Abstract
Disorders of sex development (DSDs) are congenital conditions with discrepancies between the chromosomal, gonadal, and phenotypic sex of the individual. Such disorders have historically been difficult to diagnose and cause great stress to patients and their families. Genetic analysis of human samples has been instrumental in elucidating the molecules and pathways involved in the development of the bipotential gonad into a functioning testis or ovary. However, many DSD patients still do not receive a genetic diagnosis. New genetic and genomic technologies are expanding our knowledge of the underlying mechanism of DSDs and opening new avenues for clinical diagnosis. We review the genetic technologies that have elucidated the genes that are well established in sex determination in humans, discuss findings from more recent genomic technologies, and propose a new paradigm for clinical diagnosis of DSDs.
Keywords: gonadal dysgenesis, microarray, exome sequencing, genetic diagnosis
INTRODUCTION
One of the defining moments of human lives comes early in development, when individuals embark on a male or female path. How sex is determined in humans has long been a source of fascination, and each pregnancy, with its uncertainty of the baby’s sex, is a reminder of the mysterious complexity of sexual development. Understanding this process has not only biological significance, allowing for the deciphering of the mechanisms of reproduction and sex differences in physiology and medicine, but also human significance, in tackling our own perceptions of masculinity and femininity and caring for often-ostracized conditions involving sexual organs.
Sex development includes both the determination of the gonads from the bipotential precursor and the subsequent differentiation of reproductive organs resulting from the sex hormones (reviewed in 6). Disorders of sex development (DSDs) are defined as “congenital conditions in which development of chromosomal, gonadal or anatomical sex is atypical” (46) and encompass a range of phenotypes, including ambiguous genitalia, androgen insensitivity, and a gonadal and external sex opposite to the genetic sex, a situation sometimes referred to as sex reversal (46). DSDs are caused either by variants in the pathway responsible for determining gonadal development or by disruption of sex differentiation owing to defects in hormone production or sensitivity. The birth of a child with a DSD is extraordinarily stressful for the parents because it brings great uncertainty regarding the child’s gender and psychosexual development, an uncertainty that is enhanced by the difficulty that medical teams have in quickly providing an explanation for the situation.
Here, we focus on the genetics of sex determination and discuss the dramatic advances in genetic and genomic technologies that have resulted in the discovery of many genes involved in sex determination; we also discuss how these findings have been and continue to be translated to clinical diagnoses to provide a much-needed management guide to patients, families, and practitioners. Despite the major advances in our understanding of mammalian sex determination, many people with DSDs do not currently receive a definitive genetic diagnosis. This situation is likely to change with the revolution of fast, affordable, and clinically actionable next-generation sequencing (NGS).
GENE DISCOVERY USING POSITIONAL CLONING
Prior to the sequencing of the complete human genome (60, 118), disease genes were identified primarily through positional cloning and linkage analysis. Such studies were time consuming and difficult for genes involved in sex determination because of the small number of patients and lack of the large families useful for linkage analysis. Many of the early identifications of genes involved in sex determination were achieved by positional cloning using patient samples in which microscopically visible chromosomal alternations were identified using standard cytogenetic techniques.
SRY
The testis-determining factor was long known to be present on the Y chromosome. The SRY gene was cloned using genomic DNA from four sex-reversed XX patients who carried fragments of the Y chromosome (103). Confirmation that SRY is the testis-determining factor came from subsequent studies showing mutations in the gene in XY females with gonadal dysgenesis (16, 48) and expression of mouse Sry at the appropriate time during mouse testis development (59). Transgenic mouse studies further showed that ectopic expression of Sry in the gonad results in a male phenotype with an XX genotype (58).
The discovery of the SRY gene led to testing in DSD patients. The majority of XX males who have been tested have a functional copy of the SRY gene owing either to a translocation of the SRY-containing portion of the Y chromosome onto the X chromosome or to some level of XX/XY mosaicism (24). Thus, the majority of XX males can be explained by the presence of SRY, but the remaining 10% generally remain unexplained at the genetic level. In contrast, defects in SRY explain only approximately 15% of XY gonadal dysgenesis (24, 41, 116), and thus the majority of patients with DSDs cannot be genetically explained by variants in SRY (46, 66).
SOX9
After the cloning of SRY, it became apparent that there were other factors involved in male sex determination because the majority of XY gonadal dysgenesis patients were found to have an intact copy of SRY, and a small subset of XX males cannot be explained by the presence of the SRY gene. Deletions and translocations involving the 17q24.3–25.1 chromosomal region result in a large percentage of sex reversal owing to gonadal dysgenesis in XY individuals and also cause campomelic dysplasia, a usually lethal skeletal malformation syndrome (43). Thus, this region of chromosome 17 was presumed to also include a gene (or genes) necessary for testis development (114). The sex-reversal locus on chromosome 17 is homologous with mouse chromosome 11, where an SRY-related gene, Sox9, had been identified (125), thus suggesting that a human homolog within the chromosome 17 locus is responsible for the XY gonadal dysgenesis observed in campomelic dysplasia. Cloning and mutation analysis of human SOX9 by two independent groups showed that loss-of-function mutations in the gene were responsible for both campomelic dysplasia and XY gonadal dysgenesis (36, 120). Because inactivating variants of SOX9 cause such an obvious phenotype in the bowing of the long bones, this gene is not usually tested in isolated cases of XY gonadal dysgenesis.
NR0B1/DAX1
For a long time, female sexual development was thought to be the default, passive pathway because it occurs in the absence of any gonadal tissue, as first demonstrated by the classic experiments of Jost (53–55). In recent years, several genes have been described that actively promote ovarian development or suppress aspects of the male pathway. Duplication of the short arm of the X chromosome was observed in XY females (15), suggesting the presence of a dosage-sensitive gene on the X chromosome that promotes female development. The locus was initially named DSS (for dosage-sensitive sex reversal) (11), and when the gene within the DSS locus was cloned, it was named DAX1 (for dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1), now termed NR0B1 (for nuclear receptor subfamily 0, group B, member 1) (128). Increased dosage of NR0B1 can cause female development despite the presence of the wild-type SRY gene (9, 124). Although suspected from the human clinical findings, this gene’s effect was confirmed in a mouse model: When Nr0b1 is overexpressed in mice that have naturally low levels of Sry, the XY mice develop as females (106).
NR0B1 is especially interesting because inactivating variants—the converse situation to the duplications described above—cause X-linked adrenal hypoplasia congenita (AHC), a severe form of adrenal insufficiency (128). AHC results from the developmental failure of the adrenal cortex associated with hypogonadotropic hypogonadism, a DSD caused by a combined deficiency of GnRH (gonadotropin-releasing hormone) secretion and pituitary responsiveness to GnRH (47). In AHC, the initial determination of testis development is normal and no sex reversal is observed (47). NR0B1 genetic testing is available for clinical diagnosis—both deletion/duplication analysis for cases of XY gonadal dysgenesis and full gene sequencing when AHC is suspected.
DMRT1
Another sex determination gene identified by positional cloning using human clinical samples is DMRT1. Terminal 9p deletions detected by karyotype are associated with XY sex reversal (14, 34, 40, 117). Many case reports of XY sex-reversed individuals with a 9p deletion allowed the region to be more finely mapped to the region distal of 9p24.3 (34, 40, 117). The human DMRT1 gene was initially identified on the basis of similarity with the doublesex gene in Drosophila and the mab-3 gene in Caenorhabditis elegans, both of which are required for sex determination (95). Shortly after its identification, the human DMRT1 gene was found to map within the 9p locus at 9p24.3 (94). A related sequence, DMRT2, was also located in the same region, and both of these genes are deleted in even the smallest 9p deletions associated with sex reversal; however, it is still unclear whether either or both genes are required for normal sex determination in humans (68). No single-gene variants in either DMRT1 or DMRT2 have yet been identified in human DSD samples, although there is one case report of a partial deletion of DMRT1 (exons 3 and 4) in a case of XY ovotesticular DSD (62). Recent research on mice has shown that Dmrt1 is necessary for maintenance of testicular determination (74) and that Dmrt2 is not necessary for gonadal development (101).
WT1
Wilms’s tumor is a childhood cancer of the kidney that is sometimes associated with other developmental syndromes caused by WT1 mutations, such as WAGR (Wilms’s tumor, aniridia, genitourinary anomalies, and mental retardation) (91), Denys-Drash syndrome (22, 26, 90), and Frasier syndrome (10), all of which share features of genitourinary abnormalities, particularly XY gonadal dysgenesis (99). Cytogenetic analysis showed that patients with these conditions often have chromosomal deletions that contain the 11p13 region, and the WT1 Wilms’s tumor predisposition gene was mapped within this locus (23, 38). WT1 is expressed during human gonadal development (93), and, as noted above, WT1 mutations are associated with Denys-Drash syndrome (22, 26, 90).
Genetic testing by sequence determination or duplication/deletion analysis is clinically available for all of the genes described above except DMRT1. SOX9 and WT1 variants are invariably associated with complex disease phenotypes where the DSD may not be the major clinical concern. SRY analysis by gene sequencing and fluorescence in situ hybridization (FISH) is often indicated if the sexual phenotype does not match the chromosome complement. Compared with the testing of other sex determination genes identified by positional cloning, SRY testing has a significant diagnostic yield.
GENE DISCOVERY FROM LINKAGE ANALYSIS
Linkage analysis is commonly used to identify genes associated with diseases that occur in families. This approach has not been used much in identifying genes associated with DSDs because there are not many families with multiple DSD cases. However, a few notable DSD genes have been identified through family studies.
MAP3K1
A large family with multiple cases of XY gonadal dysgenesis was used to identify a locus on the long arm of chromosome 5 that is associated with this phenotype (51). The 5-Mb region contains more than 30 candidate genes, 2 of which were known to be expressed in mouse gonads around the time of sex determination (89). These genes, MAP3K1 and MIER3, were sequenced in the affected individuals of the original family, and a splice-site variant in MAP3K1 was found to segregate with the disease phenotype but was not observed in 100 matched controls or in the 1000 Genomes Project (89). Subsequent analysis in an additional family and a number of sporadic cases identified additional MAP3K1 variants, all of which correlated with the phenotype and had not previously been reported. Functional in vitro analysis found that the MAP3K1 variants disrupted signaling and resulted in increased activation of downstream targets (89). The confluence of linkage analysis and functional data in mice firmly implicates MAP3K1 as required for normal human sex determination.
RSPO1
RSPO1 (R-spondin-1), a female-specific sex determination factor, was identified on the basis of linkage analysis in a large consanguineous family (87). This family had four 46,XX brothers who also had a skin condition known as palmoplantar hyperkeratosis (PPK) that segregated with the sex reversal in a recessive inheritance pattern. The tight linkage between PPK and sex reversal allowed the gene to be localized to the 1p34–35 chromosomal region, very close to the position of WNT4 (87) (see below). Sequencing of a large number of genes from this region identified homozygous variants in RSPO1. An additional, unrelated 46,XX male with PPK was found to have a homozygous deletion that included exon 4 of RSPO1. RSPO1 is expressed in developing human gonads around the time that sex is determined (113), and an RSPO1 variant predicted to produce a protein product with partial activity was identified in an XX ovotesticular patient (112). The data from human cases provide convincing evidence that RSPO1 is required for ovarian development. Additional data from mouse models showed that the ovarian defects in Wnt4 and Rspo1 null mice were very similar, and it appears that Rspo1 and Wnt4 proteins interact to stabilize β-catenin (25), a critical step in ovarian development (35).
FROM MICE TO HUMANS
SF-1/NR5A1
SF-1 (steroidogenic factor 1), now known as NR5A1 (nuclear receptor subfamily 5, group A, member 1), is a steroidogenic factor that functions in cooperation with other transcription factors to regulate the expression of many different genes (49). It was cloned on the basis of its role in endocrine function (86), but the knockout mouse displayed combined adrenal and XY gonadal dysgenesis phenotypes similar to those seen in some human patients. The first demonstration of SF-1 variants in human DSD patients was direct sequencing of SF-1 in female patients with XY karyotypes and primary adrenal failure (2, 3). SF-1 is expressed early during mouse gonadal development, where it has roles in promoting the expression of AMH (anti-Müllerian hormone) (102) and SOX9 (100), among many other genes involved in testis development (85); it is therefore critical in the male determination program. Many SF-1 variants have been identified in 46,XY DSD patients, and SF-1 mutations have been estimated to account for up to 13% of XY gonadal dysgenesis (84).
WNT4
WNT4, a member of a family of secreted signaling molecules (42), was the first female-specific sex determination factor to be identified. Wnt4 is expressed in the developing mouse ovary but is downregulated in the developing testis (115). Mice with Wnt4 deleted show masculinization of XX animals, suggesting that this gene is required for female sexual development (115). Human WNT4 has been localized to the 1p35 chromosomal region, which was previously found to be duplicated in a number of XY DSD patients (52). In one such patient, a duplication at the distal region of chromosome 1p containing WNT4 was confirmed, and expression analysis found increased expression of the WNT4 protein (52). WNT4 mutations have subsequently been identified in multiple studies of 46,XX DSD patients (18, 19), confirming its essential role in female development.
FGF9
A significant amount of data from mouse models has shown that FGF (fibroblast growth factor) signaling is required for male sex determination. Homozygous deletion of Fgf9 results in sex reversal of XY embryos, which show a range of phenotypes, from having hypoplastic testes to being completely female (27). Multiple secreted FGFs signal through four receptors that show overlapping spatial and temporal expression throughout development (33). Further genetic deletion studies in mice showed that Fgfr2 is the receptor required for normal testis development (8, 57).
Despite the strong evidence for the role of FGF signaling in sex determination in mice, data in humans remain sparse. Variants in FGFR2 have been associated with familial hypospadias (12), but there is only one report that suggests the direct involvement of FGFR2 in human sex determination. One patient with an XY karyotype and ambiguous genitalia was found to have a deletion of 57.4 Mb that includes the FGFR2 coding sequence (107). Although this large deletion is likely to be the cause of the patient’s condition, the deleted region contains at least 620 genes in addition to FGFR2. Thus, the role of FGF signaling in human sex determination remains to be identified.
GENOME-WIDE SCANNING
A traditional karyotype has a limit of detection of approximately 5–10 Mb in the highest-resolution chromosomes. Chromosomal microarrays (CMAs) scan the entire genome to detect duplication or deletion of genomic sequences known as copy-number variants (CNVs) at a much higher resolution (reviewed in 95). Two main types of microarrays are currently in use for genetic testing. Comparative genomic hybridization (CGH, often termed array CGH) uses two DNA samples, the patient DNA and control genomic DNA, which are labeled with two different fluorophores, usually red and green. The two samples are hybridized to the array, and the relative fluorescent signal shows regions of gain or loss of genetic material in the patient in direct comparison with the control sample. This type of array is considered the best for detecting CNVs because of the direct comparison with the control DNA and because the oligonucleotides on the array are generally longer (50–70 base pairs). However, they are not capable of detecting regions of homozygosity. Single-nucleotide polymorphism (SNP) arrays were initially developed to screen for the presence of common SNPs (defined as those present in >1% of the population) and use a single fluorescently labeled DNA for hybridization. In this approach, CNVs are determined by comparing the output with an electronic reference sequence.
Manufacturers of each type of array have attempted to create single arrays that are clinically useful for detecting both CNVs and SNPs. SNPs are not evenly distributed throughout the genome, so additional oligonucleotides have been added to SNP arrays to extend the genomic coverage, allowing comprehensive detection of both CNVs and regions of heterozygosity, which are useful in the detection of consanguinity and screening for uniparental disomy. CGH arrays have added oligonucleotides covering common SNPs to facilitate identification of regions of heterozygosity.
Both types of array are used in commercial and research labs to detect CNVs, so to avoid having to repeatedly distinguish between the two techniques, we refer to both as CMAs. Two of the most notable cases where CMA analysis of human patients has confirmed previous findings from mouse models are SOX3 and GATA4.
SOX3
There are at least 20 members of the SOX (for SRY-related HMG-box-containing) gene family. SOX3 encodes the protein that is most similar to that encoded by SRY, with 67% identity and 90% similarity at the amino acid level (21). In addition to SRY, SOX8, –9, and –10 are expressed in the developing male gonad. SOX3 is not expressed in the gonads and neither mice nor humans with mutations in SOX3 show defects in sex determination, indicating that this gene is not necessary for correct sex determination (122). However, transgenic mice that express ectopic SOX3 in the gonad show sex reversal, with XX animals developing as phenotypic males (105).
Following the development of the SOX3 transgenic mice, 16 human SRY-negative XX males were screened using CMAs. Two patients had duplications of the SOX3 coding region: One had a microduplication containing the entire SOX3 coding region and little else, and the other had a large duplication that contained at least 18 genes in addition to SOX3 (105). A recent case report also found an X chromosome duplication in an XX male individual that covered the SOX3 coding region (78). These independent reports of duplications in human XX DSD patients are consistent with the hypothesis that increased expression of SOX3 can replace SRY and is sufficient to induce the male determination pathway. A third patient had a microdeletion immediately upstream of the SOX3 coding region that could affect regulatory sequences (105). Taken together, these are compelling data that overexpression of SOX3 in the developing gonad is sufficient to trigger the male determination pathway in the absence of SRY.
GATA4
GATA4 is a member of a conserved family of transcription factors that contain two zinc-finger DNA-binding domains. It interacts with other transcription factors such as FOG2, which in mice is required for gonadal development (109). Gata4 knockout results in early embryonic lethality owing to abnormal cardiac development before sex determination is initiated (110). To determine whether mutations in Gata4 also affect sex determination in mice, a knock-in model was developed that expresses a mutated Gata4 that cannot bind to Fog2, an obligatory partner for Gata4. The Gata4ki mice show abnormal testis development, demonstrating that Gata4 is required for normal testis development in mice (70).
Human mutations in GATA4 have been associated with congenital heart defects (92) but until recently had not been reported in association with any other developmental abnormalities. Two studies in humans have provided supporting evidence for the role of GATA4 in human testis development. Lourenco and colleagues (64) used direct sequencing of a number of target genes in a family with congenital heart defects and DSDs and identified a heterozygous mutation in GATA4 that segregated with disease phenotype. In vitro studies found that the GATA4 variant disrupts the activation of AMH, which would be expected to result in decreased AMH expression in vivo. An independent study using a CMA to look for CNVs in XY gonadal dysgenesis identified one case with a deletion immediately downstream of GATA4. The deletion does not affect the GATA4 coding sequence but could result in disrupted regulation of GATA4 expression (124). Taken together, these data suggest that inactivating mutations of GATA4 may be a minor cause of XY gonadal dysgenesis in humans.
Novel Gene Discovery from Chromosomal Microarray Studies
In addition to confirming findings from mouse models, several studies using CMA analysis of human DSD patients have identified novel genes potentially involved in DSDs. A recent case report found an exonic deletion in WWOX through CMA analysis in an XY DSD patient with ambiguous genitalia (123). WWOX had not previously been associated with human DSDs, but a knockout mouse model showed abnormal gonadal development (4, 65). In this case, the published data from the mouse model lent diagnostic power to the CMA analysis of the patient.
In a large study of DSD samples, three patients with XY gonadal dysgenesis were found to have deletions in the 9p23–24 chromosomal region that showed a minimal overlap of 260 kb (107). This region includes DMRT1, which is known to be involved in human sex development (94). However, deletion of DMRT1 alone is not associated with human XY sex reversal (see above), suggesting that other genes in the deleted region may also be involved. Another gene in the region, KANK1, was proposed as a likely candidate. KANK1 is highly expressed during embryonic genital development and has been shown to interact with β-catenin (121), another gene known to be important in gonadal development (35, 67). Two additional genes, DNAJC15 and CAMK1D, have also been proposed as candidate genes in human DSDs on the basis of human CMA data coupled with sexually dimorphic expression in the developing mouse gonad (124).
A number of novel chromosomal loci have been linked with specific human phenotypes: Deletions at 12p13 and 16p11.2 are associated with hypospadias, deletions at 10p14 and Xq28 are associated with cryptorchidism, and deletions at 1p36.3, 9p24.3, and 19q12–13.1 are associated with ambiguous genitalia (107). Thus, in addition to clinical diagnosis of DSDs, CMAs have generated many candidate and chromosomal loci for further investigation of their potential roles in sex determination. However, when a CMA is used to diagnose DSD patients, such research findings can be challenging to interpret in deciding whether the results for a particular individual are causative.
Identification of Novel Regulatory Regions
CMA analysis of DSD patient samples has also identified CNVs that are outside of coding areas but close to genes known to be important in disease development. These variants potentially affect previously unrecognized regulatory sequences and are increasing our understanding of how alterations in noncoding regions may affect gene expression. Mutations in the coding region of SOX9 are associated with campomelic dysplasia and XY gonadal dysgenesis (described above), but approximately 10% of cases show only the gonadal dysgenesis in a condition known as acampomelic campomelic dysplasia. A high-density oligonucleotide array study of a family was recently described in which a deletion upstream of the SOX9 region appeared to cause XY gonadal dysgenesis in the absence of campomelic dysplasia (61). A CMA analysis of XY gonadal dysgenesis led to a similar finding of a deletion upstream of SOX9 in one patient (124). Thus, it appears likely that there are testis-specific regulators of SOX9 that are required for correct expression in the testis but not for normal bone development.
In contrast to inactivating mutations, SOX9 duplication has been associated with XX testicular and ovotesticular DSDs (45), suggesting dosage effects. Recent microarray studies have found structural changes upstream of SOX9 in such patients, suggesting a gain of function involving potential regulatory regions. A translocation of chromosomes 12 and 17 was identified in an XX male in which part of the sequence upstream of SOX9 was replaced by regulatory elements of a pseudogene (96). Two independent studies of XX males have found duplications upstream of SOX9 (13, 29), and another study identified a triplication in one family of XX males (119). These data strongly suggest that increased SOX9 expression in the gonad can be caused by alterations in regulatory regions of the gene and further support the hypothesis that elevated SOX9 expression is sufficient to induce the male sex determination pathway.
Partial duplications of the X chromosome that include the region containing the NR0B1 gene cause XY gonadal dysgenesis, whereas deletions of NR0B1 result in adrenal insufficiency, as described above. In one case of an XY female with an intact SRY gene, a deletion was identified upstream of the NR0B1 locus (104). Strikingly, this patient did not exhibit any adrenal insufficiency, suggesting that the deletion did not result in decreased DAX1 expression and instead likely caused an increased expression that would explain the sex reversal.
Diagnostic Yield from Chromosomal Microarrays
In many genetic clinics, CMA analysis is becoming standard for many patients, especially in cases of developmental delay and autism, where many chromosomal rearrangements have been identified in affected individuals (69, 76, 77). The diagnostic yield from such testing ranges from 10% to 20% (77, 98). Several studies have investigated the diagnostic yield of CMA specifically in DSD cases. One of the largest examined 116 children born with some level of DSD along with 8,951 control individuals (107). The categorization of DSDs in this study was very broad, ranging from minor genital abnormalities, such as hypospadias, all the way to complete discordance between sex chromosomes and phenotype. Chromosomal imbalances were detected in 21.5% of the affected population, most of which would not have been visible on a standard karyotype. However, this cannot be counted as a clinical diagnostic yield, because many of the CNVs identified did not contain any genes known to be involved in DSDs, and the significance of these variants at the clinical level remains uncertain. In an independent study of 23 patients with XY gonadal dysgenesis, likely diagnostic CNVs were identified in 3 patients, giving a success rate of 13% (124). This study also identified additional CNVs that did not contain known DSD genes. Although the clinical significance of novel genes for the diagnosis of DSDs is not yet known, these genes provide useful information for further investigation in additional patient cohorts.
NEXT-GENERATION SEQUENCING
Genome-wide scanning using CMAs is extremely useful for detecting submicroscopic chromosomal abnormalities but does not detect small insertions or deletions (indels) (usually defined as those <1 kb) or single-nucleotide variants (SNVs). Until recently, the detection of small indels and SNVs was the domain of Sanger sequencing, and for diagnostic purposes it was limited to a small number of candidate genes. NGS has created a paradigm shift in the use of sequencing in genetic diagnosis (72). Current NGS technologies use a sequencing-by-synthesis approach to generate sequence from a vast number of small DNA fragments simultaneously (73). NGS can sequence an entire human genome within a few weeks and also allows the rapid sequencing of smaller selections of DNA by targeted capture of either a specific set of genes or the entire subset of the genome that is expressed, i.e., exome sequencing.
The first use of NGS to sequence the entire human exome was published in 2009 (82), and these technologies are changing the genetic diagnostic landscape. By 2012, several institutions were offering whole-exome sequencing (WES) on a clinical basis. The ability to examine the entire exome sequence instead of being limited to a small number of target genes or diagnostic tests is revolutionizing the diagnosis of multigenic disorders. A recent series of papers showed that rare de novo mutations can cause autism (80, 83, 97), and a study of patients with severe intellectual disability found a diagnostic variant in 16% of cases (30). Other multigenic disorders such as deafness/hardness of hearing and cardiomyopathies are also increasingly being diagnosed using NGS (39, 63, 71).
We have reported the first use of NGS in diagnosis of DSDs (5). In this study, we used a targeted approach in which we developed a custom capture kit to isolate the coding sequences of 35 known genes of sex development and then performed NGS. We tested the targeted capture using a group of patients who had already received a genetic diagnosis (n = 7) and an additional group where the genetic cause was unknown (n = 5). We confirmed the genetic diagnosis in 100% of the samples from patients where we already knew the molecular cause of the disorder. Importantly, several of these diagnoses had previously been made only on a research basis because only a few DSD genes are available for clinical testing in Clinical Laboratory Improvement Amendments–accredited labs. Furthermore, we identified a molecular cause in two of the five patients without a previously identified genetic cause. Although this was a small study, it points to the efficacy of testing a large panel of genes in generating a genetic diagnosis.
Although we piloted the targeted capture array for DSDs, we now believe that WES is currently the most effective test for genetic diagnosis of DSDs. As new genes are identified, adding them to a targeted capture panel would require developing additional capture reagents, and the test would have to be continually revalidated to ensure analytic validity. Part of the rationale for a targeted capture panel was to ensure that the costs remain low by maximizing the amount of sample pooling for the sequencing portion of the test. However, costs associated with NGS have been falling rapidly, whereas the cost of capture has remained relatively constant. Thus, it is more economical to use WES with standard capture kits.
THE FUTURE OF GENETIC DIAGNOSIS OF DISORDERS OF SEX DEVELOPMENT
When a patient presents in a clinic with a genital malformation or phenotype that does not match the chromosomal sex, the current trend of care is for the physician to search for additional phenotypic information using metabolic and endocrine testing, imaging studies (including ultrasounds and MRI), and genetic analysis. CMAs to detect chromosomal abnormalities are available clinically and can dramatically assist diagnosis when CNVs are detected in regions with known DSD genes. However, this method does not offer clear diagnostic help when CNVs are found in regions of unknown involvement with DSDs. Furthermore, reporting requirements mean that CNVs smaller than 50 kb are unlikely to be included in a clinical lab report. The costs for such tests vary widely by the type of test and institution, but a karyotype costs between $500 and $1,000, CMA analysis costs approximately $1,500, and the endocrine and imaging studies can cost far more. From the array of testing results, the clinician may suspect the involvement of a particular gene associated with sex determination. Although we now know of many genes involved in sex determination, only a limited number are available for clinical testing, and a single gene sequence can cost up to $1,500, depending on the length and complexity of the target. Therefore, the current standard for genetic diagnosis of DSDs is limited to genotyping one or two genes chosen as likely candidates based on disease phenotype. Because of this narrow scope and the exclusion of unknown genes, large numbers of patients (perhaps still the majority) do not receive a clinical molecular diagnosis. This extensive testing is also time consuming, invasive, and costly, which adds stress for patients and their families.
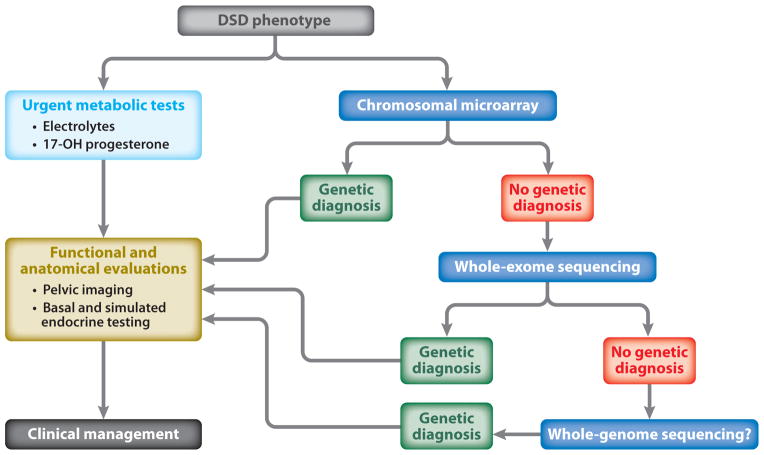
We propose a shift in the diagnostic approach to DSDs to use WES as a first-line clinical test, which will lead to faster and more accurate clinical and genetic diagnosis (Figure 1). At the University of California, Los Angeles (UCLA), the WES test can be ordered in two ways. When only a proband sample is available, we use a gene list approach in which we analyze the sequence data specifically for variants in known DSD genes. We have generated a list of well-annotated genes involved in both sex determination and differentiation (Table 1) as well as a secondary list of genes that either are more loosely associated with sexual development [e.g., the OMIM (Online Mendelian Inheritance of Man) description contains the word sex] or have data only from animal models. This allows for the rapid identification of any variants in known DSD genes in an individual sample, but when no variants are identified from one of these gene lists, this approach does not allow for any further clinical analysis and would generate a negative clinical report.
Figure 1.
Suggested clinical testing for genetic diagnosis of disorders of sex development (DSDs).
Table 1.
Primary gene list for whole-exome variant analysis of disorders of sex development (DSDs)
| Gene name | Alternative gene name(s) | Exome coverage | Associated condition(s) |
|---|---|---|---|
| Genes associated with sex determination (e.g., gonadal dysgenesis, testicular DSD, and ovotesticular DSD) | |||
| RSPO1 | RSPONDIN | 100% | 46,XX DSD and palmoplantar hyperkeratosis |
| SOX9 | SRA1 | 100% | 46,XX DSD and campomelic dysplasia |
| SRY | TDF | 100% | 46,XX testicular DSD and 46,XY ovarian DSD |
| CBX2 | CDCA6 | 99% | 46,XY DSD |
| NR0B1 | DAX1/AHCH | 98% | 46,XY DSD |
| NR5A1 | SF1 | 97% | 46,XY DSD and 46,XX premature ovarian failure |
| WWOX | 95% | 46,XY DSD | |
| DMRT1 | DMT1 | 93% | 46,XY gonadal dysgenesis |
| WNT4 | 92% | 46,XX DSD | |
| MAP3K1 | MEKK | 89% | 46,XY gonadal dysgenesis |
| DHH | HHG | 85% | 46,XY partial or complete gonadal dysgenesis |
| LHX1 | 85% | Mayer-Rokitansky-Küster-Hauser syndrome | |
| SOX3 | PHP | 78% | 46,XX and 46,XY DSD |
| WT1 | AWT1/WAGR | 77% | WAGR (Wilms’s tumor, aniridia, genital anomalies, and mental retardation) |
| DMRT2 | 76% | 46,XY gonadal dysgenesis | |
| GATA4 | 64% | 46,XY ambiguous genitalia or gonadal dysgenesis | |
| AMH | MIS | 59% | Persistent Müllerian duct syndrome |
| Genes associated with differentiation (e.g., steroid synthesis/receptors) | |||
| AKR1C4 | 3-alpha-HSD/C11/CDR/DD4/HAKRA | 100% | 46,XY DSD |
| AMHR2 | MISR2 | 100% | Persistent Müllerian duct syndrome |
| ATRX | RAD54 | 100% | Alpha-thalassemia X-linked intellectual disability syndrome |
| CYP11A1 | P450SCC | 100% | Congenital adrenal hyperplasia |
| CYP17A1 | 100% | 17-Alpha-hydroxylase-deficient congenital adrenal hyperplasia | |
| FGFR2 | 100% | Apert syndrome | |
| HSD17B3 | SDR12C2 | 100% | 17-Beta-hydroxysteroid dehydrogenase III deficiency |
| HSD3B2 | SDR11E2 | 100% | 3-Beta-hydroxysteroid dehydrogenase–deficient congenital adrenal hyperplasia |
| POR | 100% | Cytochrome P450 oxidoreductase deficiency | |
| SRD5A2 | 100% | Steroid 5-alpha-reductase deficiency | |
| STAR | StAR/STARD1 | 100% | Cholesterol desmolase–deficient congenital adrenal hyperplasia |
| AR | AIS | 95% | Complete or partial androgen insensitivity syndrome |
| LHCGR | LCGR/LGR2/LHR/ULG5 | 92% | Leydig cell hypoplasia |
| AKR1C2 | BABP/DD/DD2/HAKRD/MCDR2 | 91% | 46,XY DSD |
| CYP21A2 | CA21H/CAH1/CPS1 | 79% | 21-Hydroxylase-deficient congenital adrenal hyperplasia |
| FOXL2 | BPES | 79% | Blepharophimosis, ptosis, and epicanthus inversus |
| MAMLD1 | CG1/F18/CXORF6 | 69% | Hypospadias |
| ARX | CT121/EIEE1/ISSX | 50% | X-linked lissencephaly with ambiguous genitalia |
| Genes found to be central causes of hypogonadism | |||
| ARL6 | BBS3 | 100% | Bardet-Biedl syndrome |
| BBS2 | 100% | Bardet-Biedl syndrome | |
| BBS5 | 100% | Bardet-Biedl syndrome | |
| BBS7 | BBS2L1/FLJ10715 | 100% | Bardet-Biedl syndrome |
| BBS9 | B1/PTHB1 | 100% | Bardet-Biedl syndrome |
| BBS10 | FLJ23560 | 100% | Bardet-Biedl syndrome |
| BBS12 | FLJ35630/FLJ41559 | 100% | Bardet-Biedl syndrome |
| CHD7 | FLJ20357/FLJ20361/KIAA1416 | 100% | Kallmann syndrome (isolated GnRH deficiency with anosmia), normosmic isolated GnRH deficiency, and CHARGE syndrome |
| GNRH1 | GNRH/GRH/LHRH | 100% | Isolated abnormality in GnRH secretion or response |
| GNRHR | LHRHR | 100% | Isolated abnormality in GnRH secretion or response |
| HESX1 | ANF/RPX | 100% | Combined pituitary hormone deficiency |
| HFE | HLA-H | 100% | Hemochromatosis |
| LEP | 100% | Morbid obesity | |
| MKKS | BBS6 | 100% | Bardet-Biedl syndrome and McKusick-Kaufman syndrome |
| PROKR2 | GPR73b/GPRg2/PKR2 | 100% | Kallmann sydrome (isolated GnRH deficiency with anosmia) and normosmic isolated GnRH deficiency |
| PROP1 | 100% | Combined pituitary hormone deficiency | |
| TAC3 | NKB/ZNEUROK1 | 100% | Isolated abnormality in GnRH secretion or response |
| TACR3 | Neurokinin beta receptor/NK3R | 100% | Isolated abnormality in GnRH secretion or response |
| TRIM32 | BBS11 | 100% | Bardet-Biedl syndrome |
| TTC8 | BBS8 | 100% | Bardet-Biedl syndrome and autosomal recessive retinitis pigmentosa |
| BBS1 | 99% | Bardet-Biedl syndrome | |
| BBS4 | 99% | Bardet-Biedl syndrome | |
| FGFR1 | BFGFR/CD331/CEK/FLG | 98% | Kallmann syndrome (isolated GnRH deficiency with anosmia), normosmic isolated GnRH deficiency, and Pfeiffer syndrome |
| PCSK1 | PC1/PC3/SPC3 | 98% | Morbid obesity |
| KAL1 | Anosmin-1/KALIG-1 | 95% | Kallmann syndrome (isolated GnRH deficiency with anosmia) |
| LEPR | CD295/OBR | 95% | Morbid obesity |
| LHX3 | 87% | Combined pituitary hormone deficiency | |
| FGF8 | AIGF | 79% | Kallmann syndrome (isolated GnRH deficiency with anosmia) and normosmic isolated GnRH deficiency |
| PROK2 | BV8/KAL4/MIT1/PK2 | 76% | Kallmann syndrome (isolated GnRH deficiency with anosmia) and normosmic isolated GnRH deficiency |
| KISS1R | AXOR12/HOT7T175 | 54% | Isolated abnormality in GnRH secretion or response |
Our preferred method is to use a trio analysis in which we sequence the patient and both unaffected parents. The reason for doing this is that each individual exome has on average 20,000 variants compared with the reference sequence (1), but the majority of these variants are inherited; thus, by having the parents’ sequences we can easily define any de novo heterozygous variants in addition to any homozygous or compound heterozygous variants, and in most cases we can discount inherited heterozygous variants from unaffected parents. In some cases, the trio approach identifies variants that are likely causative but have not previously been associated with DSDs. One of the greatest challenges of WES is the interpretation and clinical reporting of genetic variants, including variants of uncertain clinical significance and incidental or secondary findings unrelated to the primary condition. One answer that we have developed at UCLA is the establishment of a Genomic Data Board to discuss each case before it is reported. These regular meetings are attended by a number of clinical geneticists, clinicians, genetic counselors, other investigators, and (when possible) the ordering physician. This group discusses the evidence that novel variants are causative and makes a determination on the level of certainty for such variants to be included in the clinical report (Table 2). Given the paucity of our knowledge regarding the genetics of sex determination, such a trio approach not only will be invaluable for diagnostic purposes but also will aid new gene discovery, which will help in diagnosis of future patients.
Table 2.
Current reporting categories of genetic variants produced by whole-exome sequencing at UCLA
| Variant category | Reportable | |
|---|---|---|
| Children | Adults | |
| Variants in genes related to the condition of interest | ||
| All variants (including those of unknown clinical significance) in genes known to be associated with the condition of interest | Yes | Yes |
| Known variants in genes suspected to be causally related to the condition of interest | Yes | Yes |
| Incidental/secondary findings | ||
| Known variants in genes associated with predisposition to future disease for which treatment/prevention is available (e.g., MSH2 for Lynch syndrome and BRCA1 for breast cancer) | Yesa | Yes |
| Variants indicating significant potential for coexisting condition for which treatment/prevention is available (e.g., ATP7B for Wilson’s disease) | Yes | Yes |
| Variants in high-penetrance genes associated with predisposition to future disease with no effective treatment or prevention (e.g., ATXN1 for spinocerebellar ataxia) | No | Yes |
| Variants indicating carrier status for autosomal recessive or X-linked conditionsb | No | Yes |
| Variants for which association or relative risk for future disease is low or unknown | No | No |
| Nonpaternityc | No | No |
Only in cases where treatment/prevention in childhood is recommended.
Especially those for which general screening is already recommended.
When parental DNA is tested in a trio analysis.
In this review we have focused on the genetic diagnosis of DSDs caused by problems with sex determination because these are particularly difficult to diagnose genetically. DSDs caused by altered endocrine function are often more easily diagnosed from additional clinical features, such as salt wasting in congenital adrenal hyperplasia (CAH). Diagnosis of these disorders benefits from rapid metabolic testing, and genetic testing for a well-known candidate gene [e.g., CYP21A2 (encoding 21-hydroxylase) for CAH] is often indicated. However, there is often significant genetic pleiotropy in DSDs caused by disrupted endocrine function, and when initial genetic testing does not confirm the diagnosis, WES is likely the most efficient next step, particularly when parental samples are available for a trio analysis.
There are many advantages to using WES as a first-line clinical test. It is currently the most effective method to screen all the known genes involved in DSDs and, in the case of a trio analysis, may suggest additional causative variants; in many cases, it may therefore at least provide a differential diagnosis. From the practitioner’s perspective, WES has many benefits. One is to reach a definitive clinical diagnosis, providing prognostic information and an aid to monitoring for any associated risks associated with the condition, such as the risk of developing gonadal tumors. Another is to allow clinicians to better classify the patient’s condition, providing a guide to its natural history and helping decision making. From the patient perspective, a comprehensive genetic test is likely to lead to a shortcut to the “diagnostic odyssey” so often experienced by families of DSD patients (5). Many DSDs are identified in young children, so genetic counseling for future reproductive decisions is an important part of the care the family receives. Anecdotally, we and others have found that families who are actively pursuing a genetic diagnosis experience a cathartic effect when such a diagnosis is reached, because they feel that the basis for the condition has finally been explained and that they have a clearer path forward even when no additional treatment options are revealed.
We believe that WES is currently the most effective test for genetic diagnosis of DSDs, but there are downsides. Turnaround time is currently 12–16 weeks at most institutions offering the test, owing both to technical considerations and to the volume of data that needs to be analyzed to reach a conclusion in each case. In cases when an answer is needed in a short time, such as the chromosomal complement of a newborn presenting with ambiguous genitalia, a karyotype can be obtained within 48 hours. There are also cases where rapid metabolic testing is required—again, for example, in salt wasting in CAH. Additionally, WES does not capture 100% of the exons, so causal variants could be missed owing to poor or no coverage of the relevant exon. WES samples only the coding portion of the genome and therefore cannot detect changes in the noncoding portion. Despite estimates that more than 85% of Mendelian genetic traits are within the coding regions or splice sites (28), recent projects such as ENCODE (Encyclopedia of DNA Elements) are showing that noncoding DNA has far more active roles than was previously thought (31). In fact, it is now suggested that WES sequencing will be able to detect a diagnostic variant in only 50% of cases. Despite these shortcomings, WES remains the best available first-line genetic test available for clinical diagnosis of DSDs (75, 108).
FUTURE RESEARCH AND GENOMIC TRANSLATION DIRECTIONS
Use of Experimental Animal Models
Since the discovery of SRY as the testis-determining factor, there has been extensive research using mouse models to identify novel genes required for sex determination. Expression analysis of the early developing testes and ovaries in mice showed that many transcripts are differentially expressed at the earliest stages of gonadal determination (17, 20, 81). These studies confirmed that many genes are involved in the correct development of functional testes and ovaries; however, the identified genes have not been associated with human DSDs. An N-ethyl-N-nitrosurea mutagenesis screen selecting for male mice with micropenis identified an autosomal recessive mutation in the gene encoding gonadotropin-releasing hormone receptor (Gnrhr) (88). This is a useful mouse model for the study of the known variants in human GNRHR that are involved in hypogonadotropic hypogonadism, but this method has not revealed any novel genes involved in human DSDs. There are several genes that we now know to be important in human sex determination that were initially discovered in animal models. These have been identified mostly by targeted analysis of individual factors that have roles in other processes, such as SF-1, or by analysis of complex genetic crosses, such as the sex-reversal phenotype when the Y chromosome from the POSA (Ypos) mouse strain is crossed onto the C57BL/6 inbred genetic background (7). Overall, these findings suggest that mouse models are useful for elucidating the mechanism of action of genes identified in human DSDs but are not the best system for identifying novel genetic causes of DSDs.
Mutational Load
We may be reaching the limits of understanding complex disease by looking only for single-gene associations. In many cases, complex phenotypes are likely due to the sum of deleterious variants in an individual (44, 127). Over the past few years, several disease groups have been recognized from what were previously thought to be individual diseases. For instance, the RASopathies are germline disorders of genes involved in the canonical Ras-Raf-MAPK pathway and include Noonan syndrome and neurofibromatosis type I (111). Bardet-Biedl syndrome, which is associated with anomalies in genital development, is now recognized to be a ciliopathy, a group of disorders in which the cellular cilia are disrupted (126). In these conditions, variants in multiple genes in the same pathway give rise to overlapping diseases, and variants in single genes are associated with more than one disease. This suggests that disruption of pathways or cellular “modules” may be a common mechanism of disrupted development processes such as those observed in DSDs. The developmental decision of the bipotential gonad to differentiate to become a testis or ovary is controlled by small changes in expression in the network of molecules expressed in the undifferentiated gonad. When SRY is present, the balance of the network tips toward the male fate; when SRY is absent, it tips toward the female fate (79). Most of the genes known to be involved in human sex determination have protein products that interact to generate the bipotential gonad, and variants in many of them are already known to disrupt sex determination (32). Network analysis methods have been created to identify genetic interactions, termed expression quantitative trait loci (eQTLs), during development (37, 50). A recent analysis compared the eQTLs of the C57BL/6 mouse, which is susceptible to sex reversal, with those of the 129S1 line, which is resistant to it. The authors identified expression networks associated with either male or female development and used the position of a gene within the network to predict its function (79). This type of analysis in human DSD samples could prove fruitful in identifying new combinations of variants that contribute to sex determination.
CONCLUSION
It has been a long journey from the slow-moving discovery of the testis-determining factor SRY to the fast-paced advances brought by the new genomic technologies. Many new genes involved in sex determination have been identified, and the path to a diagnosis for patients with DSDs has been made considerably easier. The field of sexual development as a whole has grown up. DSDs were once viewed as rare occurrences with controversial and idiosyncratic management, vocal activists, and little medical knowledge. Current understanding has resulted in DSD diagnosis including a greater number of phenotypes, including any developmental variation of the reproductive tract or sex chromosome aneuploidy. Clinical practice in the treatment of DSDs is becoming more evidence based (with efforts to standardize diagnoses and phenotypes), more multidisciplinary (with clear guidelines about team approaches), and more consensual (with the inclusion of advocacy groups in clinical and research efforts). The National Institutes of Health recently funded a large multisite, multidisciplinary network to standardize data elements related to DSDs, including genetic, phenotypic, and psychosocial findings. This project will result in a searchable data bank to be used as an analytical and reporting platform for generating best clinical practices for the management of DSDs. The advances in genomic analysis that have allowed well-defined categorizations of patients with DSDs and therefore clear, evidence-driven outcome studies have been a catalyst in this process.
Genetic diagnosis of DSDs has become the first step on which families can build a future and address their many anxious questions. We still face challenges, starting with a less-than-perfect diagnostic ability even with the most sophisticated genome-wide techniques. Whole-genome methods and network analyses of multiple variants may provide better answers. The influence of environmental factors on sex development, which has started to be clearly identified (56), is likely to become an even greater issue. Translating both the genetic and the epigenetic information to the bedside is the future path for improving the health and welfare of patients with DSDs.
Acknowledgments
The authors are partially funded by the DSD Translational Research Network (National Institutes of Health grant 1 R01 HD068138).
Glossary
- DSD
disorder of sex development
- NGS
next-generation sequencing
- AHC
adrenal hypoplasia congenita
- FISH
fluorescence in situ hybridization
- AMH
anti-Müllerian hormone
- CMA
chromosomal microarray
- CNV
copy-number variant
- CGH
comparative genomic hybridization
- SNP
single-nucleotide polymorphism
- SNV
single-nucleotide variant
- WES
whole-exome sequencing
- CAH
congenital adrenal hyperplasia
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Ruth M. Baxter, Email: rbaxter@mednet.ucla.edu.
Eric Vilain, Email: evilain@ucla.edu.
LITERATURE CITED
- 1.1000 Genomes Proj. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achermann JC, Ito M, Hindmarsh PC, Jameson JL. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 1999;22:125–26. doi: 10.1038/9629. [DOI] [PubMed] [Google Scholar]
- 3.Achermann JC, Meeks JJ, Jameson JL. Phenotypic spectrum of mutations in DAX-1 and SF-1. Mol Cell Endocrinol. 2001;185:17–25. doi: 10.1016/s0303-7207(01)00619-0. [DOI] [PubMed] [Google Scholar]
- 4.Aqeilan RI, Hagan JP, de Bruin A, Rawahneh M, Salah Z, et al. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology. 2009;150:1530–35. doi: 10.1210/en.2008-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arboleda VA, Lee H, Sánchez FJ, Délot EC, Sandberg DE, et al. Targeted massively parallel sequencing provides comprehensive genetic diagnosis for patients with disorders of sex development. Clin Genet. 2013;83:35–43. doi: 10.1111/j.1399-0004.2012.01879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arboleda VA, Vilain E. Disorders of sex development. In: Strauss JF, Barbieri RL, editors. Yen and Jaffe’s Reproductive Endocrinology. 6 Philadelphia: Saunders; 2009. pp. 367–93. [Google Scholar]
- 7.Arboleda VA, Vilain E. The evolution of the search for novel genes in mammalian sex determination: from mice to men. Mol Genet Metab. 2011;104:67–71. doi: 10.1016/j.ymgme.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagheri-Fam S, Sim H, Bernard P, Jayakody I, Taketo MM, et al. Loss of Fgfr2 leads to partial XY sex reversal. Dev Biol. 2008;314:71–83. doi: 10.1016/j.ydbio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Barbaro M, Oscarson M, Schoumans J, Staaf J, Ivarsson SA, Wedell A. Isolated 46,XY gonadal dysgenesis in two sisters caused by a Xp21.2 interstitial duplication containing the DAX1 gene. J Clin Endocrinol Metab. 2007;92:3305–13. doi: 10.1210/jc.2007-0505. [DOI] [PubMed] [Google Scholar]
- 10.Barbaux S, Niaudet P, Gubler MC, Grunfeld JP, Jaubert F, et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet. 1997;17:467–70. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- 11.Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, et al. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- 12.Beleza-Meireles A, Lundberg F, Lagerstedt K, Zhou X, Omrani D, et al. FGFR2, FGF8, FGF10 and BMP7 as candidate genes for hypospadias. Eur J Hum Genet. 2007;15:405–10. doi: 10.1038/sj.ejhg.5201777. [DOI] [PubMed] [Google Scholar]
- 13.Benko S, Gordon CT, Mallet D, Sreenivasan R, Thauvin-Robinet C, et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet. 2011;48:825–30. doi: 10.1136/jmedgenet-2011-100255. [DOI] [PubMed] [Google Scholar]
- 14.Bennett CP, Docherty Z, Robb SA, Ramani P, Hawkins JR, Grant D. Deletion 9p and sex reversal. J Med Genet. 1993;30:518–20. doi: 10.1136/jmg.30.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein R, Jenkins T, Dawson B, Wagner J, Dewald G, et al. Female phenotype and multiple abnormalities in sibs with a Y chromosome and partial X chromosome duplication: H–Y antigen and Xg blood group findings. J Med Genet. 1980;17:291–300. doi: 10.1136/jmg.17.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, et al. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–50. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- 17.Beverdam A, Koopman P. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet. 2006;15:417–31. doi: 10.1093/hmg/ddi463. [DOI] [PubMed] [Google Scholar]
- 18.Biason-Lauber A. WNT4, RSPO1, and FOXL2 in sex development. Semin Reprod Med. 2012;30:387–95. doi: 10.1055/s-0032-1324722. [DOI] [PubMed] [Google Scholar]
- 19.Biason-Lauber A, Konrad D, Navratil F, Schoenle EJ. A WNT4 mutation associated with Müllerian-duct regression and virilization in a 46,XX woman. N Engl J Med. 2004;351:792–98. doi: 10.1056/NEJMoa040533. [DOI] [PubMed] [Google Scholar]
- 20.Bouma GJ, Hudson QJ, Washburn LL, Eicher EM. New candidate genes identified for controlling mouse gonadal sex determination and the early stages of granulosa and Sertoli cell differentiation. Biol Reprod. 2010;82:380–89. doi: 10.1095/biolreprod.109.079822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–55. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 22.Bruening W, Bardeesy N, Silverman BL, Cohn RA, Machin GA, et al. Germline intronic and exonic mutations in the Wilms’ tumour gene (WT1) affecting urogenital development. Nat Genet. 1992;1:144–48. doi: 10.1038/ng0592-144. [DOI] [PubMed] [Google Scholar]
- 23.Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell. 1990;60:509–20. doi: 10.1016/0092-8674(90)90601-a. [DOI] [PubMed] [Google Scholar]
- 24.Cameron FJ, Sinclair AH. Mutations in SRY and SOX9: testis-determining genes. Hum Mutat. 1997;9:388–95. doi: 10.1002/(SICI)1098-1004(1997)9:5<388::AID-HUMU2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 25.Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, et al. Activation of β-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet. 2008;17:1264–77. doi: 10.1093/hmg/ddn016. [DOI] [PubMed] [Google Scholar]
- 26.Clarkson PA, Davies HR, Williams DM, Chaudhary R, Hughes IA, Patterson MN. Mutational screening of the Wilms’s tumour gene, WT1, in males with genital abnormalities. J Med Genet. 1993;30:767–72. doi: 10.1136/jmg.30.9.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–89. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 28.Cooper D, Krawczak M, Antonarakis SE. The nature and mechanisms of human gene mutation. In: Scriver C, Beaudet AL, Sly W, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. New York: McGraw-Hill; 1995. pp. 259–91. [Google Scholar]
- 29.Cox JJ, Willatt L, Homfray T, Woods CG. A SOX9 duplication and familial 46,XX developmental testicular disorder. N Engl J Med. 2011;364:91–93. doi: 10.1056/NEJMc1010311. [DOI] [PubMed] [Google Scholar]
- 30.de Ligt J, Willemsen MH, van Bon BWM, Kleefstra T, Yntema HG, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–29. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 31.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggers S, Sinclair A. Mammalian sex determination-insights from humans and mice. Chromosome Res. 2012;20:215–38. doi: 10.1007/s10577-012-9274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Flejter WL, Fergestad J, Gorski J, Varvill T, Chandrasekharappa S. A gene involved in XY sex reversal is located on chromosome 9, distal to marker D9S1779. Am J Hum Genet. 1998;63:794–802. doi: 10.1086/302016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleming A, Ghahramani N, Zhu MX, Delot EC, Vilain E. Membrane β-catenin and adherens junctions in early gonadal patterning. Dev Dyn. 2012;241:1782–98. doi: 10.1002/dvdy.23870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–30. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 37.Franke L, Jansen RC. eQTL analysis in humans. Methods Mol Biol. 2009;573:311–28. doi: 10.1007/978-1-60761-247-6_17. [DOI] [PubMed] [Google Scholar]
- 38.Gessler M, Poustka A, Cavenee W, Neve RL, Orkin SH, Bruns GA. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature. 1990;343:774–78. doi: 10.1038/343774a0. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh N, Haddad H. Recent progress in the genetics of cardiomyopathy and its role in the clinical evaluation of patients with cardiomyopathy. Curr Opin Cardiol. 2011;26:155–64. doi: 10.1097/HCO.0b013e3283439797. [DOI] [PubMed] [Google Scholar]
- 40.Guioli S, Schmitt K, Critcher R, Bouzyk M, Spurr NK, et al. Molecular analysis of 9p deletions associated with XY sex reversal: refining the localization of a sex-determining gene to the tip of the chromosome. Am J Hum Genet. 1998;63:905–8. doi: 10.1086/302017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawkins JR. Mutational analysis of SRY in XY females. Hum Mutat. 1993;2:347–50. doi: 10.1002/humu.1380020504. [DOI] [PubMed] [Google Scholar]
- 42.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houston CS, Opitz JM, Spranger JW, Macpherson RI, Reed MH, et al. The campomelic syndrome: review, report of 17 cases, and follow-up on the currently 17-year-old boy first reported by Maroteaux et al in 1971. Am J Med Genet. 1983;15:3–28. doi: 10.1002/ajmg.1320150103. [DOI] [PubMed] [Google Scholar]
- 44.Howrigan DP, Simonson MA, Kamens HM, Stephens SH, Wills AG, et al. Mutational load analysis of unrelated individuals. BMC Proc. 2011;5(Suppl 9):S55. doi: 10.1186/1753-6561-5-S9-S55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet. 1999;87:349–53. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 46.Hughes IA, Houk C, Ahmed SF, Lee PA. Consensus statement on management of intersex disorders. J Pediatr Urol. 2006;2:148–62. doi: 10.1016/j.jpurol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Jadhav U, Harris RM, Jameson JL. Hypogonadotropic hypogonadism in subjects with DAX1 mutations. Mol Cell Endocrinol. 2011;346:65–73. doi: 10.1016/j.mce.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jager RJ, Anvret M, Hall K, Scherer G. A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature. 1990;348:452–54. doi: 10.1038/348452a0. [DOI] [PubMed] [Google Scholar]
- 49.Jameson JL. Of mice and men: the tale of steroidogenic factor-1. J Clin Endocrinol Metab. 2004;89:5927–29. doi: 10.1210/jc.2004-2047. [DOI] [PubMed] [Google Scholar]
- 50.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–91. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 51.Jawaheer D, Juo SH, Le Caignec C, David A, Petit C, et al. Mapping a gene for 46,XY gonadal dysgenesis by linkage analysis. Clin Genet. 2003;63:530–35. doi: 10.1034/j.1399-0004.2003.00082.x. [DOI] [PubMed] [Google Scholar]
- 52.Jordan BK, Mohammed M, Ching ST, Delot E, Chen XN, et al. Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am J Hum Genet. 2001;68:1102–9. doi: 10.1086/320125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jost A. Recherches sur la différenciation sexuelle de l’embryon de lapin. I Introduction et embryologic génitale normale. Arch Anat Microsc Morphol Exp. 1947;36:151–200. [Google Scholar]
- 54.Jost A. Recherches sur la différenciation sexuelle de l’embryon de lapin. II Action des androgènes de synthèse sur l’histogenèse génitale. Arch Anat Microsc Morphol Exp. 1947;36:242–70. [Google Scholar]
- 55.Jost A. Recherches sur la différenciation sexuelle de l’embryon de lapin. III Rôle des gonades foetales dans la différenciation sexuelle somatique. Arch Anat Microsc Morphol Exp. 1947;36:271–315. [Google Scholar]
- 56.Jurewicz J, Hanke W. Exposure to phthalates: reproductive outcome and children health. A review of epidemiological studies. Int J Occup Med Environ Health. 2011;24:115–41. doi: 10.2478/s13382-011-0022-2. [DOI] [PubMed] [Google Scholar]
- 57.Kim Y, Bingham N, Sekido R, Parker KL, Lovell-Badge R, Capel B. Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc Natl Acad Sci USA. 2007;104:16558–63. doi: 10.1073/pnas.0702581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–21. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 59.Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–52. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 60.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 61.Lecointre C, Pichon O, Hamel A, Heloury Y, Michel-Calemard L, et al. Familial acampomelic form of campomelic dysplasia caused by a 960 kb deletion upstream of SOX9. Am J Med Genet A. 2009;149A:1183–89. doi: 10.1002/ajmg.a.32830. [DOI] [PubMed] [Google Scholar]
- 62.Ledig S, Hiort O, Wunsch L, Wieacker P. Partial deletion of DMRT1 causes 46,XY ovotesticular disorder of sexual development. Eur J Endocrinol. 2012;167:119–24. doi: 10.1530/EJE-12-0136. [DOI] [PubMed] [Google Scholar]
- 63.Lin X, Tang W, Ahmad S, Lu J, Colby CC, et al. Applications of targeted gene capture and next-generation sequencing technologies in studies of human deafness and other genetic disabilities. Hear Res. 2012;288:67–76. doi: 10.1016/j.heares.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lourenco D, Brauner R, Rybczynska M, Nihoul-Fekete C, McElreavey K, Bashamboo A. Loss-of-function mutation in GATA4 causes anomalies of human testicular development. Proc Natl Acad Sci USA. 2011;108:1597–602. doi: 10.1073/pnas.1010257108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ludes-Meyers JH, Kil H, Nuñez MI, Conti CJ, Parker-Thornburg J, et al. WWOX hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chromosomes Cancer. 2007;46:1129–36. doi: 10.1002/gcc.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lux A, Kropf S, Kleinemeier E, Jürgensen M, Thyen U. Clinical evaluation study of the German network of disorders of sex development (DSD)/intersexuality: study design, description of the study population, and data quality. BMC Public Health. 2009;9:110. doi: 10.1186/1471-2458-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, Capel B. Stabilization of β-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet. 2008;17:2949–55. doi: 10.1093/hmg/ddn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machado AZ, da Silva TE, Frade Costa EM, dos Santos MG, Nishi MY, et al. Absence of inactivating mutations and deletions in the DMRT1 and FGF9 genes in a large cohort of 46,XY patients with gonadal dysgenesis. Eur J Med Genet. 2012;55:690–94. doi: 10.1016/j.ejmg.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 69.Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–45. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manuylov NL, Zhou B, Ma Q, Fox SC, Pu WT, Tevosian SG. Conditional ablation of Gata4 and Fog2 genes in mice reveals their distinct roles in mammalian sexual differentiation. Dev Biol. 2011;353:229–41. doi: 10.1016/j.ydbio.2011.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J. 2010;31:806–14. doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–41. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 74.Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–4. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mefford HC. Diagnostic exome sequencing—are we there yet? N Engl J Med. 2012;367:1951–53. doi: 10.1056/NEJMe1211659. [DOI] [PubMed] [Google Scholar]
- 76.Mefford HC, Batshaw ML, Hoffman EP. Genomics, intellectual disability, and autism. N Engl J Med. 2012;366:733–43. doi: 10.1056/NEJMra1114194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moalem S, Babul-Hirji R, Stavropolous DJ, Wherrett D, Bagli DJ, et al. XX male sex reversal with genital abnormalities associated with a de novo SOX3 gene duplication. Am J Med Genet A. 2012;158A:1759–64. doi: 10.1002/ajmg.a.35390. [DOI] [PubMed] [Google Scholar]
- 79.Munger SC, Capel B. Sex and the circuitry: progress toward a systems-level understanding of vertebrate sex determination. Wiley Interdiscip Rev Syst Biol Med. 2012;4:401–12. doi: 10.1002/wsbm.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–45. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, et al. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–77. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 82.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–76. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ostrer H. 46,XY disorder of sex development and 46,XY complete gonadal dysgenesis. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, editors. GeneReviews. Seattle: Univ. Wash; 2008. http://www.ncbi.nlm.nih.gov/books/NBK1547. [Google Scholar]
- 85.Ozisik G, Achermann JC, Jameson JL. The role of SF1 in adrenal and reproductive function: insight from naturally occurring mutations in humans. Mol Genet Metab. 2002;76:85–91. doi: 10.1016/s1096-7192(02)00032-x. [DOI] [PubMed] [Google Scholar]
- 86.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–77. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 87.Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–9. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 88.Pask AJ, Kanasaki H, Kaiser UB, Conn PM, Janovick JA, et al. A novel mouse model of hypogonadotrophic hypogonadism: N-ethyl-N-nitrosourea-induced gonadotropin-releasing hormone receptor gene mutation. Mol Endocrinol. 2005;19:972–81. doi: 10.1210/me.2004-0192. [DOI] [PubMed] [Google Scholar]
- 89.Pearlman A, Loke J, Le Caignec C, White S, Chin L, et al. Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am J Hum Genet. 2010;87:898–904. doi: 10.1016/j.ajhg.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pelletier J, Bruening W, Kashtan CE, Mauer SM, Manivel JC, et al. Germline mutations in the Wilms’ tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell. 1991;67:437–47. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 91.Pelletier J, Bruening W, Li FP, Haber DA, Glaser T, Housman DE. WT1 mutations contribute to abnormal genital system development and hereditary Wilms’ tumour. Nature. 1991;353:431–34. doi: 10.1038/353431a0. [DOI] [PubMed] [Google Scholar]
- 92.Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 93.Pritchard-Jones K, Fleming S, Davidson D, Bickmore W, Porteous D, et al. The candidate Wilms’ tumour gene is involved in genitourinary development. Nature. 1990;346:194–97. doi: 10.1038/346194a0. [DOI] [PubMed] [Google Scholar]
- 94.Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum Mol Genet. 1999;8:989–96. doi: 10.1093/hmg/8.6.989. [DOI] [PubMed] [Google Scholar]
- 95.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–95. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 96.Refai O, Friedman A, Terry L, Jewett T, Pearlman A, et al. De novo 12;17 translocation upstream of SOX9 resulting in 46,XX testicular disorder of sex development. Am J Med Genet A. 2010;152A:422–26. doi: 10.1002/ajmg.a.33201. [DOI] [PubMed] [Google Scholar]
- 97.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schaaf CP, Wiszniewska J, Beaudet AL. Copy number and SNP arrays in clinical diagnostics. Annu Rev Genomics Hum Genet. 2011;12:25–51. doi: 10.1146/annurev-genom-092010-110715. [DOI] [PubMed] [Google Scholar]
- 99.Schedl A, Hastie N. Multiple roles for the Wilms’ tumour suppressor gene, WT1 in genitourinary development. Mol Cell Endocrinol. 1998;140:65–69. doi: 10.1016/s0303-7207(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 100.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–34. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 101.Seo KW, Wang Y, Kokubo H, Kettlewell JR, Zarkower DA, Johnson RL. Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev Biol. 2006;290:200–10. doi: 10.1016/j.ydbio.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 102.Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the Müllerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–61. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 103.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–44. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 104.Smyk M, Berg JS, Pursley A, Curtis FK, Fernandez BA, et al. Male-to-female sex reversal associated with an approximately 250 kb deletion upstream of NR0B1 (DAX1) Hum Genet. 2007;122:63–70. doi: 10.1007/s00439-007-0373-8. [DOI] [PubMed] [Google Scholar]
- 105.Sutton E, Hughes J, White S, Sekido R, Tan J, et al. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Investig. 2011;121:328–41. doi: 10.1172/JCI42580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R. Dax1 antagonizes Sry action in mammalian sex determination. Nature. 1998;391:761–67. doi: 10.1038/35799. [DOI] [PubMed] [Google Scholar]
- 107.Tannour-Louet M, Han S, Corbett ST, Louet JF, Yatsenko S, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS ONE. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19:R145–51. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development. 2002;129:4627–34. doi: 10.1242/dev.129.19.4627. [DOI] [PubMed] [Google Scholar]
- 110.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–39. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 111.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–36. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tomaselli S, Megiorni F, De Bernardo C, Felici A, Marrocco G, et al. Syndromic true hermaphroditism due to an R-spondin1 (RSPO1) homozygous mutation. Hum Mutat. 2008;29:220–26. doi: 10.1002/humu.20665. [DOI] [PubMed] [Google Scholar]
- 113.Tomaselli S, Megiorni F, Lin L, Mazzilli MC, Gerrelli D, et al. Human RSPO1/R-spondin1 is expressed during early ovary development and augments β-catenin signaling. PLoS ONE. 2011;6:e16366. doi: 10.1371/journal.pone.0016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tommerup N, Schempp W, Meinecke P, Pedersen S, Bolund L, et al. Assignment of an autosomal sex reversal locus (SRA1) and campomelic dysplasia (CMPD1) to 17q24.3–q25.1. Nat Genet. 1993;4:170–74. doi: 10.1038/ng0693-170. [DOI] [PubMed] [Google Scholar]
- 115.Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–9. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 116.Veitia R, Ion A, Barbaux S, Jobling MA, Souleyreau N, et al. Mutations and sequence variants in the testis-determining region of the Y chromosome in individuals with a 46,XY female phenotype. Hum Genet. 1997;99:648–52. doi: 10.1007/s004390050422. [DOI] [PubMed] [Google Scholar]
- 117.Veitia R, Nunes M, Brauner R, Doco-Fenzy M, Joanny-Flinois O, et al. Deletions of distal 9p associated with 46,XY male to female sex reversal: definition of the breakpoints at 9p23.3–p24.1. Genomics. 1997;41:271–74. doi: 10.1006/geno.1997.4648. [DOI] [PubMed] [Google Scholar]
- 118.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 119.Vetro A, Ciccone R, Giorda R, Patricelli MG, Della Mina E, et al. XX males SRY negative: a confirmed cause of infertility. J Med Genet. 2011;48:710–12. doi: 10.1136/jmedgenet-2011-100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wagner T, Wirth J, Meyer J, Zabel B, Held M, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–20. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Kakinuma N, Zhu Y, Kiyama R. Nucleo-cytoplasmic shuttling of human Kank protein accompanies intracellular translocation of β-catenin. J Cell Sci. 2006;119:4002–10. doi: 10.1242/jcs.03169. [DOI] [PubMed] [Google Scholar]
- 122.Weiss J, Meeks JJ, Hurley L, Raverot G, Frassetto A, Jameson JL. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol. 2003;23:8084–91. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.White S, Hewitt J, Turbitt E, van der Zwan Y, Hersmus R, et al. A multi-exon deletion within WWOX is associated with a 46,XY disorder of sex development. Eur J Hum Genet. 2012;20:348–51. doi: 10.1038/ejhg.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.White S, Ohnesorg T, Notini A, Roeszler K, Hewitt J, et al. Copy number variation in patients with disorders of sex development due to 46,XY gonadal dysgenesis. PLoS ONE. 2011;6:e17793. doi: 10.1371/journal.pone.0017793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wright EM, Snopek B, Koopman P. Seven new members of the Sox gene family expressed during mouse development. Nucleic Acids Res. 1993;21:744. doi: 10.1093/nar/21.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Investig. 2009;119:428–37. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zaghloul NA, Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. 2010;26:168–76. doi: 10.1016/j.tig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, et al. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–41. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]