Abstract
According to previous investigations, CD14 is suggested to play a pivotal role in initiating and perpetuating the pro-inflammatory response during sepsis. A functional polymorphism within the CD14 gene, rs2569190, has been shown to impact the pro-inflammatory response upon stimulation with lipopolysaccharide, a central mediator of inflammation in sepsis. In this study, we hypothesized that the strong pro-inflammatory response induced by the TT genotype of CD14 rs2569190 may have a beneficial effect on survival (30-day) in patients with sepsis. A total of 417 adult patients with sepsis (and of western European descent) were enrolled into this observational study. Blood samples were collected for rs2569190 genotyping. Patients were followed over the course of their stay in the ICU, and the 30-day mortality risk was recorded as the primary outcome parameter. Sepsis-related organ failure assessment (SOFA) scores were quantified at sepsis onset and throughout the observational period to monitor organ failure as a secondary variable. Moreover, organ support-free days were evaluated as a secondary outcome parameter. TT-homozygous patients were compared to C-allele carriers. Kaplan-Meier survival analysis revealed a higher 30-day mortality risk among C-allele carriers compared with T homozygotes (p = 0.0261). To exclude the effect of potential confounders (age, gender, BMI and type of infection) and covariates that varied at baseline with a p-value < 0.2 (e.g., comorbidities), we performed multivariate Cox regression analysis to examine the survival time. The CD14 rs2569190 C allele remained a significant covariate for the 30-day mortality risk in the multivariate analysis (hazard ratio, 2.11; 95% CI, 1.08-4.12; p = 0.0282). The 30-day mortality rate among C allele carriers was 23%, whereas the T homozygotes had a mortality rate of 13%. Additionally, an analysis of organ-specific SOFA scores revealed a significantly higher SOFA-Central nervous system score among patients carrying the C allele compared with T-homozygous patients (1.9±1.1 and 1.6±1.0, respectively; p = 0.0311). In conclusion, CD14 rs2569190 may act as a prognostic variable for the short-term outcome (30-day survival) in patients with sepsis.
Introduction
Worldwide, sepsis is one of the most frequent complications due to infection among critically ill patients and is increasing in prevalence [1–3]. Although according to the Surviving Sepsis Campaign, mortality from sepsis has decreased due to improved supportive care and evidence-based guidelines for diagnosis and timely intervention [4, 5], mortality remains at approximately 30% [4], with higher mortality rates in developing countries [6, 7]. Lipopolysaccharide (LPS) or endotoxin, the major component of the outer membrane of gram-negative bacteria, plays a major role in initiating the pro-inflammatory response associated with sepsis [8]. LPS triggers inflammation in gram-negative sepsis, as well as in gram-positive and fungal sepsis, when excessive amounts of gut-derived LPS are released during intestinal hypoperfusion [9, 10]. LPS binds membrane or soluble CD14 (sCD14) and the myeloid differentiation-2 (MD-2)-TLR4 complex, resulting in NF-kB activation and the production of IL-6, IL-1b, TNF and type I interferons [11–13]. Although CD14 was initially described as the essential co-receptor mediating the LPS activation of monocytes, subsequent investigations have shown that it also participates in immune cell activation by gram-positive cell-wall components, such as peptidoglycan [14]. Findings of numerous investigations suggest a pivotal role for CD14 in initiating and perpetuating the pro-inflammatory response during the course of sepsis [14–16]. The pro-inflammatory response is essential to eradicate primary infections and prevent the acquisition of secondary infections in patients with sepsis [17].
A functional polymorphism located at position -159 in the promoter region of the CD14 gene rs2569190 has been shown to influence the pro-inflammatory response upon stimulation in human leucocytes [18], is associated with transcriptional activity of the promoter and can also affect the production of sCD14 and tumor necrosis factor α [19]. Furthermore, the TT genotype of rs2569190 has been associated with a stronger pro-inflammatory response and higher CD14 transcriptional activity [18, 19]. Several studies have previously investigated the association between CD14 rs2569190 and the susceptibility to sepsis, as well as the outcome of sepsis, and found controversial results [16]. Given that CD14 may play an essential role in the pro-inflammatory response, we hypothesized that the strong pro-inflammatory response induced by the TT genotype of CD14 rs2569190 may improve survival (30-day) in patients with sepsis.
The aim of the study was achieved; in accordance with our hypothesis, patients with the TT genotype showed an improved 30-day survival compared with C-allele carriers.
Materials and Methods
Patients
Between April 2012 and June 2014, adult patients of European descent admitted to the intensive care units (ICUs) at the University Medical Center Goettingen (UMG) were screened daily for sepsis, severe sepsis and septic shock according to standard criteria [20]. European descent was determined by questioning the patients, their next of kin or their legal representatives. The patient exclusion criteria have been described previously elsewhere [21, 22] and comprised pre-existing diseases, immunosuppressive status and medications that may modulate the inflammatory response in patients with sepsis. The study was approved by the University of Goettingen ethics committee, Goettingen, Germany (15/1/12) and conformed to the Declaration of Helsinki ethical principles (Seoul, 2008). Written informed consent was obtained either from patients or their legal representatives.
Data collection
Mortality risk within 30 days of sepsis onset was recorded as the primary outcome parameter. Secondary outcome variables included organ dysfunction, which was evaluated using the Sequential Organ Failure Assessment (SOFA) score during the 28-day observational period in the ICU [23]. Ventilator-free days, vasopressor-free days and dialysis-free days were also recorded as secondary variables. Patients were further followed up for a maximum of 90 days, and the 90-day mortality was recorded as a secondary outcome variable. The length of ICU stay, several laboratory parameters and microbiological findings were also recorded as secondary endpoints. Relevant clinical data were collected from the electronic patient record system (IntelliSpace Critical Care and Anesthesia (ICCA); Philips Healthcare, Andover, Massachusetts, USA); all relevant medical records, including microbiological findings and medications, are available in this system. Preexisting conditions and medical histories were determined by examining the physicians’ notes; questioning the patients, their next of kin or their legal representatives; and by consulting the patient’s family doctor.
CD14 rs2569190 genotyping
DNA was extracted by automated solid phase extraction from 350 μl EDTA whole blood using an EZ1 DNA Blood Kit in BioRobot EZ1 or from PBMCs using an AllPrep DNA Mini Kit according to the manufacturer’s instructions (all from Qiagen, Hilden, Germany). The DNA quantity and quality were determined spectrophotometrically. Genotyping was performed using the pre-designed TaqMan SNP genotyping assay C__16043997_10 according to the manufacturer’s instructions (Life Technology, Darmstadt, Germany).
A total of 15% of the samples were genotyped in duplicate, yielding results that showed complete concordance. The observed genotypes were in Hardy-Weinberg equilibrium. The identity of the DNA samples was controlled by sex-typing and showed 100% concordance between the initially documented and the genetically determined sex [24].
Statistical analyses
The Hardy-Weinberg equilibrium was performed using a chi-square test. Statistical analyses were performed with Statistica (version 10, StatSoft, Tulsa, Oklahoma, USA). The significance was based on contingency tables and calculated using Fisher’s exact or chi-square tests, as appropriate. Two continuous variables were compared using the Mann-Whitney test. Time-to-event data were compared using Cox's F-test from the Statistica function survival. P-values less than 0.05 were considered significant. To exclude the effects of potential confounders (age, gender, BMI and type of infection: gram-negative, gram-positive and fungal) and covariates that varied at baseline with a p-value < 0.2 (e.g., comorbidities), we performed a multivariate Cox regression analysis to examine the survival time.
Results
Patients
A total of 417 adult patients of western European descent with sepsis were enrolled in this prospective investigation. Of these patients, 279 were male (67%), and 138 were female (33%) (Table 1). The patient median age was 65. The genotype distribution of CD14 rs2569190 was 117:225:75 (CC:CT:TT), which is consistent with Hardy–Weinberg equilibrium (p = 0.1833). The minor allele frequency was 45%. The CD14 rs2569190 CC and CT genotypes were pooled together to explore the clinical impact of the TT genotype compared with that of C-allele carriers in accordance with our a priori hypothesis (Table 1). The sepsis subtypes included sepsis/severe sepsis (39%) and septic shock (61%). The morbidity scores, SOFA and APACHE II, at baseline were 9.2±4.0 and 21.5±7.3, respectively (Table 1). The comorbidities included hypertension, a history of myocardial infarction, chronic obstructive pulmonary disease (COPD), renal dysfunction, diabetes mellitus, chronic liver diseases, a history of cancer, and a history of stroke (Table 1). Moreover, recent surgical history, site of infection and organ support (mechanical ventilation, vasopressor therapy and renal-replacement therapy) were also recorded at baseline (Table 1).
Table 1. Patient baseline characteristics according to CD14 rs2569190 genotype.
| All(n = 417) | CT/CC(n = 342) | TT(n = 75) | P value | |
|---|---|---|---|---|
| Age [years] | 63±15 | 63±15 | 64±15 | 0.4123 |
| Male, % | 67 | 67 | 68 | 0.8241 |
| Body-mass index | 28±7 | 28±7 | 28±8 | 0.9366 |
| Severity of sepsis | ||||
| Sepsis/severe sepsis, % | 39 | 39 | 43 | 0.5169 |
| Septic shock, % | 61 | 61 | 57 | 0.5169 |
| SOFA score | 9.2±4.0 | 9.3±4.0 | 8.7±4.2 | 0.3340 |
| APACHE II score | 21.5±7.3 | 21.6±7.3 | 21.0±7.3 | 0.6236 |
| Comorbidities, % | ||||
| Hypertension | 58 | 59 | 53 | 0.3877 |
| History of myocardial infarction | 7 | 6 | 11 | 0.1987 |
| COPD | 17 | 16 | 23 | 0.1324 |
| Renal dysfunction | 12 | 11 | 12 | 0.8834 |
| Diabetes mellitus (NIDDM) | 10 | 10 | 8 | 0.5562 |
| Diabetes mellitus (IDDM) | 12 | 11 | 16 | 0.2712 |
| Chronic liver diseases | 7 | 7 | 7 | 0.9138 |
| History of cancer | 19 | 18 | 23 | 0.3312 |
| History of stroke | 6 | 6 | 7 | 0.7867 |
| Recent surgical history, % | 0.3618 | |||
| Elective surgery | 30 | 28 | 36 | |
| Emergency surgery | 51 | 52 | 48 | |
| No history of surgery | 19 | 20 | 16 | |
| Site of infection, % | 0.4953 | |||
| Lung | 54 | 55 | 48 | |
| Abdomen | 27 | 26 | 29 | |
| Bone or soft tissue | 5 | 5 | 5 | |
| Surgical wound | 2 | 1 | 4 | |
| Urogenital | 2 | 2 | 4 | |
| Primary bacteremia | 7 | 7 | 4 | |
| Other | 4 | 4 | 5 | |
| Organ support, % | ||||
| Mechanical ventilation | 84 | 84 | 84 | 0.9860 |
| Use of vasopressor | 61 | 61 | 57 | 0.5134 |
| Renal-replacement therapy | 10 | 9 | 11 | 0.7271 |
The data are presented as the means ± SD or percentages
Disease severity at baseline
No differences in age, gender, or body mass index were found between the two groups of study subjects. Furthermore, no differences were found in the SOFA and APACHE II scores at the onset of sepsis according to the CD14 rs2569190 genotype. Additionally, the comorbidities were equally distributed between TT patients and C-allele carriers. Moreover, there was no difference with respect to organ support (mechanical ventilation, vasopressor therapy and renal-replacement therapy) between the two groups (Table 1).
Mortality analysis
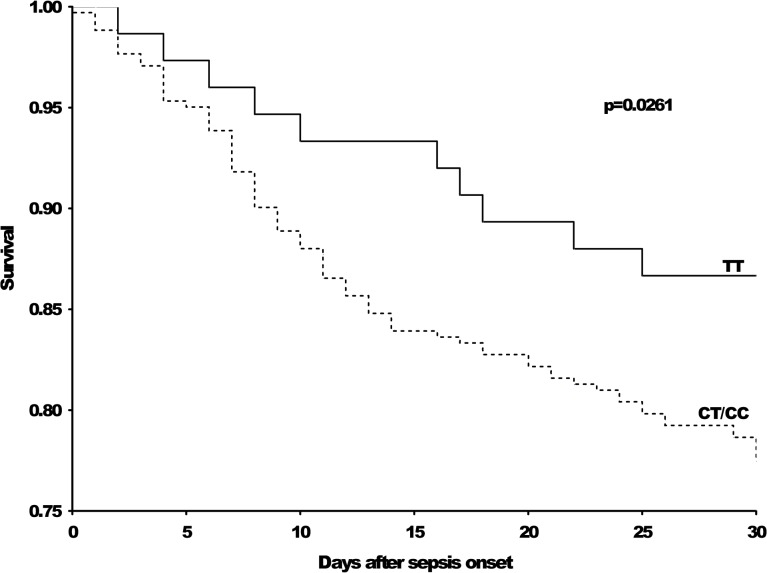
Kaplan-Meier survival analysis revealed a higher 30-day mortality risk among C-allele carriers compared with T-homozygous patients (p = 0.0261) (Fig 1). Within the observational period of 30 days after sepsis onset, 77% of the C-allele carriers had survived, whereas the T homozygotes had a survival rate of 87% (Table 2). An analysis of 90-day mortality revealed no significant difference between the two groups (p = 0.2297) (Table 2).
Fig 1. Kaplan-Meier survival analysis.
The Kaplan-Meier curve shows the survival curves censored at day 30 for the CD14 rs2569190 TT and CT/CC genotypes. Within this patient sample, the mortality risk was higher among CT/CC patients than among patients with a TT genotype (p = 0.0261, Cox's F-test).
Table 2. Disease severity over the course of sepsis.
| All(n = 417) | CT/CC(n = 342) | TT(n = 75) | P value | |
|---|---|---|---|---|
| SOFA score | 6.8±3.6 | 7.0±3.7 | 6.0±3.1 | 0.0918 |
| SOFA-Respiratory score | 1.9±0.8 | 1.9±0.8 | 1.8±0.8 | 0.2215 |
| SOFA-Cardiovascular score | 1.5±1.0 | 1.5±1.0 | 1.3±0.8 | 0.2468 |
| SOFA-Central nervous system score | 1.9±1.1 | 1.9±1.1 | 1.6±1.0 | 0.0311 |
| SOFA-Renal score | 0.8±1.2 | 0.9±1.2 | 0.7±1.2 | 0.5202 |
| SOFA-Coagulation score | 0.3±0.6 | 0.4±0.6 | 0.3±0.5 | 0.5071 |
| SOFA-Hepatic score | 0.4±0.7 | 0.4±0.7 | 0.3±0.6 | 0.5180 |
| Mortality analysis, % | ||||
| Death at day 30 | 21 | 23 | 13 | 0.0490 |
| Death at day 90 | 31 | 32 | 27 | 0.2297 |
| Length of ICU stay (days) | 17±15 | 17±15 | 18±17 | 0.6259 |
| Organ support-free days: | ||||
| Vasopressor-free (days) | 11±7 | 10±7 | 11±7 | 0.1734 |
| Ventilator-free (days) | 5±5 | 5±5 | 6±6 | 0.1547 |
| Dialysis-free (days) | 14±8 | 14±8 | 14±8 | 0.7254 |
| ECMO-free (days) | 15±9 | 15±9 | 15±8 | 0.5240 |
| Inflammatory values | ||||
| Leucocytes (1000/μl) | 14±5 | 14±5 | 14±5 | 0.9162 |
| CRP (mg/l) (n) | 151±84 (208) | 148±85 (169) | 165±81 (39) | 0.2097 |
| Procalcitonin (ng/dl) (n) | 4.5±10.7 (361) | 4.1±10.3 (293) | 6.0±12.6 (68) | 0.0654 |
| Laboratory values | ||||
| Lactate (mmol/l) | 1.7±1.1 | 1.7±1.2 | 1.6±0.6 | 0.2092 |
| Bilirubin (mg/dl) | 1.3±2.2 | 1.3±2.3 | 0.9±0.9 | 0.1844 |
| GOT (IU/l) | 209±725 | 238±804 | 90±116 | 0.1961 |
| GPT (IU/l) | 106±224 | 113±243 | 73±87 | 0.2341 |
| Kidney values | ||||
| Creatinine (mg/dl) | 1.3±1.0 | 1.3±1.0 | 1.2±0.8 | 0.8254 |
| Creatinine clearance | 103±68 | 102±65 | 108±81 | 0.8614 |
The data are presented as the means ± SD or percentages.
Multivariate analysis
To exclude the effects of potential confounders (age, gender, BMI and type of infection: gram-negative, gram-positive and fungal) and covariates that varied at baseline with a p-value < 0.2 (e.g., comorbidities), we performed a multivariate Cox regression analysis to examine the survival time. The CD14 rs2569190 C allele remained a significant covariate for 30-day mortality risk in the multivariate analysis (hazard ratio, 2.11; 95% CI, 1.08–4.12; p = 0.0282) (Table 3). This finding indicates that, despite baseline differences in certain variables (i.e., history of myocardial infarction and COPD) and in other potential confounders (age, gender, BMI and type of infection: gram-negative, gram-positive and fungal), the CD14 rs2569190 C allele remained a prognostic variable with a significant effect on the short-term outcome (30-day survival) in our cohort of sepsis patients (Table 3).
Table 3. Cox regression analysis.
| Variable | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| Age | 1.03 | 1.01–1.05 | P<0.0001 |
| Gender, Male | 1.63 | 0.99–2.67 | 0.0515 |
| BMI | 1.02 | 0.98–1.05 | 0.2283 |
| History of myocardial infarction | 0.94 | 0.43–2.08 | 0.8959 |
| COPD | 1.17 | 0.68–1.98 | 0.5596 |
| Gram-negative infection | 0.68 | 0.43–1.06 | 0.0919 |
| Gram-positive infection | 0.74 | 0.44–1.24 | 0.2612 |
| Fungal infection | 0.74 | 0.48–1.15 | 0.1920 |
| CD14 rs2569190 C allele | 2.11 | 1.08–4.12 | 0.0282 |
Disease severity
During their stay in the ICU, C-allele carriers presented higher mean SOFA scores than did T homozygotes (7.0±3.7 and 6.0±3.1, respectively; p = 0.0918) (Table 2). An analysis of organ-specific SOFA scores revealed a significantly higher SOFA-Central nervous system score among patients carrying the C allele than among T-homozygous patients (1.9±±1.1 and 1.6±1.0, respectively; p = 0.0311) (Table 2). The remaining organ-specific SOFA scores did not differ between the two groups. In addition, an analysis of the organ support-free days (ventilator-free, vasopressor-free and dialysis-free days) showed no differences between C-allele carriers and T homozygotes (Table 2). Moreover, the distribution of infection type (gram-negative, gram-positive, fungal and viral) did not vary between the two studied groups (Table 4). Infection type was determined based upon confirmed culture results.
Table 4. Infection types over the observational period.
| CT/CC(n = 342) | TT(n = 75) | P value | |
|---|---|---|---|
| Infection type | |||
| Gram-negative | 68% | 59% | 0.1393 |
| Gram-positive | 81% | 76% | 0.3450 |
| Fungal | 56% | 53% | 0.7979 |
| Viral | 11% | 7% | 0.3000 |
Inflammatory values
An analysis of inflammatory values revealed higher c reactive protein (CRP) levels among the TT patients compared with the C-allele carriers (165±81 and 148±85, respectively; p = 0.2097). Furthermore, procalcitonin levels were higher among the TT homozygotes compared with the C-allele carriers (6.0±12.6 and 4.1±10.3, respectively; p = 0.0654).
Discussion
This investigation addresses the hypothesis that sepsis patients with the TT genotype of CD14 rs2569190 may have an improved 30-day survival compared with patients carrying the C allele. Our observation of a significantly improved 30-day survival among CD14 rs2569190 TT patients confirms our hypothesis. The 30-day mortality rate among C allele carriers was 23%, whereas the T homozygotes had a mortality rate of 13%. To the best of our knowledge, these findings underscore for the first time the beneficial potential clinical impact of the CD14 rs2569190 TT genotype on 30-day survival in a cohort of septic patients of exclusively western European descent while remaining consistent with observations from Brazilian cohorts that also revealed a beneficial effect of the TT genotype on the survival of critically ill patients [25–27]. Similarly, our findings are in accordance with studies that showed an increased risk of death among CD14 rs2569190 CC critically ill patients with burn injury [28, 29].
A particular strength of this observation lies in the fact that, despite baseline differences in certain variables (i.e., history of myocardial infarction and COPD) and in other potential confounders (age, gender, BMI and type of infection: gram-negative, gram-positive and fungal), the CD14 rs2569190 C allele remained a prognostic variable with a significant effect on the short-term outcome (30-day survival) in our cohort of sepsis patients. After including the morbidity scores APACHE II and SOFA into the multivariate Cox-regression analysis, the CD14 rs2569190 C allele remained a significant covariate for 30-day mortality risk (hazard ratio, 2.16; 95% CI, 1.09–4.26; p = 0.0265). The resulted hazard ratio (hazard ratio, 2.16) for the C allele is marginally higher than the respective hazard ratio without APACHE II and SOFA scores (hazard ratio, 2.11; 95% CI, 1.08–4.12; p = 0.0282).
The biological correlation of this beneficial clinical association of the TT genotype with the 30-day survival could be attributed to previous functional observations demonstrating a significantly stronger pro-inflammatory response among TT patients compared with C-allele carriers [18, 19]. Although statistical significance was not reached, this argument is supported by the observation of a trend of higher inflammatory values (CRP and procalcitonin) among TT patients compared with C-allele carriers during the ICU stay. This assumption is in conformance with the increasingly prevailing opinion among researchers that the major problem faced by sepsis patients is a predominant state of immunosuppression characterized by a reduced pro-inflammatory status and increased anti-inflammatory response [17]. However, our results are not consistent with previous investigations demonstrating a beneficial effect of the CC genotype on septic shock survival and other studies that found no association between the CD14 rs2569190 genotype and survival among patients with sepsis [15, 16]. An explanation for this inconsistency may be that most of the previous studies had a sample size that was too small to detect the impact of the TT genotype on the survival of patients with sepsis.
The lack of a significant association between the CD14 rs2569190 TT genotype and 90-day mortality suggest little or no biological impact of the TT genotype on long-term mortality. As shown by our group, the 90-day mortality risk among patients with sepsis is related to the functional programmed cell death 1 genetic polymorphism rs11568821 GG genotype [30], which has been shown to influence transcriptional activity [31] and increase PD-1 expression [32]; PD-1 is thought to play an important role in the so-called sepsis-induced immunosuppression [17].
A major advantage of our study was that we report the first investigations of organ-specific dysfunction over the clinical course of disease with regard to CD14 rs2569190 genotypes using organ-specific SOFA scores. The significantly higher SOFA-Central nervous system scores among C-allele patients indicate a pronounced neurological or consciousness impairment in this group. This effect might be attributed to the fact that C-allele carriers, who have been shown to express less LPS-neutralizing sCD14 [33], may have compromised LPS-clearance compared with T homozygotes, making the C-allele carriers more susceptible to neurological dysfunction due to higher serum LPS levels. This explanation is in line with reports in animal models showing that the peripheral injection of LPS induces alterations in neurobehavioral performance and cognitive dysfunction [34, 35]. However, this finding has to be interpreted carefully because the SOFA-Central nervous system score can be affected by the sedative medications given to critically ill patients, which is a potential confounding factor.
One possible limitation to this study is that it focused on the clinical impact of CD14 rs2569190 exclusively; therefore, we cannot exclude that other functional SNPs located in nearby genes and in linkage disequilibrium with the CD14 rs2569190 SNP are responsible for the observed phenotypic effects on the 30-day mortality risk. A further potential limitation of this study is measurement bias, e.g., several clinical parameters (e.g., blood pressure, heart rate, and respiratory rate) are automatically recorded in the electronic patient record system, and we cannot exclude measurement errors with absolute certainty. However, all clinical records were repeatedly checked for plausibility before beginning the statistical analysis.
In conclusion, this is the first study to show a beneficial association of the CD14 rs2569190 TT genotype with short-term survival (30-day) in a cohort of sepsis patients of exclusively western European descent. According to our observations, it is worthwhile to consider CD14 rs2569190 genetic variants in future studies that investigate the genetic risk stratification for short-term mortality in patients with sepsis. Further validation in other ethnic groups is recommended.
Acknowledgments
The authors thank the staff of the ICUs of the Department of Anesthesiology and the Department of General and Visceral Surgery, all of whom were involved in patient care and monitoring. The authors also thank Luisa von Gruben, Simon Wilmers, Yvonne Klee, Sebastian Gerber, Chang Ho Hong and Evelyn Mulwande for their help with data acquisition. This study was supported by the German Research Foundation (DFG) and the Open Access Publication Funds of Göttingen University
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This study was supported by the German Research Foundation (DFG) and the Open Access Publication Funds of Göttingen University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Critical care medicine. 2001;29(7):1303–10. Epub 2001/07/11. PubMed . [DOI] [PubMed] [Google Scholar]
- 2. Hotchkiss RS, Opal S. Immunotherapy for sepsis—a new approach against an ancient foe. The New England journal of medicine. 2010;363(1):87–9. Epub 2010/07/02. 10.1056/NEJMcibr1004371 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lagu T, Rothberg MB, Shieh MS, Pekow PS, Steingrub JS, Lindenauer PK. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Critical care medicine. 2012;40(3):754–61. Epub 2011/10/04. 10.1097/CCM.0b013e318232db65 PubMed . [DOI] [PubMed] [Google Scholar]
- 4. Levy MM, Dellinger RP, Townsend SR, Linde-Zwirble WT, Marshall JC, Bion J, et al. The Surviving Sepsis Campaign: results of an international guideline-based performance improvement program targeting severe sepsis. Critical care medicine. 2010;38(2):367–74. Epub 2009/12/26. 10.1097/CCM.0b013e3181cb0cdc PubMed . [DOI] [PubMed] [Google Scholar]
- 5. Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Critical care medicine. 2013;41(2):580–637. Epub 2013/01/29. 10.1097/CCM.0b013e31827e83af PubMed . [DOI] [PubMed] [Google Scholar]
- 6. Siddiqui S. Not "surviving sepsis" in the developing countries. Journal of the Indian Medical Association. 2007;105(4):221 Epub 2007/09/08. PubMed . [PubMed] [Google Scholar]
- 7. Silva E, Pedro Mde A, Sogayar AC, Mohovic T, Silva CL, Janiszewski M, et al. Brazilian Sepsis Epidemiological Study (BASES study). Critical care (London, England). 2004;8(4):R251–60. Epub 2004/08/18. 10.1186/cc2892 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beutler BA. TLRs and innate immunity. Blood. 2009;113(7):1399–407. Epub 2008/09/02. 10.1182/blood-2008-07-019307 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Opal SM, Scannon PJ, Vincent JL, White M, Carroll SF, Palardy JE, et al. Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. The Journal of infectious diseases. 1999;180(5):1584–9. Epub 1999/10/09. 10.1086/315093 PubMed . [DOI] [PubMed] [Google Scholar]
- 10. Marshall JC, Foster D, Vincent JL, Cook DJ, Cohen J, Dellinger RP, et al. Diagnostic and prognostic implications of endotoxemia in critical illness: results of the MEDIC study. The Journal of infectious diseases. 2004;190(3):527–34. Epub 2004/07/10. 10.1086/422254 PubMed . [DOI] [PubMed] [Google Scholar]
- 11. Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunologic research. 2007;39(1–3):249–60. Epub 2007/10/06. PubMed . [DOI] [PubMed] [Google Scholar]
- 12. Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nature reviews Microbiology. 2005;3(1):36–46. Epub 2004/12/21. 10.1038/nrmicro1068 PubMed . [DOI] [PubMed] [Google Scholar]
- 13. Miyake K. Roles for accessory molecules in microbial recognition by Toll-like receptors. Journal of endotoxin research. 2006;12(4):195–204. Epub 2006/09/07. 10.1179/096805106x118807 PubMed . [DOI] [PubMed] [Google Scholar]
- 14. Pugin J, Heumann ID, Tomasz A, Kravchenko VV, Akamatsu Y, Nishijima M, et al. CD14 is a pattern recognition receptor. Immunity. 1994;1(6):509–16. Epub 1994/09/01. PubMed . [DOI] [PubMed] [Google Scholar]
- 15. Gibot S, Cariou A, Drouet L, Rossignol M, Ripoll L. Association between a genomic polymorphism within the CD14 locus and septic shock susceptibility and mortality rate. Critical care medicine. 2002;30(5):969–73. Epub 2002/05/15. PubMed . [DOI] [PubMed] [Google Scholar]
- 16. Zhang AQ, Yue CL, Gu W, Du J, Wang HY, Jiang J. Association between CD14 promoter -159C/T polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PloS one. 2013;8(8):e71237 Epub 2013/08/31. 10.1371/journal.pone.0071237 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nature reviews Immunology. 2013;13(12):862–74. Epub 2013/11/16. 10.1038/nri3552 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin J, Yao YM, Yu Y, Chai JK, Huang ZH, Dong N, et al. Effects of CD14-159 C/T polymorphism on CD14 expression and the balance between proinflammatory and anti-inflammatory cytokines in whole blood culture. Shock (Augusta, Ga). 2007;28(2):148–53. Epub 2007/05/23. 10.1097/SHK.0b013e3180341d35 PubMed . [DOI] [PubMed] [Google Scholar]
- 19. Gu W, Dong H, Jiang DP, Zhou J, Du DY, Gao JM, et al. Functional significance of CD14 promoter polymorphisms and their clinical relevance in a Chinese Han population. Critical care medicine. 2008;36(8):2274–80. Epub 2008/07/04. 10.1097/CCM.0b013e318180b1ed PubMed . [DOI] [PubMed] [Google Scholar]
- 20. Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Critical care medicine. 2003;31(4):1250–6. Epub 2003/04/12. 10.1097/01.ccm.0000050454.01978.3b PubMed . [DOI] [PubMed] [Google Scholar]
- 21. Mansur A, von Gruben L, Popov AF, Steinau M, Bergmann I, Ross D, et al. The regulatory toll-like receptor 4 genetic polymorphism rs11536889 is associated with renal, coagulation and hepatic organ failure in sepsis patients. Journal of translational medicine. 2014;12:177 Epub 2014/06/22. 10.1186/1479-5876-12-177 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mansur A, Klee Y, Popov AF, Erlenwein J, Ghadimi M, Beissbarth T, et al. Primary bacteraemia is associated with a higher mortality risk compared with pulmonary and intra-abdominal infections in patients with sepsis: a prospective observational cohort study. BMJ open. 2015;5(1):e006616 Epub 2015/01/08. 10.1136/bmjopen-2014-006616 PubMed . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vincent JL, de Mendonca A, Cantraine F, Moreno R, Takala J, Suter PM, et al. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on "sepsis-related problems" of the European Society of Intensive Care Medicine. Critical care medicine. 1998;26(11):1793–800. Epub 1998/11/21. PubMed . [DOI] [PubMed] [Google Scholar]
- 24. Tzvetkov MV, Meineke I, Sehrt D, Vormfelde SV, Brockmoller J. Amelogenin-based sex identification as a strategy to control the identity of DNA samples in genetic association studies. Pharmacogenomics. 2010;11(3):449–57. Epub 2010/03/20. 10.2217/pgs.10.14 PubMed . [DOI] [PubMed] [Google Scholar]
- 25. Fallavena PR, Borges TJ, Paskulin DD, Paludo FJ, Goetze TB, de Oliveira JR, et al. The influences of CD14 -260C>T polymorphism on survival in ICU critically ill patients. Immunological investigations. 2009;38(8):797–811. Epub 2009/10/29. 10.3109/08820130903258818 PubMed . [DOI] [PubMed] [Google Scholar]
- 26. Fallavena PR, de Jesus Borges T, Paskulin DD, Thurow HS, de Oliveira Paludo FJ, Dos Santos Froes C, et al. The synergy of -260T T CD14 and -308GG TNF-alpha genotypes in survival of critically ill patients. Scandinavian journal of immunology. 2013;77(1):62–8. Epub 2012/10/06. 10.1111/sji.12002 PubMed . [DOI] [PubMed] [Google Scholar]
- 27. D'Avila LC, Albarus MH, Franco CR, Aguiar BB, Oliveira JR, Dias FS, et al. Effect of CD14 -260C>T polymorphism on the mortality of critically ill patients. Immunology and cell biology. 2006;84(4):342–8. Epub 2006/03/03. 10.1111/j.1440-1711.2006.01432.x PubMed . [DOI] [PubMed] [Google Scholar]
- 28. Barber RC, Aragaki CC, Chang LY, Purdue GF, Hunt JL, Arnoldo BD, et al. CD14-159 C allele is associated with increased risk of mortality after burn injury. Shock (Augusta, Ga). 2007;27(3):232–7. Epub 2007/02/17. 10.1097/01.shk.0000239770.10528.9a PubMed . [DOI] [PubMed] [Google Scholar]
- 29. Barber RC, Chang LY, Arnoldo BD, Purdue GF, Hunt JL, Horton JW, et al. Innate immunity SNPs are associated with risk for severe sepsis after burn injury. Clinical medicine & research. 2006;4(4):250–5. Epub 2007/01/11. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mansur A, Hinz J, Hillebrecht B, Bergmann I, Popov AF, Ghadimi M, et al. Ninety-day survival rate of patients with sepsis relates to programmed cell death 1 genetic polymorphism rs11568821. Journal of investigative medicine: the official publication of the American Federation for Clinical Research. 2014;62(3):638–43. Epub 2014/01/28. 10.231/jim.0000000000000059 PubMed . [DOI] [PubMed] [Google Scholar]
- 31. Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature genetics. 2002;32(4):666–9. Epub 2002/10/29. 10.1038/ng1020 PubMed . [DOI] [PubMed] [Google Scholar]
- 32. Kristjansdottir H, Steinsson K, Gunnarsson I, Grondal G, Erlendsson K, Alarcon-Riquelme ME. Lower expression levels of the programmed death 1 receptor on CD4+CD25+ T cells and correlation with the PD-1.3A genotype in patients with systemic lupus erythematosus. Arthritis and rheumatism. 2010;62(6):1702–11. Epub 2010/02/24. 10.1002/art.27417 PubMed . [DOI] [PubMed] [Google Scholar]
- 33. Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A Polymorphism* in the 5' flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. American journal of respiratory cell and molecular biology. 1999;20(5):976–83. Epub 1999/05/05. 10.1165/ajrcmb.20.5.3494 PubMed . [DOI] [PubMed] [Google Scholar]
- 34. Fan LW, Pang Y, Lin S, Tien LT, Ma T, Rhodes PG, et al. Minocycline reduces lipopolysaccharide-induced neurological dysfunction and brain injury in the neonatal rat. Journal of neuroscience research. 2005;82(1):71–82. Epub 2005/08/25. 10.1002/jnr.20623 PubMed . [DOI] [PubMed] [Google Scholar]
- 35. Hsieh PF, Chia LG, Ni DR, Cheng LJ, Ho YP, Tzeng SF, et al. Behavior, neurochemistry and histology after intranigral lipopolysaccharide injection. Neuroreport. 2002;13(3):277–80. Epub 2002/04/04. PubMed . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.