Abstract
Deterioration of adult stem cells accounts for much of aging-associated compromised tissue maintenance. How stem cells maintain metabolic homeostasis remains elusive. Here, we identified a regulatory branch of the mitochondrial unfolded protein response (UPRmt), which is mediated by the interplay of SIRT7 and NRF1 and is coupled to cellular energy metabolism and proliferation. SIRT7 inactivation caused reduced quiescence, increased mitochondrial protein folding stress (PFSmt), and compromised regenerative capacity of hematopoietic stem cells (HSCs). SIRT7 expression was reduced in aged HSCs, and SIRT7 up-regulation improved the regenerative capacity of aged HSCs. These findings define the deregulation of a UPRmt-mediated metabolic checkpoint as a reversible contributing factor for HSC aging.
Aging is characterized by physiological decline and increased susceptibility to pathologies and mortality. The rate of aging is controlled by evolutionarily conserved genetic pathways (1, 2). The general cause of aging is the chronic accumulation of cellular damage (2, 3). This conceptual framework raises fundamental questions about aging. What are the origins of aging-causing damage? What is the cell or tissue specificity for sensing or responding to such damage? Are the effects of cellular damage on physiological aging reversible?
Adult stem cells mostly reside in a metabolically inactive quiescent state to preserve their integrity but convert to a metabolically active proliferative state to replenish the tissue (4–6). The signals that trigger stem cells to exit the cell cycle and enter quiescence, and the signal transduction leading to the transition, remain elusive.
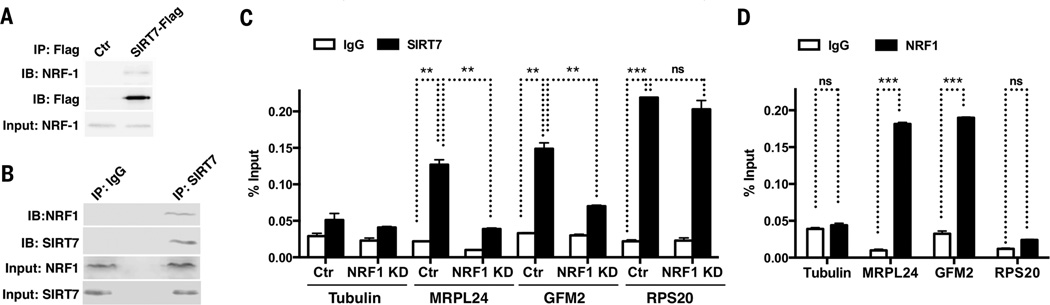
SIRT7 is a histone deacetylase that is recruited to its target promoters by interactions with transcription factors for transcriptional repression (7). We took a proteomic approach to identify SIRT7-interacting transcription factors. We transfected human embryonic kidney–293T (HEK-293T) cells with Flag-tagged SIRT7, affinity-purified the Flag-tagged SIRT7 interactome, and identified SIRT7-interacting proteins by mass spectrometry. Among the potential SIRT7-interacting proteins was nuclear respiratory factor 1 (NRF1), a master regulator of mitochondria (8). Transfected Flag-SIRT7 and endogenous SIRT7 interacted with NRF1 in HEK-293T cells (Fig. 1, A and B).
Fig. 1. NRF1 stabilizes SIRT7 at the promoters of mitochondrial translational machinery components.
(A and B) Coimmunoprecipitation of transfected Flag-tagged or endogenous SIRT7 with endogenous NRF1 from HEK-293T cells. (C and D) ChIP followed by quantitative real-time polymerase chain reaction (ChIP-qPCR) showing SIRT7 (C) and NRF1 (D) occupancy at gene promoters. Error bars, mean ± SE. **P < 0.01; ***P < 0.001; ns, P > 0.05. Student’s t test.
SIRT7 bound the proximal promoters of mitochondrial ribosomal proteins (mRPs) and mitochondrial translation factors (mTFs) but not other NRF1 targets (Fig. 1C and fig. S1A) (7).NRF1 bound the same regions as SIRT7 at the proximal promoters of mRPs and mTFs but not RPS20 (Fig. 1D and fig. S1B), where SIRT7 binding is mediated through Myc (9). SIRT7 binding sites were found adjacent to NRF1 consensus binding motifs at the promoters of mRPs and mTFs (fig. S1C). NRF1 knockdown (KD) using small interfering RNA (siRNA) reduced SIRT7 occupancy at the promoters of mRPs and mTFs but not RPS20 (Fig. 1C and fig. S2). SIRT7 KD using short hairpin RNAs led to increased expression of mRPs and mTFs, which was abrogated by NRF1 siRNA (Fig. 2, A and B, and fig. S3). Thus,NRF1 targets SIRT7 specifically to the promoters of mRPs and mTFs for transcriptional repression.
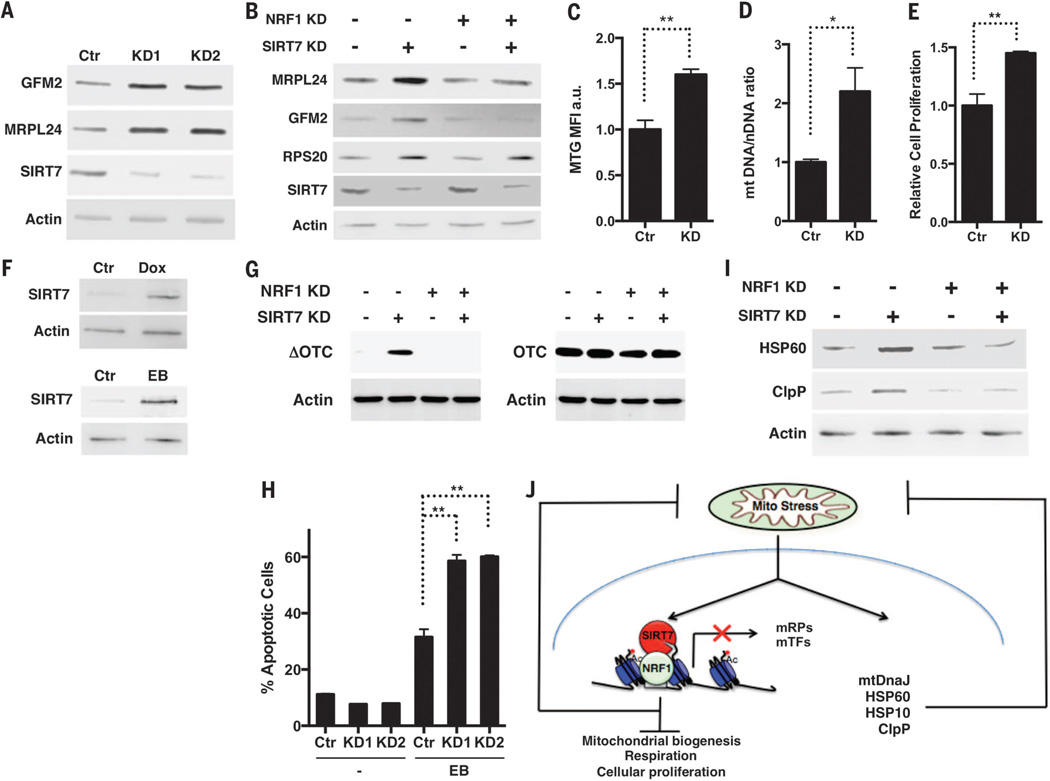
Fig. 2. SIRT7 limits mitochondrial activity, proliferation, and PFSmt.
(A and B) Increased expression of GFM2 and MRPL24 in SIRT7 KD cells is abrogated by NRF1 siRNA. (C and D) Increased mitochondrial mass in SIRT7 KD cells determined by MTG staining and mitochondrial DNA quantification. n = 3. (E) Increased proliferation in SIRT7 KD cells. n = 3. (F) PFSmt induces SIRT7 expression. Dox, doxycycline. EB, ethidium bromide. (G) Increased accumulation of misfolded ΔOTC in SIRT7 KD cells is rescued by NRF1 siRNA. OTC is used as a control. (H) Increased apoptosis in SIRT7 KD cells treated with EB. n = 3. (I) NRF1 siRNA abrogates increased PFSmt in SIRT7 KD cells. (J) A proposed model. Error bars, mean ± SE. *P < 0.05. **P < 0.01. Student’s t test.
Transcriptional repression of mitochondrial and cytosolic (7, 9) translation machinery by SIRT7 suggests that SIRT7 might suppress mitochondrial activity and proliferation. SIRT7 KD cells had increased mitochondrial mass, citrate synthase activity, adenosine triphosphate levels, respiration, and proliferation, whereas cells overexpressing wild type (WT) but not a catalytically inactive SIRT7 mutant (H187Y) showed reduced mitochondrial mass, respiration, and proliferation (Fig. 2, C to E, and fig. S4, A to G) (10, 11). NRF1 siRNA abrogated the increased mitochondrial activity and proliferation of SIRT7 KD cells (fig. S4, H to J). Thus, SIRT7 represses NRF1 activity to suppress mitochondrial activity and proliferation.
Sirtuins are increasingly recognized as stress resistance genes (12–14). Nutrient deprivation induced SIRT7 expression (fig. S5A). Upon nutrient deprivation stress, cells reduce mitochondrial activity, growth, and proliferation to prevent cell death (15, 16). When cultured in nutrient deprived medium, cells overexpressing SIRT7 showed increased survival, whereas SIRT7 KD cells showed reduced survival, which was improved by NRF1 siRNA (fig. S5). Thus, SIRT7 suppresses NRF1 activity to promote nutritional stress resistance.
Perturbation of mitochondrial proteostasis, a form of mitochondrial stress, activates the mitochondrial unfolded protein response (UPRmt), a retrograde signaling pathway leading to transcriptional up-regulation of mitochondrial chaperones and stress relief (17, 18). Mitochondrial dysfunction results in attenuated translation, which helps restore mitochondrial homeostasis (19). SIRT7-mediated transcriptional repression of the translation machinery suggests that SIRT7 may alleviate mitochondrial protein folding stress (PFSmt). PFSmt induced SIRT7 expression (Fig. 2F). Induction of PFSmt by overexpression of an aggregation-prone mutant mitochondrial protein, ornithine transcarbamylase (ΔOTC), results in UPRmt activation and efficient clearance of misfolded ΔOTC (18). In SIRT7 KD cells, misfolded ΔOTC accumulated to a higher level (Fig. 2G). SIRT7 KD cells displayed increased apoptosis upon PFSmt (Fig. 2H) but are not prone to general apoptosis (9). Thus, SIRT7 alleviates PFSmt and promotes PFSmt resistance. Consistently, mitochondrial dysfunction is manifested in the metabolic tissues of SIRT7-deficient mice (20).
PFSmt induced the expression of canonical UPRmt genes in SIRT7-deficient cells (fig. S6, A and B), indicating that induction of SIRT7 and canonical UPRmt genes is in separate branches of the UPRmt. Untreated SIRT7 KD cells displayed increased expression of canonical UPRmt genes (fig. S6, A and B), but SIRT7 did not bind to their promoters (fig. S1A) (7), suggesting that SIRT7 deficiency results in constitutive PFSmt and compensatory induction of canonical UPRmt genes. NRF1 siRNA abrogated increased PFSmt, but not endoplasmic reticulum stress, in SIRT7 KD cells (Fig. 2I and fig. S6C). Thus, the interplay between SIRT7 and NRF1 constitutes a regulatory branch of UPRmt, functioning as the nexus of reduced mitochondrial translation for homeostasis reestablishment and repressed energy metabolism and proliferation (Fig. 2J).
What is the physiological relevance of the SIRT7-mediated UPRmt? In Caenorhabditis elegans, the UPRmt is activated during a developmental stage when a burst of mitochondrial biogenesis takes place and is attenuated when mitochondrial biogenesis subsides (17). Thus, the SIRT7-mediated UPRmt may be important for cells that experience bursts of mitochondrial biogenesis and convert between growth states with markedly different bioenergetic demands and proliferative potentials, such as stem cells. Quiescent adult stem cells have low mitochondrial content, but mitochondrial biogenesis increases during proliferation and differentiation (4).
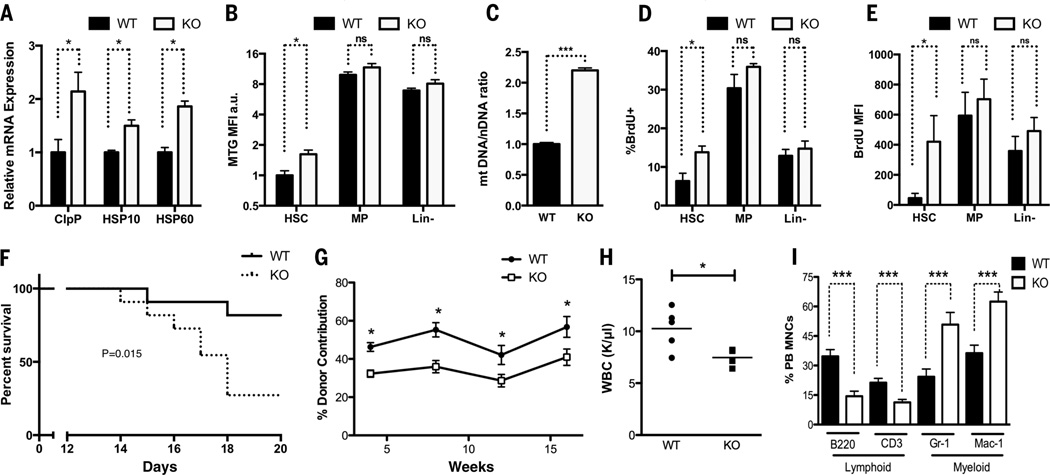
Because SIRT7 is highly expressed in the hematopoietic system, we focused on hematopoietic stem cells (HSCs) isolated from SIRT7+/+ and SIRT7−/− mice (fig. S7). SIRT7−/− HSCs had increased expression of UPRmt genes, indicative of increased PFSmt (Fig. 3A). Mitotracker green (MTG) staining, mitochondrial DNA quantification, and electron microscopy revealed increased mitochondrial numbers in SIRT7−/− HSCs (Fig. 3, B and C, and fig. S8). In vivo bromodeoxyuridine (BrdU) incorporation showed increased proliferation of SIRT7−/− HSCs (Fig. 3D). SIRT7−/− HSCs also exhibited an increased propensity to enter the cell cycle upon ex vivo culture with cytokines (Fig. 3E). However, there was no difference in mitochondrial number or proliferation in the differentiated subpopulations of these two genotypes (Fig. 3, B, D, and E). Animals with loss of HSC quiescence are sensitive to the myeloablative drug 5-fluorouracil (21). Upon 5-fluorouracil treatment, mice reconstituted with SIRT7−/− bone marrow cells (BMCs) died sooner than SIRT7+/+ controls (Fig. 3F). Thus, HSCs require SIRT7 to limit PFSmt, mitochondrial mass, and proliferation.
Fig. 3. SIRT7 ensures HSC maintenance.
(A) qPCR showing increased PFSmt in SIRT7−/− HSCs. n = 4. (B and C) MTG staining and mitochondrial DNA quantification showing increased mitochondrial mass in SIRT7−/− HSCs. n = 4. MP, myeloid progenitor cells. Lin-, lineage-negative cells. (D) In vivo BrdU incorporation showing increased proliferation of SIRT7−/− HSCs. n = 4. (E) Increased propensity of SIRT7−/− HSCs to cycle upon ex vivo culture with cytokines, indicated by BrdU pulse. n =4. (F) Mice reconstituted with SIRT7−/− BMCs have increased sensitivity to 5-fluorouracil. n = 12. Log-rank test. (G) Competitive transplantation using SIRT7+/+ and SIRT7−/− BMCs as donors showing reduced reconstitution capacity of SIRT7−/− HSCs. n = 15. Representative of five transplants. (H) Reduced white blood cell count in SIRT7−/− mice. (I)Myeloid-biased differentiation in the peripheral blood (PB) of SIRT7−/− mice. MNCs, mononuclear cells. n = 7. Error bars, mean ± SE. *P < 0.05. ***P < 0.001. ns, P > 0.05. Student’s t test.
Reduced HSC quiescence causes compromised regenerative function (21). SIRT7−/− BMCs or purified HSCs displayed a 40%reduction in long-term reconstitution of the recipients’ hematopoietic system compared with their SIRT7+/+ counterparts (Fig. 3G and fig. S9A). SIRT7−/− mice also had reduced numbers of white blood cells (Fig. 3H). Although SIRT7+/+ and SIRT7−/− mice had comparable HSC frequency in the bone marrow under steady-state conditions, there was a 50% reduction in the frequency of SIRT7−/− HSCs upon transplantation (fig. S9, B and C). SIRT7−/− HSCs showed increased apoptosis upon transplantation (fig. S9D), which may account for compromised HSC engraftment. HSCs differentiate into lymphoid and myeloid lineages. Myeloid-biased differentiation was apparent in SIRT7−/− mice or in mice reconstituted with SIRT7−/− HSCs (Fig. 3I and fig. S9E). Thus, SIRT7 promotes HSC maintenance and prevents myeloid-biased differentiation.
Reintroduction of SIRT7 in SIRT7−/− HSCs improved reconstitution capacity and rescued myeloid-biased differentiation (fig. S9, F and G), indicating that SIRT7 regulates HSC maintenance cell-autonomously. NRF1 inactivation in SIRT7−/− HSCs reduced PFSmt; improved HSC quiescence, engraftment, and reconstitution; and rescued myeloid-biased differentiation (figs. S9C and S10). Thus, SIRT7 represses NRF1 activity to alleviate PFSmt and ensure HSC maintenance.
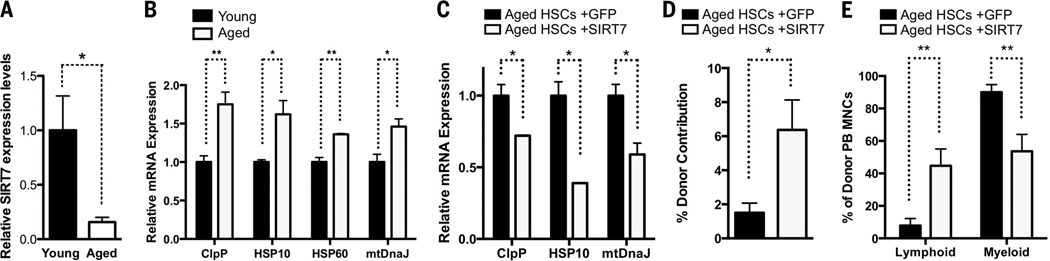
SIRT7 expression was reduced in aged HSCs (Fig. 4A) (22). Notably, defects manifested in SIRT7−/− HSCs (increased PFSmt and apoptosis, loss of quiescence, decreased reconstitution capacity, and myeloid-biased differentiation) resemble aspects of aged HSCs (23–25) (Fig. 4B). SIRT7 overexpression or NRF1 inactivation in aged HSCs reduced PFSmt, improved reconstitution capacity, and rescued myeloid-biased differentiation (Fig. 4, C to E, and fig. S11). Thus, SIRT7 down-regulation results in increased PFSmt in aged HSCs, contributing to their functional decline.
Fig. 4. HSC aging is regulated by SIRT7.
(A) qPCR showing reduced SIRT7 expression in aged HSCs. n = 3. (B and C) qPCR showing that increased PFSmt in aged HSCs is rescued by SIRT7 overexpression. n = 3. (D and E) Competitive transplantation using aged HSCs transduced with SIRT7 or control lentivirus as donors. SIRT7 overexpression increases reconstitution capacity and reverses myeloid-biased differentiation of aged HSCs. n =7. Error bars, mean ± SE. *P < 0.05. **P < 0.01. Student’s t test.
Collectively, our results highlight PFSmt as a trigger of a metabolic checkpoint that regulates HSC quiescence and establish the deregulation of UPRmt as a contributing factor for HSC aging. Using a stress signal as a messenger to return to quiescence may ensure the integrity of HSCs, which persist throughout the entire life span for tissue maintenance. The interplay between SIRT7, which is induced upon PFSmt, andNRF1, a master regulator of mitochondria, is uniquely positioned to integrate PFSmt to metabolic checkpoint regulation.
SIRT7 represses NRF1 activity to reduce the expression of the mitochondrial translation machinery and to alleviate PFSmt (Figs. 1 and 2). In vivo gene expression studies cannot distinguish direct versus indirect effects. In this regard, chromatin immunoprecipitation (ChIP) sequencing studies are informative in identifying direct SIRT7 targets (7). Although gene expression changes in the metabolic tissues of SIRT7−/− mice are likely reflective of severe mitochondrial and metabolic defects (20), transiently knocking down SIRT7 in cultured cells can capture the direct effect of SIRT7 on its targets (7, 9) (Fig. 2) and may account for different gene expression changes. In contrast to the severe defects in metabolic tissues of SIRT7−/− mice, SIRT7−/− HSCs have increased mitochondria number and proliferation under homeostatic conditions but do not farewell upon transplantation stress (Fig. 3). These observations are consistent with the notion that while metabolic tissues have a large number of mitochondria, HSCs have very few mitochondria under homeostatic conditions and increase mitochondrial biogenesis upon transplantation. The combined power of biochemistry, cell culture, and mouse genetics is necessary to tease out direct and indirect effects of SIRT7 under various physiological conditions. Our proposed model (Fig. 2J) is consistent with the functions of SIRT7 in chromatin remodeling and gene repression (7), stress responses (9) (Fig. 2), and mitochondrial maintenance (20) (Fig. 2).
Reintroduction of SIRT7 in aged HSCs reduces PFSmt and improves their regenerative capacity. Thus, PFSmt-induced HSC aging is reversible. It appears that HSC aging is not due to the passive chronic accumulation of cellular damage over the lifetime but to the regulated repression of cellular protective programs, giving hope for targeting the dysregulated cellular protective programs to reverse HSC aging and rejuvenate tissue homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Nolla, A. Valeros, and K. Heydari for cell sorting; R. Zalpuri for electron microscopy; U. Andersen for mass spectrometry; J. Kim for technical assistance; the Chua laboratory for SIRT7 antibody and constructs; and the Hoogenraad laboratory for OTC constructs. This work was supported by NIH R01 AG040990 (D.C.), R01AG040061 (C.M.H.), T32 AG000266 (M.M.), Ellison Medical Foundation (D.C.), Glenn Foundation (D.C.), NSF Graduate Research Fellowship Program (J.S.), and Siebel Stem Cell Institute (D.C. and M.M.).
Footnotes
The supplementary materials contain additional data.
www.sciencemag.org/content/347/6228/1374/suppl/DC1
Materials and Methods
Figs. S1 to S11
Tables S1 to S3
References (26–29)
REFERENCES AND NOTES
- 1.Kenyon CJ. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 2.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijg J, Campisi J. Nature. 2008;454:1065–1071. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, et al. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webb AE, et al. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber MF, et al. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpulla RC. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 9.Shin J, et al. Cell Rep. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu JY, Zhang Y, Shen YF. Acta Academiae Medicinae Sinicae. 2009;31:724–727. doi: 10.3881/j.issn.1000-503X.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. J. Physiol. Pharmacol. 2008;59(suppl. 9):201–212. [PubMed] [Google Scholar]
- 12.Brown K, et al. Cell Rep. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giblin W, Skinner ME, Lombard DB. Trends Genet. 2014;30:271–286. doi: 10.1016/j.tig.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Inoki K, Zhu T, Guan KL. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 16.Laplante M, Sabatini DM. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haynes CM, Ron D. J. Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, et al. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haynes CM, Fiorese CJ, Lin YF. Trends Cell Biol. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu D, et al. Cell Metab. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto K, et al. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Chambers SM, et al. PLOS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janzen V, et al. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 24.Oh J, Lee YD, Wagers AJ. Nat. Med. 2014;20:870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Signer RA, Morrison SJ. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.