gene. A single miRNA is able to regulate over 100 different target transcripts (Lim et al., 2005), suggesting that whole-genome transcriptome analysis in response to miRNA expression, correlated with the in silico analysis presented here, has the potential to fully characterize the target genes responsible for the associated phenotypes.
4.0 Conclusion
Here we have demonstrated that the A. gambiae miRNA biogenesis pathway regulates resistance to P. falciparum infection. P. falciparum infection results in the regulation of mosquito miRNAs, of which miRNA aga-miR-305 is particularly noteworthy: It negatively regulates the A. gambiae anti-Plasmodium and antibacterial defenses and is therefore likely to post-transcriptionally regulate a variety of immune genes. Inhibition of aga-miR-305 by antagomir-mediated depletion results in a greater resistance to infection, and, conversely, injection of the aga-miR-305 mimic results in an increased susceptibility to infection. We have also presented a comprehensive target analysis of aga-miR-305 that may provide information on the target genes that are responsible for this miRNA's influence on P. falciparum development. These data provide a foundation for further investigation of the role of specific miRNAs in regulating the A. gambiae immune response to malaria parasites and bacteria.
There is growing evidence that miRNAs are involved in negatively regulating the immune response to prevent an over-activation of the immune system. For example, in mammals, bacterially induced miRNAs negatively regulate Toll-like receptor (TLR) signaling (Nahid et al., 2011). Several insect miRNAs have previously been shown to negatively regulate innate immune pathways and to determine susceptibility to infection (Choi and Hyun, 2012; Etebari and Asgari, 2013; Hussain et al., 2013; Lee and Hyun, 2014). Most notably, an Ae. aegypti blood meal-inducible miRNA negative regulates the TOLL pathway through up-regulation of Cactus and down-regulation of REL1 (Hussain et al., 2013). We hypothesize that P. falciparum infection results in the up-regulation of aga-miR-305, which then fine-tunes the immune response through the targeting of immune genes. By this process, the mosquito's immune response is tempered to prevent over-allocation of resources to the host defense response.
Transgenic insects can be generated to express tandem repeats of miRNA binding sites (miRNA sponges) and thereby effectively inhibit the expression of endogenous miRNAs (Loya et al., 2009). miRNA sponge vectors result in a greater degree of miRNA inhibition than does the antagomir approach used in this study (Bak et al., 2013), and transgenic direct intracellular production of the miRNA sponge would be expected to enhance and/or prolong inhibition when compared to antagomir injection into the hemolymph. miRNA sponge methodology could be used to develop P. falciparum-resistant transgenic mosquito lines with blood meal-inducible depletion of parasite agonist miRNAs, such as aga-miR-305, in the midgut and fat body tissues. We have previously demonstrated that transgenic overexpression of the A. gambiae IMD pathway-controlled transcription factor REL2 can lead to a near-refractory phenotype to P. falciparum infection (Dong et al., 2011). We hypothesize that the sponge methodology could be combined with such existing Rel2-overexpressing transgenic lines, thereby potentiating the resistance phenotype. Over 3 billion people worldwide are at risk of malaria infection, and the disease remains a constant burden, calling for novel control strategies. Further investigation of A. gambiae immune-responsive miRNAs may contribute toward the development of novel malaria control strategies based on Plasmodium-resistant transgenic mosquitoes (Dong et al., 2011).
Supplementary Material
Figure S1. The red dot indicates the miRNA considered to be statistically significant. Black dots indicate miRNAs not considered statistically significant. The horizontal axis represents the log2 FC, and vertical axis the –log10 P-value (in a moderated t-test) for differences in expression following normalization to the 75th percentile.
Figure S2: RNA from P. falciparum-infected and naïve mosquitoes that were used for miRNA microarray analyses was used for qRT-PCR analysis of (A) aga-miR-989 18-hr expression and (B) aga-miR-305 24-hr expression. Expression was normalized to the geometric mean of reference miRNAs that displayed equal abundance across infected and control samples (A: aga-miR-2, aga-miR-279, and aga-miR-9b; B: aga-miR-2, aga-miR-1175, and aga-miR-9b). Reference miRNAs were selected following NormFinder analysis of miRNA microarray expression data.
Table S1: Predicted targets of the TOLL pathway and antimicrobial peptides (AMPs). Potential TOLL and AMP target genes were identified using in silico analysis of 66 A. gambiae miRNAs (accessed from miRBase http://www.mirbase.org/) using RNAhybrid, miRanda and PITA software. miRNAs predicted by two software programs are highlighted in red. The software parameters used are detailed in Materials and Methods. A. gambiae immune gene names, accession numbers, and miRbase miRNA names are given in the table.
Table S2: Primers used for dsRNA synthesis, qRT-PCR and expression profiling of miRNAs.
Table S3: Statistical analyses of oocyst loads in A. gambiae P. falciparum infection experiments.
Highlights.
A. gambiae miRNAs can potentially postranscriptionally regulate immune genes
Inhibiting the A. gambiae miRNA biogenesis pathway reduces P. falciparum infection prevalence
Two A. gambiae miRNAs are differentially expressed upon P. falciparum infection
aga-miR-305 acts as a negative regulator of the A. gambiae immune response
aga-miR-305 potentially regulates known A. gambiae anti-Plasmodium effector genes.
Acknowledgments
We thank the insectary personnel at the Johns Hopkins Malaria Research Institute for assistance with mosquito rearing. This work has been supported by the National Institutes of Health/National Institute for Allergy and Infectious Disease R01AI061576 and the Calvin A. and Helen H. Lang fellowship (to N.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo Negatively Regulates Antimicrobial Response in the Drosophila Respiratory Epithelium. PLoS Pathog. 2011;7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bak RO, Hollensen AK, Primo MN, Sorensen CD, Mikkelsen JG. Potent microRNA suppression by RNA Pol II-transcribed 'Tough Decoy' inhibitors. RNA-Publ. RNA Soc. 2013;19:280–293. doi: 10.1261/rna.034850.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukova I, Ye T, Levashina EA. Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics. 2014;15:557. doi: 10.1186/1471-2164-15-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg BJ, Trop S, Das S, Dimopoulos G. Bacteria- and IMD Pathway-Independent Immune Defenses against Plasmodium falciparum in Anopheles gambiae. PLoS One. 2013;8:e72130. doi: 10.1371/journal.pone.0072130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-target recognition. PLoS. Biol. 2005;3:404–418. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. PNAS. 2010;107:22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Harrison T, Hess AM, Ebel GD. MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 2014;23:132–139. doi: 10.1111/imb.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Schaffert S, Fragoso R, Loh C. Regulation of immune responses and tolerance: the microRNA perspective. Immunol. Rev. 2013;253:112–128. doi: 10.1111/imr.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IK, Hyun S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev. Comp. Immunol. 2012;37:50–54. doi: 10.1016/j.dci.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Clayton AM, Dong YM, Dimopoulos G. The Anopheles Innate Immune System in the Defense against Malaria Infection. J. Innate Immun. 2014;6:169–181. doi: 10.1159/000353602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, Kafatos FC. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection. PNAS. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YM, Aguilar R, Xi ZY, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:513–525. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YM, Das S, Cirimotich C, Souza-Neto JA, McLean KJ, Dimopoulos G. Engineered Anopheles Immunity to Plasmodium Infection. PLoS Pathog. 2011;7:e1002458. doi: 10.1371/journal.ppat.1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YM, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biology. 2003;5 doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etebari K, Asgari S. Conserved microRNA miR-8 blocks activation of the Toll pathway by upregulating Serpin 27 transcripts. RNA Biology. 2013;10:1358–1366. doi: 10.4161/rna.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiture M, Baxter RHG, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, Hoffmann JA, Blandin SA, Levashina EA. Two Mosquito LRR Proteins Function as Complement Control Factors in the TEP1-Mediated Killing of Plasmodium. Cell Host Microbe. 2009;5:273–284. doi: 10.1016/j.chom.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Fullaondo A, Lee SY. Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 2012;36:267–273. doi: 10.1016/j.dci.2011.03.034. [DOI] [PubMed] [Google Scholar]
- Garver LS, Bahia AC, Das S, Souza-Neto JA, Shiao J, Dong YM, Dimopoulos G. Anopheles Imd Pathway Factors and Effectors in Infection Intensity-Dependent Anti-Plasmodium Action. PLoS Pathog. 2012;8:e1002737. doi: 10.1371/journal.ppat.1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu BW. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lazaro M, Dinglasan RR, Hernandez-Hernandez FD, Rodriguez MH, Laclaustra M, Jacobs-Lorena M, Flores-Romo L. Anopheles gambiae Croquemort SCRBQ2, expression profile in the mosquito and its potential interaction with the malaria parasite Plasmodium berghei. Insect Biochemistry and Molecular Biology. 2009;39:395–402. doi: 10.1016/j.ibmb.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd H, Taylor PJ, Adams D, Underhill A, Eggleston P. Evaluating the costs of mosquito resistance to malaria parasites. Evolution. 2005;59:2560–2572. [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Walker T, O'Neill SL, Asgari S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochemistry and Molecular Biology. 2013;43:146–152. doi: 10.1016/j.ibmb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jain S, Rana V, Shrinet J, Sharma A, Tridibes A, Sunil S, Bhatnagar RK. Blood Feeding and Plasmodium infection alters the miRNome of Anopheles stephensi. PLoS One. 2014;9:e98402. doi: 10.1371/journal.pone.0098402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nature Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Koufaris C, Wright J, Currie RA, Gooderham NJ. Hepatic MicroRNA Profiles Offer Predictive and Mechanistic Insights After Exposure to Genotoxic and Epigenetic Hepatocarcinogens. Toxicol. Sci. 2012;128:532–543. doi: 10.1093/toxsci/kfs170. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee G, Hyun S. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Drosophila. Dev. Comp. Immunol. 2014 doi: 10.1016/j.dci.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han JJ, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han JJ, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nature Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas K, Raikhel AS. Insect MicroRNAs: Biogenesis, expression profiling and biological functions. Insect Biochemistry and Molecular Biology. 2013;43:24–38. doi: 10.1016/j.ibmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead E, Li M, Tu Z, Zhu J. Translational regulation of Anopheles gambiae mRNAs in the midgut during Plasmodium falciparum infection. BMC Genomics. 2012;13:366. doi: 10.1186/1471-2164-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S, Agianian B, Turlure F, Relogio A, Morlais I, Kafatos FC, Christophides GK. Anopheles gambiae PGRPLC-Mediated Defense against Bacteria Modulates Infections with Malaria Parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri C, Jacques JC, Thiery I, Riehle MM, Xu JN, Bischoff E, Morlais I, Nsango SE, Vernick KD, Bourgouin C. Fine Pathogen Discrimination within the APL1 Gene Family Protects Anopheles gambiae against Human and Rodent Malaria Species. PLoS Pathog. 2009;5:e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid MA, Satoh M, Chan EKL. MicroRNA in TLR signaling and endotoxin tolerance. Cell. Mol. Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC. [June 2014];VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-Rich Repeat Protein Complex Activates Mosquito Complement in Defense Against Plasmodium Parasites. Science. 2009;324:258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyakunnon S, Yao YY, Li YJ, Gu JB, Peng HJ, Chen XG. Functional characterization of three MicroRNAs of the Asian Tiger Mosquito, Aedes albopictus. Parasites Vectors. 2013;6:230. doi: 10.1186/1756-3305-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA-Publ. RNA Soc. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle MM, Markianos K, Niare O, Xu JN, Li J, Toure AM, Podiougou B, Oduol F, Diawara S, Diallo M, Coulibaly B, Ouatara A, Kruglyak L, Traore SF, Vernick KD. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- Stone WJR, Eldering M, van Gemert GJ, Lanke KHW, Grignard L, van de Vegte-Bolmer MG, Siebelink-Stoter R, Graumans W, Roeffen WFG, Drakeley CJ, Sauerwein RW, Bousema T. The relevance and applicability of oocyst prevalence as a read-out for mosquito feeding assays. Sci Rep. 2013;3:3418. doi: 10.1038/srep03418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz J, Muller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8:1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNAPubl. RNA Soc. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The red dot indicates the miRNA considered to be statistically significant. Black dots indicate miRNAs not considered statistically significant. The horizontal axis represents the log2 FC, and vertical axis the –log10 P-value (in a moderated t-test) for differences in expression following normalization to the 75th percentile.
Figure S2: RNA from P. falciparum-infected and naïve mosquitoes that were used for miRNA microarray analyses was used for qRT-PCR analysis of (A) aga-miR-989 18-hr expression and (B) aga-miR-305 24-hr expression. Expression was normalized to the geometric mean of reference miRNAs that displayed equal abundance across infected and control samples (A: aga-miR-2, aga-miR-279, and aga-miR-9b; B: aga-miR-2, aga-miR-1175, and aga-miR-9b). Reference miRNAs were selected following NormFinder analysis of miRNA microarray expression data.
Table S1: Predicted targets of the TOLL pathway and antimicrobial peptides (AMPs). Potential TOLL and AMP target genes were identified using in silico analysis of 66 A. gambiae miRNAs (accessed from miRBase http://www.mirbase.org/) using RNAhybrid, miRanda and PITA software. miRNAs predicted by two software programs are highlighted in red. The software parameters used are detailed in Materials and Methods. A. gambiae immune gene names, accession numbers, and miRbase miRNA names are given in the table.
Table S2: Primers used for dsRNA synthesis, qRT-PCR and expression profiling of miRNAs.
Table S3: Statistical analyses of oocyst loads in A. gambiae P. falciparum infection experiments.


